Abstract
Background
The runt-related transcription factor 2 (Runx2) is a cell-fate-determining factor that controls osteoblast differentiation and bone formation. It has been previously demonstrated that microRNAs (miRNAs) play important roles in osteogenesis. However, the Runx2-regulated miRNAs that have been reported thus far are limited. In this study, we pursued to identify these miRNAs in Tet-on stable C2C12 cell line (C2C12/Runx2Dox subline).
Results
Microarray analysis revealed that alterations in miRNA expression occur with 54 miRNAs. Among these miRNAs, miR-690 was identified as a positive regulator of Runx2-induced osteogenic differentiation of C2C12 cells through gain- and loss-of-function assays. Expression of miR-690 is induced by Runx2, which binds directly to the putative promoter of mir-690 (Mirn690). The miR-690 proceeds to inhibit translation of the messenger RNA encoding the nuclear factor kappa B (NF-κB) subunit p65 whose overexpression inhibits Runx2-induced osteogenic differentiation of C2C12 cells. Interleukin-6 (IL-6), a downstream target of NF-κB pathway, is upregulated by p65 overexpression but significantly downregulated during this differentiation process. Furthermore, overexpression of IL-6 impedes the expression of osteocalcin, a defined marker of late osteoblast differentiation.
Conclusions
Together, our results suggest that the miR-690 transactivated by Runx2 acts as a positive regulator of Runx2-induced osteogenic differentiation by inactivating the NF-κB pathway via the downregulation of the subunit p65.
Similar content being viewed by others
Background
The proper differentiation of mesenchymal progenitor cells into osteoblast lineage is pivotal for bone development in vivo. Osteoblast differentiation is governed by numerous regulatory pathways involving transcription factors, signaling molecules, and chromatin modifiers [1–3]. In addition, posttranscriptional mechanisms including miRNA-mediated regulation also play important roles in regulating osteogenic differentiation.
MicroRNAs (miRNAs) are small (approximately 22nt), functional, highly conserved, endogenous noncoding RNA. They regulate protein translation or mRNA stability by imperfect binding to the 3′ untranslated region (3′UTR) of their target genes. These small RNAs have emerged as key regulators of almost every biological process in eukaryotes, including early development, growth and differentiation, and cell apoptosis. The impact of miRNAs on osteoblast differentiation has been investigated in several studies. It has been shown that miR-135 targets the intracellular receptor SMAD family member 5 and inhibits bone morphogenetic protein 2(BMP-2)-induced osteogenic differentiation of C2C12 cells [4]. MiR-141 and its homolog miR-200a are involved in BMP-2-induced preosteoblast differentiation by regulating their common target distal-less homeobox 5 [5]. In primary mouse osteoblasts, miR-93 has been found to target osterix and then inhibits osteoblast mineralization [6]. MiR-204 and its homolog miR-211 stimulate adipocyte differentiation but inhibit osteoblast differentiation of bone marrow stromal cells through targeting runt-related transcription factor 2 (Runx2) [7]. In addition, a panel of 11 miRNAs, including miR-204, has been found to control osteogenic lineage progression by targeting Runx2 [8]. These results suggest that the expression level of osteogenesis-related factors could be tightly controlled by miRNAs, which is an effective method for regulating osteogenesis. Additionally, the miRNAs controlled by these factors (especially the osteoblast-specific transcription factors) are also indispensable for regulating osteogenesis.
Runx2 (also known as Cbfa1, AML-3, and PEBP2aA), an osteoblast-specific transcription factor belonging to the runt-domain gene family [9], is expressed in mesenchymal condensations during early embryonic development [10]. Many previous studies have revealed that during both embryogenesis and postnatal life, Runx2 plays a key role in regulating osteoblast and hypertrophic chondrocyte differentiation, and bone formation [11–13]. The underlying mechanism by which Runx2 regulates these processes is still unclear, and the identification of Runx2-regulated miRNAs can provide new insight into the mechanism. However, Runx2-controlled miRNAs that have been identified thus far are limited.
In the current study, we explored Runx2-regulated miRNAs by using C2C12/Runx2Dox subline previously described [14], and obtained 54 differentially expressed miRNAs during Runx2-induced osteogenic differentiation, and identified miR-690 as a positive regulator during this differentiation process. Furthermore, we demonstrated that miR-690 is directly induced by Runx2. The nuclear factor kappa B (NF-κB) subunit p65 was identified as a direct target of miR-690, and its overexpression inhibits Runx2-induced osteogenic differentiation. Interleukin-6 (IL-6), a downstream target of NF-κB pathway [15], is upregulated by p65 overexpression but significantly downregulated during this differentiation process. Furthermore, overexpression of IL-6 impedes Runx2-induced late osteoblast differentiation. Together, our findings suggest that Runx2 promotes osteogenic differentiation of C2C12 cells by inactivating NF-κB pathway via the upregulation of miR-690.
Results
Identification of differentially expressed miRNAs during Runx2-induced osteogenic differentiation
Elucidating Runx2-regulated miRNAs could provide new insight into the mechanism whereby Runx2 functions in osteogenesis. Here, we used the C2C12/Runx2Dox subline previously described [14] to explore this question. In C2C12/Runx2Dox cells, the expression of Flag-Runx2 can be tightly regulated by doxycycline (Dox) treatment. In the absence of Dox, the C2C12/Runx2Dox cells cultured in medium containing 2 % horse serum can be induced into multinucleated myotubes (Additional file 1: Figure S1A). Additionally, C2C12/Runx2Dox cells also can be induced into osteoblasts by BMP-2 treatment in the absence of Dox (Additional file 1: Figure S1B). These results suggest that the differentiation characteristics of C2C12/Runx2Dox cells is similar to that of C2C12 myogenic progenitor cells. Taken together, we believed that the C2C12/Runx2Dox cells are actually the wild type C2C12 myogenic progenitor cells in the absence of Dox.
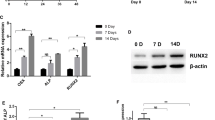
Osteogenic differentiation of C2C12 cells was evidenced by increased expression of genes associated with osteoblast differentiation, alkaline phosphatase (Alp) and osteocalcin (OC), at the indicated times after Dox treatment (Fig. 1a). The expression of mRNA and protein for Flag-Runx2 was increased in a time-dependent manner (Fig. 1a, Additional file 2: Figure S2A). In immunocytochemistry analysis, Flag-Runx2 was found to be mainly expressed in the nuclei of Dox-treated C2C12/Runx2Dox cells, and the percentage of cells overexpressing Flag-Runx2 was also increased in a time-dependent manner (Additional file 3: Figure S3). The osteoblast phenotype was confirmed by demonstration of increased ALP activity and Alizarin Red staining for matrix mineralization (Fig. 1b). These results suggest that the C2C12/Runx2Dox subline works well. Moreover, the cell line harboring the empty vector (C2C12-empty vector Tet-on cell line, C2C12/vectorDox cells) was used to investigate and exclude the influence of Dox. As shown in Additional file 4: Figure S4, the ALP activity and matrix mineralization were absent in Dox-treated C2C12/vectorDox cells. These results suggest that Dox itself has no effect on the osteogenic differentiation of C2C12 cells.
A program of miRNAs is expressed during Runx2-induced osteogenic differentiation of C2C12 cells. a C2C12/Runx2Dox cells were treated with Dox for the times indicated (0–72 h). The mRNA level of Runx2, Alp, and OC was determined by real-time qPCR. All data are presented as the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01 compared with untreated cells (0 h). b ALP and matrix mineralizing activity were measured by ALP and Alizarin red staining at days 7 and 28, respectively. Similar results were obtained in three independent experiments. c Differentially expressed miRNAs from miRNA microarray data. 54 miRNAs whose expression changed by ≥ 1.5-fold at least one time point were chosen as the differentially expressed miRNAs by comparing with untreated cells (0 h), and these miRNAs were arranged by unsupervised hierarchical clustering. Data value displayed as red and green represent elevated (high) and reduced expression (low), respectively
To detect miRNAs related to Runx2-induced osteogenic differentiation, miRNA profiling was performed by using total RNA from Dox-treated C2C12/Runx2Dox cells collected at 0, 3, 6, 9, 12, 24, 48, and 72 h. The values of the miRNA expression level at each time point were clustered and graphically illustrated (Additional file 5: Figure S5). Expression levels of 54 miRNAs were altered during this differentiation process (Fig. 1c). Of these, 13 miRNAs were upregulated in the early induction phase (6–24 h), and declined to basal levels in the late induction phase (48 and 72 h) (blue box in Fig. 1c). As shown in Fig. 1a and Additional file 2: Figure S2, C2C12/Runx2Dox cells were induced into osteoblasts by 12 h based on expression of muscle and bone phenotypic genes, suggesting the crucial role of early induction phase in Runx2-induced transdifferentiation of myoblasts (C2C12 cells) into the osteoblastic phenotype. Thus, for osteogenesis to proceed, there is a requirement for upregulation of the 13 miRNAs in the dataset responsive to Runx2 overexpression to convert the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. We then focused our attention on these miRNAs.
Effect of miR-690 on Runx2-induced osteogenic differentiation
To evaluate the biological effect of the above-mentioned 13 miRNAs, anti-miRs and miRNA mimics for these miRNAs were used in a functional screening for their impact on Runx2-induced osteogenic differentiation. Here, we identified miR-690 as a potential positive regulator of Runx2-induced osteogenic differentiation. Expression level of miR-690 was measured at different time points during this differentiation process. MiR-690 was significantly upregulated at hour 6, and remained upregulated up to hour 24, and declined to basal levels at hour 48 and 72 respectively (Fig. 2a). Additionally, Dox itself also has no effect on the expression of miR-690 (Additional file 6: Figure S6). The miR-690 expression change assayed by real-time quantitative PCR (qPCR) was generally similar to the change in miR-690 expression as determined by miRNA microarray (Fig. 1c).
miR-690 synergistically potentiates Runx2-induced osteogenic differentiation. a Real-time qPCR analysis of miR-690 was performed in Dox-treated samples used for miRNA profiling analysis. b, c Effect of miR-690 on expression of osteoblastic markers. C2C12/Runx2Dox cells were transfected with miR control (b), miR-690 (b), anti-miR control (c), and anti-miR-690 (c), respectively, and then treated with Dox for 3 days. The mRNA level of Alp and OC was measured by real-time qPCR. *P < 0.05 compared with miR control or anti-miR control samples. d Effect of miR-690 on ALP activity. C2C12/Runx2Dox cells were transfected with miR control and miR-690, respectively, and then treated with Dox for 7 days. The ALP activity was determined by ALP staining. Similar results were obtained in three independent experiments
To study the effect of miR-690 on Runx2-induced osteogenic differentiation, we transfected miR control, miR-690, anti-miR control, and anti-miR-690 into C2C12/Runx2Dox cells, respectively, and found that transfection of miR-690 increased expression of Alp and OC (Fig. 2b), whereas transfection of anti-miR-690 reduced their expression (Fig. 2c). Moreover, ALP activity was slightly increased in miR-690-transfected C2C12/Runx2Dox cells (Fig. 2d). These findings demonstrate that miR-690 synergistically potentiates Runx2-induced osteogenic differentiation of C2C12 cells.
Mir-690 is a direct transcriptional target of Runx2
Mir-690 expression can be rapidly induced following activation of Runx2 (Fig. 2a), raising the possibility that mir-690 is a direct target of Runx2. To test this hypothesis, we first examined the −1 kb genomic sequence upstream of the mir-690 stem-loop to search for potential Runx2-binding site using TFSEARCH database [16] and ConSite program [17]. Indeed, we identified four potential sites at −313 to −305 (site 4, Runx2-4), −298 to −290 (site 3, Runx2-3), −246 to −238 (site 2, Runx2-2), and −64 to −56 (site 1, Runx2-1), respectively (Fig. 3a). With an electrophoretic mobility shift assay (EMSA), the labeled oligonucleotide containing Runx2-2 produced a significant shift (BS) in the Dox-treated cells nuclear extract (Fig. 3b, BS in lane 1), but the labeled oligonucleotides containing Runx2-1, Runx2-3, or Runx2-4 failed to do so (data not shown). No apparent shift was produced by the oligonucleotide containing Runx2-2 in the extract from non-treated cells (Fig. 3b, lane 8). The observed shift was abolished by 10- to 50-fold molar excess of unlabeled oligonucleotides (Fig. 3b, lanes 2–4). The labeled oligonucleotide contained a 9 bp mutation also abolished the shift (Fig. 3b, lane 7). Furthermore, the anti-Flag antibody, not the mouse IgG antibody, supershifted the complex (BS) (Fig. 3b, lanes 5 and 6), suggesting the specificity of the interaction. Thus, the EMSA assay demonstrated that the mir-690 promoter contains one Runx2-binding site, Runx2-2.
miR-690 is directly regulated by Runx2. a Schematic representation of mouse mir-690 genomic locus, which indicates the locations of the four potential Runx2-binding sites. The italic letters indicate the core sequence of each site. b EMSA shows the interaction between mir-690 site (Runx2-2) and Runx2 in Dox-treated cells. The bottom arrow indicates the DNA–protein complex (BS). The top arrow indicates the Runx2-supershifed complex. WT wild-type, Mut mutant. c ChIP analysis was performed to confirm the interaction of Runx2 with mir-690 promoter in vivo. PCR was performed with primer-1, which was designed to amplify a fragment of the mir-690 promoter flanking the Runx2-2 site. The primer-2 for an unrelated part (−1800/− 1653) in the distal 5′ flanking region of the Runx2-2 site was utilized for the control reaction in this ChIP analysis
Chromatin immunoprecipitation (ChIP) assay was performed to examine whether Runx2 interacts with mir-690 promoter in vivo. As shown in Fig. 3c, the anti-Flag antibody specifically enriched the regions containing Runx2-2 (primer-1). By contrast, no positive signals were detected in various negative controls, including the PCR control (H2O) or immunoprecipitation control. Additionally, Runx2 did not associate with the unrelated part in the distal 5′ flanking region of the Runx2-2 site (primer-2). These results indicate that Runx2 binds to the mir-690 promoter in vivo. Together, these results suggest that Runx2 acts as a transcriptional activator on mir-690.
MiR-690 directly targets p65 through translational inhibition
To understand the molecular mechanism that underlies miR-690-mediated regulation, we search for potential targets of miR-690 implicated in Runx2-induced osteogenic differentiation using the miRNA target prediction algorithms DIANA-microT, miRDB, and miRWalk [18–20]. More than 100 genes were predicted to be potential target genes for miR-690. Among the predicted genes, we identified p65, a NF-κB subunit with a negative role in osteoblast differentiation [21]. According to in silicon analysis, p65 has two potential binding sites for miR-690 within its 3′UTR (Fig. 4a). To investigate whether p65 can be directly targeted by miR-690, we engineered a luciferase reporter that has wild-type 3′UTR of p65 (Fig. 4b). Co-transfection of the p65 3′UTR luciferase reporter with miR-690 resulted in downregulation of the luciferase activity compared with the miR control (Fig. 4c). In comparison, miR-690-Mut had no effect on p65 3′UTR luciferase reporter (Fig. 4c), implying that p65 is a direct target of miR-690. Thus, we further examined how miR-690 repressed p65 expression. Real-time qPCR results revealed no significant change in p65 mRNA level when miR-690 was transfected into C2C12/Runx2Dox cells (Fig. 4d). However, Western blot result showed reduced p65 protein level in the miR-690-transfected cells compared with miR control sample (Fig. 4e), indicating that miR-690 mediates translational repression but not mRNA degradation of p65. Additionally, the expression of p65 was slightly increased at the protein but not the mRNA level in the anti-miR-690-transfected cells compared with anti-miR control sample (Additional file 7: Figure S7). Together, these results indicate that miR-690 directly targets p65 through translational inhibition.
miR-690 directly targets p65. a Schematic representing miR-690 target sites in p65 3′UTR and base-pairing of miR-690 sequences with the 3′UTR. b Schematic diagram illustrating the design of luciferase reporter with p65 3′UTR. The sequences of miR-690 and miR-690-Mut are also shown. The underlined sequences represent wild-type and mutated seed sequences of miR-690. Rluc Renilla luciferase, Luc Firefly luciferase, pA polyadenylation signal. c miR control, miR-690, or miR-690-mut was co-transfected with psiCHECK-2-p65 3′UTR reporter into C2C12/Runx2Dox cells. After 48 h, cells were harvested and luciferase activities were measured. Luciferase activity = (Renilla/Firefly) ratio. Renilla, raw renilla luciferase activity; Firefly, firefly luciferase activity. Data are presented as mean ± SD (n = 3). *P < 0.05 compared with miR control sample. d, e Relative mRNA level of p65 (d) and protein level of p65 (e) in miR control- or miR-690-transfected cells
p65 exerts its effect on Runx2-induced osteogenic differentiation partially through IL-6 upregulation
Since p65 is directly targeted by miR-690, it was necessary to examine whether p65 exerts an opposite effect on Runx2-induced osteogenic differentiation as compared with miR-690. p65 expression vector was used for transfection of C2C12/Runx2Dox cells, followed by Dox treatment for 3 days. Real-time qPCR data revealed that p65 overexpression attenuates the expression of Alp and OC (Fig. 5a), suggesting that p65 is a negative regulator of Runx2-induced osteogenic differentiation. Additionally, the mRNA level of p65 was unchanged during this differentiation process (Additional file 8: Figure S8A). However, the protein level of p65 was decreased in Dox-treated C2C12/Runx2Dox cells at hour 12, possibly resulting from the high level of miR-690 at this time point (Additional file 8: Figure S8B). This finding shows that p65 is indeed a direct target of miR-690 during this differentiation process.
p65 exerts its effect on Runx2-induced osteogenic differentiation partially through IL-6 upregulation. a, b The empty vector (pcDNA3.1) or p65 expression vector (pcDNA3.1-p65) was transfected into C2C12/Runx2Dox cells. The transfected cells were treated with Dox for 3 days and subjected to real-time qPCR analysis for Alp (a), OC (a) and IL-6 (b) mRNAs, respectively. Data are presented as mean ± SD (n = 3). *P < 0.05 compared with pcDNA3.1-transfected cells. Western blot was performed to assess the overexpression level of p65 (a, inset). c Real-time qPCR results of IL-6 in Dox-treated cells. Data are presented as mean ± SD (n = 3). *P < 0.05, **P < 0.01 compared with untreated cells (0 h). d The empty vector (pcDNA3.1) or IL-6 expression vector (pcDNA3.1-IL-6) was transfected into C2C12/Runx2Dox cells. The transfected cells were treated with Dox for 3 days and subjected to real-time qPCR analysis for Alp and OC mRNAs. Data are presented as mean ± SD (n = 3). *P < 0.05 compared with pcDNA3.1-transfected cells
Previous studies have shown that IL-6 is a direct transcriptional target of NF-κB [15], and that osteoblast differentiation can be negatively regulated by IL-6 [22]. This observation raised the possibility that p65 may exert its negative effect on Runx2-induced osteogenic differentiation by upregulating IL-6. We therefore performed a series of experiments to investigate this possibility. IL-6 expression was found to be upregulated in C2C12/Runx2Dox cells transfected with p65 expression vector in the presence of Dox for 3 days (Fig. 5b). In the absence of Dox, IL-6 expression was also upregulated by p65 overexpression (Additional file 9: Figure S9). These results suggest that IL-6 could be activated by p65 during this differentiation process. To further verify the above-mentioned possibility, we also examined the mRNA level of IL-6 in Dox-treated C2C12/Runx2Dox cells, and found that the expression of IL-6 was significantly downregulated (Fig. 5c), implying the negative effect of IL-6 on Runx2-induced osteogenic differentiation. In addition, the role of IL-6 in the differentiation process of C2C12/Runx2Dox cells was examined by transfection of IL-6 expression vector. As shown in Fig. 5d, the expression of OC, a defined marker of late osteoblast differentiation, was significantly downregulated by IL-6 overexpression. However, the expression of Alp, a marker of early osteoblast differentiation, was not affected. Additionally, the negative effect of p65 on the expression of OC was attenuated in si-IL-6-transfected C2C12/Runx2Dox cells compared with si-NC sample (Additional file 10: Figure S10). Taken together, these results indicate that p65 exerts its negative effect on Runx2-induced osteogenic differentiation partially through IL-6 upregulation.
Discussion
In the present study, we reported an extensive genomic-wide profiling of miRNA expression during Runx2-induced osteogenic differentiation, and identified 54 differentially expressed miRNAs. MiR-690, a newly identified Runx2-targeted miRNA, was found to promote Runx2-induced osteogenic differentiation through translational inhibition of its target p65, permitting the downregulation of the expression of p65 downstream targets (IL-6 and other undisclosed targets); this favours, in turn, Runx2-induced osteogenic differentiation (Fig. 6).
Balint et al. have demonstrated that induction of osteogenic differentiation of C2C12 cells by BMP-2 could be separated into several distinct stages, and that the 8–12 h window may make the point of osteoblast phenotypic determination, and that the bone phenotype can be further established from 16 to 24 h [23]. These results are consistent with the finding reported by Li et al. [4]. In our study, the commitment of C2C12 myoblasts to the osteogenic phenotype is recognized by 12 h but not by 8 h (Fig. 1a). The discrepancy between BMP-2- and Runx2-induced systems might arise from the different osteogenic effect of BMP-2 and Runx2 on C2C12 cells. Taken together, we concluded that the period from 12 to 24 h appears to be a point of osteoblast phenotypic determination, and that the period from 24 to 72 h might be used for further establishing bone phenotype during Runx2-induced osteogenesis.
Many researches have shown that miRNAs play an important role in regulating osteoblast differentiation and bone formation [24]. A panel of miRNAs has been reported to control osteogenesis by targeting the cell-fate-determining transcription factor Runx2 [8], but the Runx2-regulated miRNAs that have been identified are still limited (e.g., miR-2861-3960 cluster) [25]. In this study, we performed a miRNA microarray by using total RNA from Dox-treated C2C12/Runx2Dox cells, and identified 54 differentially expressed miRNAs regulated by Runx2. During Runx2-induced osteogenic differentiation of C2C12 cells, the microarray data could reveal the followings: First, 19 miRNAs were upregulated and 35 miRNAs were downregulated during osteogenic differentiation from 609 mouse miRNAs on the array. Second, the 35 downregulated miRNAs were predicted to target mainly regulatory factors that function to promote myogenesis and inhibit osteogenesis independently. MiR-206, a significantly downregulated miRNA in microarray data (Fig. 1c), was reported to be a muscle-specific miRNA and to promote myoblast differentiation by directly targeting paired box 7 [26, 27]. MiR-466f-3p and miR-125a-5p, whose expression were also downregulated (Fig. 1c), were found to be significantly downregulated during osteogenic differentiation of bone marrow- and adipose tissue-derived mesenchymal stem cells [28, 29], suggesting the general role of these miRNAs in the inhibition of osteoblast differentiation. However, the bona fide targets of these miRNAs are still unclear during osteogenesis. Third, the 19 upregulated miRNAs could be divided into two categories: 13 miRNAs whose expression was upregulated in the early induction phase (6-24 h) but declined to basal levels in the late induction phase (48 and 72 h) (blue box in Fig. 1c), and 6 miRNAs whose expression was only significantly upregulated in the late induction phase (48 and 72 h) (red box in Fig. 1c). The distinct expression pattern of 19 upregulated miRNAs revealed that these miRNAs might exert different effect on Runx2-induced osteogenic differentiation. MiR-21a-5p, miR-762, and miR-1224, belonging to the 6 upregulated miRNAs, were reported to be involved in bone matrix mineralization [30–32], suggesting that the 6 upregulated miRNAs might participate in the regulation of Runx2-induced late osteoblast differentiation.
Here, we concluded that the 13 upregulated miRNAs might play a key role in the commitment of C2C12 cells into osteoblastic lineage. To evaluate the biological effect of the 13 upregulated miRNAs on Runx2-induced osteogenic differentiation, anti-miRs and miRNA mimics for these miRNAs were used in a functional screening. Finally, the screening identified miR-690 as a candidate regulator of Runx2-induced osteogenic differentiaition. Previous study has reported that miR-690 might play an important role in glucose regulation of β-cell function [33]. MiR-690 is highly overexpressed in functional myeloid-derived suppressor cells (MDSCs) and manipulates MDSC activity by repressing transcription factor CCAAT/enhancer-binding protein α in inflammatory diseases as well as cancer [34, 35]. MiR-690 can also regulate fibroblast migration and dermal wound repair by directly targeting Versican [36]. In addition, miR-690 was found to be involved in granulopoiesis [37], glutamate-induced excitotoxicity [38], and regulating testosterone signaling in liver [39]. However, the function of miR-690 in regulating osteogenic differentiation was not clear. In this study, data obtained from in vitro experiments revealed that miR-690 overexpression enhances Runx2-induced osteogenic differentiation of C2C12 cells, whereas knockdown of miR-690 leads to the opposite effect (Fig. 2b–d). These findings suggest that miR-690 is an osteogenesis-related miRNA and positively regulates Runx2-induced osteogenic differentiation.
As shown in Fig. 2a, miR-690 expression was rapidly induced following activation of Runx2, suggesting that the transcriptional activation of mir-690 is dependent upon Runx2 expression. It further suggested that mir-690 may be a direct target of Runx2. To define the molecular mechanism by which Runx2 regulates the expression of miR-690, we performed in silico analysis and identified four potential Runx2-binding sites in the −1 kb genomic sequence upstream of the mir-690 stem loop (Fig. 3a). EMSA analysis using nuclear extract from Dox-treated C2C12/Runx2Dox cells demonstrated specific binding of Runx2 for the Runx2 motif at position −246 (site2, Runx2-2)(Fig. 3b). However, the data obtained from these artificial in vitro systems may not necessarily reflect the regulation and function of the endogenous gene. To address this concern, we used ChIP analysis to corroborate the EMSA data. This study clearly demonstrated that Runx2 is specifically associated with the putative promoter of mir-690 during osteogenic differentiation of C2C12 cells (Fig. 3c). Together, our findings reveal that mir-690 could be a direct target of Runx2 during osteogeneis.
To study the molecular mechanism by which miR-690 regulates Runx2-induced osteogenic differentiation, we searched for potential target genes that have an established function in inhibiting osteogenesis. Interestingly, the 3′UTR of p65 possesses two match sites to the miR-690 seed region. p65, a subunit of NF-κB, which can function either as a homodimer or as a classical heterodimer with the p50 subunit of NF-κB [40]. Indeed, p65 3′UTR luciferase reporter assays confirmed that p65 is a direct target of miR-690 (Fig. 4c). We showed that miR-690 mediates translational repression but not mRNA degradation of p65 (Fig. 4d, e). Overexpression of p65 inhibits Runx2-induced osteogenic differentiation (Fig. 5a), and the effect of p65 on Runx2-induced late osteoblast differentiation is partially mediated through upregulating the NF-κB downstream target IL-6 (Figs. 5, 6). In this study, the target that mediates the effect of p65 on Runx2-induced early osteoblast differentiation was not investigated, and other previously reported (e.g. IL-8, ICAM-1, miR-17, miR-21, miR-27b, and miR-30b) [41–43] or undisclosed targets might participate in this process.
It has been reported that p65 can inhibit BMP-2-induced osteogenic differentiation by decreasing the DNA binding of the Smad complex [21]. Tumor necrosis factor alpha (TNFα), a proinflammatory cytokine, can exert its negative effect on osteoblast differentiation and bone formation through the NF-κB p50/p65 heterodimer [40]. Runx2 stability can be reduced by TNFα through upregulation of E3 ubiquitin ligases Smad ubiquitin regulatory factor 1 [44]. WW domain containing E3 ubiquitin protein ligase 1, which was significantly upregulated in mesenchymal stem cells (MSCs) from TNFα-transgenic mice [45], was found to be recruited by Schnurri-3 and to mediate Runx2 degradation in a ubiquitin–proteasome-dependent manner [46]. MiR-3077-5p, a miRNA upregulated by TNFα in MSCs from osteoporosis bone marrow of mice, was demonstrated to directly target Runx2 and to inhibit Runx2 translation [47]. These findings raise the possibility that Runx2 function could be negatively regulated by the NF-κB p50/p65 heterodimer. Thus, it is necessary to inactivate the canonical NF-κB pathway by Runx2 through miR-690-mediated downregulation of p65 during this differentiation process.
In addition, other undisclosed miR-690 targets may contribute to the phenotypic effects observed on inhibition or overexpression of miR-690. There are more than 100 predicted targets for miR-690 in DIANA-microT, miRDB, and miRWalk [18–20], some of which are very relevant in the process of osteoblast differentiation and bone formation (e.g., Bcl2, Igfbp5, Smad7, Crem, Stat3, and Mmp28). The involvement of other potential osteogenic targets should be elucidated in future studies.
Conclusions
Taken together, our study provides a comprehensive profiling of Runx2-regulated miRNAs during osteogenic differentiation of C2C12 myogenic progenitor cells. MiR-690, a newly identified Runx2-targeted miRNA, exerts its positive effect on Runx2-induced osteogenic differentiation by inactivating the NF-κB pathway via the downregulation of the NF-κB subunit p65. Our findings enrich the knowledge of the Runx2-centered regulatory network that functions in osteoblast differentiation and bone formation, and may give some tips for treating human osseous defects.
Methods
Primers and probes
All primers and probes are available in Additional file 11: Tables S1–S7.
Cell culture
The establishment of C2C12/Runx2Dox sub-line has been described previously [14]. C2C12/Runx2Dox cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % Tet System Approved fetal bovine serum (FBS) (Clontech, Mountain View, CA, USA). Hygromycin B and G418 were purchased from Invitrogen and added into the medium at the concentration of 500 μg/ml and 800 μg/ml, respectively. For osteogenic differentiation, the C2C12/Runx2Dox cells were cultured in medium containing Dox (10 μg/ml) (Clontech) and BMP-2 (100 ng/ml) (R&D Systems, Minneapolis, MN, USA) respectively. For myogenic differentiation, C2C12/Runx2Dox cells were grown in growth medium consisting of high-glucose DMEM supplemented with 10 % FBS (Invitrogen). At 80–90 % confluence, myogenic differentiation was induced by replacing the growth medium with differentiation medium consisting of DMEM supplemented with 2 % horse serum (Invitrogen). Additionally, the establishment and culture of C2C12/VectorDox cells were similar to that of C2C12/Runx2Dox cells.
MiRNA microarray analysis
Total RNA containing small RNA was extracted from Dox-treated C2C12/Runx2Dox cells by using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). MiRNA profiling was performed using GeneChip miRNA Array version 1.0 (Affymetrix, Santa Clara, CA, USA). The array comprised 6703 mature microRNA sequences of 71 organisms from the Sanger miRNA database (V.11) and an additional 922 encompassed human snoRNAs and scaRNAs (from Ensembl database and snoRNABase).
Microarray experiments were performed according to the manufacturer’s instructions. Briefly, 1 μg total RNA was labeled with Biotin FlashTag Biotin Labeling Kit (Affymetrix). The labeling reaction was hybridized on the miRNA Array in Affymetrix Hybridization Oven 640 (Affymetrix) at 48 °C for 16 h. The arrays were stained with Fluidics Station 450 using fluidics script FS450_0003 (Affymetrix), and then scanned on a GeneChip microarray scanner (Affymetrix).
Microarray data analysis
MiRNA probe outliers were defined as per the manufacturer’s instructions (Affymetrix) and further analyzed for data summarization, normalization and quality control by using the miRNA QC Tool software (www.affymetrix.com). To select the differentially expressed miRNAs, we used threshold values of ≥1.5 and ≤−1.5-fold change and a FDR significance level of <5 %. The data was Log2 transformed and median centered by miRNAs using the Adjust Data function of CLUSTER 3.0 software (Michiel de Hoon, Human Genome Center, University of Tokyo, Tokyo, Japan), then further analyzed with hierarchical clustering with average linkage. Finally, we performed tree visualization by using Java Treeview (Stanford University School of Medicine, Stanford, CA, USA).
Reverse transcription PCR
Total RNA was extracted by Trizol reagent (Invitrogen). A 20 μl reaction mixture containing 1 μg of total RNA was reversely transcribed to cDNA using RT Ace reverse transcriptase (Toyobo, Osaka, Japan). Real-time qPCR was performed on the cDNA to examine the expression levels of mRNAs and miRNAs. The PCR reaction mix was prepared using SYBR Green PCR Master Mix (Takara, Shiga, Japan). PCR conditions were as follows: 95 °C for 30 s, 40 cycles of denaturation at 95 °C for 5 s, annealing at 60 °C for 30 s, and extension at 72 °C for 15 s. PCR was carried out using the Real-Time PCR Detection System Rotor-Gene 6000 (Corbett Research, Mortlake, NSW, Australia). Relative expression levels were calculated as ratios normalized against those of 18S rRNA or U6 snRNA. The data from real-time qPCR were analyzed by the ΔCt method, and the ΔCt value was determined by subtracting the 18S rRNA Ct value from the target gene Ct value or the U6 snRNA Ct value from the target miRNA Ct value. The ΔCt of the treated cells (ΔCts) was subtracted from the ΔCt of the untreated cells (ΔCtu) (ΔΔCt = ΔCts – ΔCtu), and the expression level for a target gene or miRNA in the treated cells compared with the level in the untreated cells was calculated as follows: x-fold of untreated control = \(2^{{ - {\Delta \Delta }{\text{C}}_{\text{t}} }}\).
Western blot analysis
Cells were washed with PBS and lysed with mammalian protein extraction reagent from M-PER (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with protease inhibitor mixture tablet (Roche, Indianapolis, IN, USA). Protein concentrations were determined by BCA protein assay (Thermo Fisher Scientific). For western blots, total proteins (30–50 μg) were resolved by 12 % SDS-PAGE gel and transferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). Membranes were blocked with 5 % non-fat milk for 1 h at room temperature and incubated with primary antibody diluted in 2 % non-fat milk at 4 °C overnight, followed by incubation with the second antibody conjugated to horseradish peroxidase (HRP) for 1 h at room temperature and measured with the enhanced chemiluminescence system (Perkin-Elmer Life and Analytical Sciences, Waltham, MA, USA) and recorded on X-ray films (Fuji, Tokyo, Japan). Protein expression was normalized by β-actin. Primary antibodies used for blotting were anti-p65 (Cell Signaling Technology, Beverly, MA, USA), anti-Flag (Sigma, St Louis, MO, USA), and anti-β-actin (Cell Signaling Technology). Anti-rabbit IgG-HRP (Cell Signaling Technology) were used to detect primary antibodies.
Immunofluorescence
Cells were washed with PBS, then fixed with 4 % formaldehyde in PBS for 20 min, and finally permeabilized with 0.2 % Triton X-100 solution for 5 min. Cells were incubated with anti-Flag antibody for 1 h, washed three times with PBS, and then incubated with appropriate Alexa Fluor 555-conjugated secondary antibody and washed again three times in PBS. Coverslips were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen), and examined using a Leica fluorescence microscope (Leica, Wetzlar, Germany).
In vitro osteoblastogenesis and cell staining
Cells (C2C12/Runx2Dox and C2C12/VectorDox cells) were cultured in DMEM containing Dox, β-glycerophosphate disodium (10 mM) and ascorbic acid (50 μg/ml). The medium was changed every 3 days. For ALP staining and Alizarin red staining, the detailed procedures were described previously [48, 49].
Transfection
To examine the effect of miR-690 on Runx2-induced osteogenic differentiation, the miR-690 mimics and anti-miR-690 oligos purchased from GenePharma (Shanghai, China) were used to promote and inhibit miR-690 activity, respectively. Negative controls (miR and anti-miR control) were used for both reactions. To examine the effect of p65 and IL-6 on Runx2-induced osteogenic differentiation, the cDNAs encoding mouse full-length p65 and IL-6 were obtained by PCR and cloned into the expression vector pcDNA3.1 (Invitrogen), and the IL-6-targeting siRNA oligos (si-IL-6) was purchased from Sigma. Negative controls (pcDNA3.1 and si-NC oligos) were also used for these reactions. For transfection, Lipofectamine 2000 (Invitrogen) was mixed with above-mentioned RNAs (50 nM/100 nM) or vectors (2.5 μg/well) according to the manufacturer’s instructions, and these solutions were directly mixed with C2C12/Runx2Dox cells in 6-well culture plates. For osteogenic differentiation, the medium was replaced with fresh medium containing 10 % FBS and 10 μg/ml of Dox at 4 h after transfection.
Luciferase reporter assay
A 697-bp fragment of the p65 3′UTR containing the predicted binding sites for miR-690 was amplified from mouse genomic DNA using specific primers. Amplicon was cleaved with XhoI and NotI (MBI Fermentas, Burlington, ON, Canada) and cloned in between the XhoI and NotI cleavage sites of psiCHECK-2 vector (Promega, Madison, WI, USA) downstream of the Renilla luciferase reporter gene. The resulting construct was named psiCHECK-2-p65. MiR-690 (50 nM), miR-690-mut (mimics containing a mutated miR-690 seed sequence, 50 nM) (GenePharma), or miR control (50 nM) was co-transfected with psiCHECK-2-p65 reporter (200 ng) into C2C12/Runx2Dox cells using Lipofectamine 2000. After 48 h, firefly and renilla luciferase activity were measured in cell lysates using a Dual-Luciferase Reporter Assay System (Promega) on a Fusion plate reader (Perkin-Elmer life and Analytical Sciences). Firefly luciferase activity was used for normalization and as an internal control for transfection efficiency.
EMSA
Nuclear extracts were prepared from Dox-treated C2C12/Runx2Dox cells according to the manufacturer’s instructions (Active Motif, Carlsbad, CA, USA). EMSA was performed using the EMSA kit (Promega). For supershift assays, an antibody against Flag-Runx2 (anti-Flag antibody) (Sigma) or normal mouse IgG (Sigma) was added to the reaction mixture and incubated 25 min before the addition of the labeled oligonucleotide. All DNA–protein complexes were resolved by electrophoresis on 5 % native polyacrylamide gels and transmembrane to immobilon-Ny + (Millipore).
ChIP assay
ChIP assay was performed according to the manufacturer’s instructions (Active Motif). Briefly, the crosslinked protein-DNA complexes prepared from Dox-treated C2C12/Runx2Dox cells were incubated with 4 μg of the anti-Flag antibody (Sigma) or normal mouse IgG (Sigma) and rotated at 4 °C overnight, and then were incubated with protein G beads at 4 °C for 4 h. The complexes were eluted with buffer containing 1 % SDS and 0.1 M NaHCO3, and crosslinks were reversed at 65 °C. DNA was recovered by phenol–chloroform extraction and ethanol precipitation and then subjected to PCR analysis. Amplified products were electrophoresed through 2 % agarose gel and visualized by Goldview staining.
Statistical analysis
All data were expressed as the mean ± standard deviation (SD) (n = 3), and statistical analysis was performed using SPSS software (version 16.0, Chicago, USA). Differences between two groups were analyzed by the unpaired Student’s t test, and differences between multiple groups were analyzed by ANOVA. P < 0.05 was considered statistically significant.
Abbreviations
- Runx2:
-
runt-related transcription factor 2
- miRNAs:
-
microRNAs
- NF-κB:
-
nuclear factor kappa B
- OC:
-
osteocalcin
- 3′UTR:
-
3′ untranslated region
- BMP-2:
-
bone morphogenetic protein 2
- IL-6:
-
interleukin-6
- ALP:
-
alkaline phosphatase
- Dox:
-
doxycycline
- qPCR:
-
quantitative PCR
- EMSA:
-
electrophoretic mobility shift assay
- ChIP:
-
chromatin immunoprecipitation
- MDSCs:
-
myeloid-derived suppressor cells
- TNFα:
-
tumor necrosis factor alpha
- MSCs:
-
mesenchymal stem cells
- DMEM:
-
Dulbecco’s modified Eagle medium
- FBS:
-
fetal bovine serum
- HRP:
-
horseradish peroxidase
- SD:
-
standard deviation
References
Jensen ED, Gopalakrishnan R, Westendorf JJ. Regulation of gene expression in osteoblasts. BioFactors. 2010;36:25–32.
Soltanoff CS, Yang S, Chen W, Li YP. Signaling networks that control the lineage commitment and differentiation of bone cells. Crit Rev Eukaryot Gene Expr. 2009;19:1–46.
Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, et al. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–9.
Li Z, Hassan MQ, Volinia S, van Wijnen AJ, Stein JL, Croce CM, et al. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc Natl Acad Sci USA. 2008;105:13906–11.
Itoh T, Nozawa Y, Akao Y. MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem. 2009;284:19272–9.
Yang L, Cheng P, Chen C, He HB, Xie GQ, Zhou HD, et al. miR-93/Sp7 function loop mediates osteoblast mineralization. J Bone Miner Res. 2012;27:1598–606.
Huang J, Zhao L, Xing L, Chen D. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells. 2010;28:357–64.
Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor Runx2. Proc Natl Acad Sci USA. 2011;108:9863–8.
Kania MA, Bonner AS, Duffy JB, Gergen JP. The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes Dev. 1990;4:1701–13.
Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–54.
Enomoto H, Enomoto-Iwamoto M, Iwamoto M, Nomura S, Himeno M, Kitamura Y, et al. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J Biol Chem. 2000;275:8695–702.
Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–64.
Banerjee C, McCabe LR, Choi JY, Hiebert SW, Stein JL, Stein GS, et al. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J Cell Biochem. 1997;66:1–8.
Yu S, Geng Q, Sun F, Yu Y, Pan Q, Hong A. Osteogenic differentiation of C2C12 myogenic progenitor cells requires the Fos-related antigen Fra-1—a novel target of Runx2. Biochem Biophys Res Commun. 2013;430:173–8.
Libermann TA, Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol Cell Biol. 1990;10:2327–34.
Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, et al. Databases on transcriptional regulation: TRANSFAC: TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–7.
Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–52.
Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, et al. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37:W273–6.
Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–7.
Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–47.
Hirata-Tsuchiya S, Fukushima H, Katagiri T, Ohte S, Shin M, Nagano K, et al. Inhibition of BMP2-induced bone formation by the p65 subunit of NF-κB via an interaction with Smad4. Mol Endocrinol. 2014;28:1460–70.
Peruzzi B, Cappariello A, Del Fattore A, Rucci N, De Benedetti F, Teti A. c-Src and IL-6 inhibit osteoblast differentiation and integrate IGFBP5 signalling. Nat Commun. 2012;3:630.
Balint E, Lapointe D, Drissi H, van der Meijden C, Young DW, van Wijnen AJ, et al. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J Cell Biochem. 2003;89:401–26.
Fang S, Deng Y, Gu P, Fan X. MicroRNAs regulate bone development and regeneration. Int J Mol Sci. 2015;16:8227–53.
Hu R, Liu W, Li H, Yang L, Chen C, Xia ZY, et al. A Runx2/miR-3960/miR-2861 regulatory feedback loop during mouse osteoblast differentiation. J Biol Chem. 2011;286:12328–39.
Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87.
Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–14.
Gong Y, Xu F, Zhang L, Qian Y, Chen J, Huang H, et al. MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol Cell Biochem. 2014;387:227–39.
Zhang ZJ, Zhang H, Kang Y, Sheng PY, Ma YC, Yang ZB, et al. miRNA expression profile during osteogenic differentiation of human adipose-derived stem cells. J Cell Biochem. 2012;113:888–98.
Yang N, Wang G, Hu C, Shi Y, Liao L, Shi S, et al. Tumor necrosis factor α suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis. J Bone Miner Res. 2013;28:559–73.
Mo X, Lu Y, Han J. Effects of targeted modulation of miR-762 on expression of the IFITM5 gene in Saos-2 cells. Intractable Rare Dis Res. 2014;3:12–8.
Sehic A, Risnes S, Khuu C, Khan QE, Osmundsen H. Effects of in vivo transfection with anti-miR-214 on gene expression in murine molar tooth germ. Physiol Genomics. 2011;43:488–98.
Tang X, Muniappan L, Tang G, Ozcan S. Identification of glucose-regulated miRNAs from pancreatic β cells reveals a role for miR-30d in insulin transcription. RNA. 2009;15:287–93.
Hegde VL, Tomar S, Jackson A, Rao R, Yang X, Singh UP, et al. Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Δ9-tetrahydrocannabinol in vivo: regulation of CCAAT/enhancer-binding protein α by microRNA-690. J Biol Chem. 2013;288:36810–26.
Chen S, Zhang Y, Kuzel TM, Zhang B. Regulating Tumor Myeloid-Derived Suppressor Cells by MicroRNAs. Cancer Cell Microenviron. 2015;2:e637.
Yang W, Yee AJ. Versican 3′-untranslated region (3′UTR) promotes dermal wound repair and fibroblast migration by regulating miRNA activity. Biochim Biophys Acta. 2014;1843:1373–85.
Wong JJ, Ritchie W, Gao D, Lau KA, Gonzalez M, Choudhary A, et al. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J Hematol Oncol. 2014;7:42.
Hunsberger JG, Fessler EB, Chibane FL, Leng Y, Maric D, Elkahloun AG, et al. Mood stabilizer-regulated miRNAs in neuropsychiatric and neurodegenerative diseases: identifying associations and functions. Am J Transl Res. 2013;5:450–64.
Delić D, Grosser C, Dkhil M, Al-Quraishy S, Wunderlich F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids. 2010;75:998–1004.
Soysa NS, Alles N. NF-kappaB functions in osteoclasts. Biochem Biophys Res Commun. 2009;378:1–5.
Fritz EA, Glant TT, Vermes C, Jacobs JJ, Roebuck KA. Titanium particles induce the immediate early stress responsive chemokines IL-8 and MCP-1 in osteoblasts. J Orthop Res. 2002;20:490–8.
Kurokouchi K, Kambe F, Yasukawa K, Izumi R, Ishiguro N, Iwata H, et al. TNF-alpha increases expression of IL-6 and ICAM-1 genes through activation of NF-kappaB in osteoblast-like ROS17/2.8 cells. J Bone Miner Res. 1998;13:1290–9.
Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–32.
Kaneki H, Guo R, Chen D, Yao Z, Schwarz EM, Zhang YE, et al. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem. 2006;281:4326–33.
Zhao L, Huang J, Zhang H, Wang Y, Matesic LE, Takahata M, et al. Tumor necrosis factor inhibits mesenchymal stem cell differentiation into osteoblasts via the ubiquitin E3 ligase Wwp1. Stem Cells. 2011;29:1601–10.
Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–7.
Liao L, Yang X, Su X, Hu C, Zhu X, Yang N, et al. Redundant miR-3077-5p and miR-705 mediate the shift of mesenchymal stem cell lineage commitment to adipocyte in osteoporosis bone marrow. Cell Death Dis. 2013;4:e600.
Nakamura T, Toita H, Yoshimoto A, Nishimura D, Takagi T, Ogawa T, et al. Potential involvement of Twist2 and Erk in the regulation of osteoblastogenesis by HB-EGF-EGFR signaling. Cell Struct Funct. 2010;35:53–61.
Sun F, Xie Q, Ma J, Yang S, Chen Q, Hong A. Nuclear factor Y is required for basal activation and chromatin accessibility of fibroblast growth factor receptor 2 promoter in osteoblast-like cells. J Biol Chem. 2009;284:3136–47.
Authors’ contributions
SY, QG, and QP: conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing; ZL: collection and/or assembly of data, data analysis, and interpretation; SD: data analysis, and interpretation; QX: collection and/or assembly of data; FS and CW: collection of data, and manuscript revision; YH: data analysis and interpretation, manuscript revision, and administrative support; AH: conception and design, data analysis and interpretation, manuscript writing, and financial support of manuscript. All authors read and approved the final manuscript.
Acknowledgements
We wish to express our gratitude to Beijing CapitalBio Corporation (Beijing, China) for its assistance in obtaining miRNA profiling data. This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 90919050, 81371913, 31400824), the National Key New Drug Creation of China (2012ZX09103301-034), the Guangdong Province Higher Vocational Colleges and Schools Pearl River Scholar Funded Scheme (2012).
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Shouhe Yu, Qianqian Geng and Qiuhui Pan contributed equally to this work
Additional files
13578_2016_73_MOESM1_ESM.tif
Additional file 1: Figure S1. C2C12/Runx2Dox cells are actually the wild type C2C12 myogenic progenitor cells in the absence of Dox. (A) C2C12/Runx2Dox cells were cultured in medium containing 2 % horse serum for different times. Total RNA prepared from cells at the indicated times was subjected to real-time qPCR analysis. Data are presented as mean ± SD (n = 3). *P < 0.05 compared with untreated cells (0 d). (B) C2C12/Runx2Dox cells were treated with Dox and BMP-2 for 9 days respectively, and the ALP activity was determined by ALP staining. Similar results were obtained in three independent experiments.
13578_2016_73_MOESM2_ESM.tif
Additional file 2: Figure S2. The expression of Flag-Runx2 and Myog during Runx2-induced osteogenic differentiation. (A) C2C12/Runx2Dox cells were treated with Dox for the times indicated and subjected to western blot analysis with anti-Flag antibody to detect Flag-Runx2. Similar results were obtained in three independent experiments. (B) C2C12/Runx2Dox cells were treated with Dox for the times indicated and subjected to real-time qPCR analysis for Myog mRNA. Data are presented as mean ± SD (n = 3). *P < 0.05 compared with untreated cells (0 h).
13578_2016_73_MOESM3_ESM.tif
Additional file 3: Figure S3. Detection of Flag-Runx2 by immunofluorescence microscopy in Dox-treated C2C12/Runx2Dox cells at short (A) and long (B) incubation times, respectively. Immunofluorescence staining for Flag-Runx2 (red) revealed the nuclear distribution (arrow) of Runx2. Nuclei were visualized by DAPI (blue). Scale bar in micrograph represents 20 μm.
13578_2016_73_MOESM4_ESM.tif
Additional file 4: Figure S4. Dox itself has no effect on the osteogenic differentiation of C2C12 cells. In Dox-treated C2C12/vectorDox cells, ALP and matrix mineralizing activity were measured by ALP and Alizarin red staining at days 9 and 28, respectively. Similar results were obtained in three independent experiments.
13578_2016_73_MOESM5_ESM.tif
Additional file 5: Figure S5. Clustered miRNA expression patterns of 609 miRNAs are shown graphically. Each row represents a different miRNA, and each column displays miRNA expression at each time point (0, 3, 6, 9, 12, 24, 48, and 72 h). Data values displayed as yellow and blue represent elevated and reduced expression, respectively.
13578_2016_73_MOESM6_ESM.tif
Additional file 6: Figure S6. Dox itself has no effect on the expression of miR-690. Real-time qPCR results of miR-690 in Dox-treated C2C12/vectorDox cells. Data are presented as mean ± SD (n = 3).
13578_2016_73_MOESM7_ESM.tif
Additional file 7: Figure S7. Effect of miR-690 inhibitor on the mRNA and protein levels of p65. The anti-miR control (100 nM) or anti-miR-690 (100 nM) was transfected into C2C12/Runx2Dox cells, and then the mRNA (A) and protein (B) levels of p65 were examined by real-time qPCR and western blot respectively. The mRNA level of p65 was normalized to 18S rRNA. Beta-actin expression was used as a loading control. Similar results were obtained in three independent experiments.
13578_2016_73_MOESM8_ESM.tif
Additional file 8: Figure S8. The expression of p65 is decreased at the protein but not the mRNA level during Runx2-induced osteogenic differentiation. C2C12/Runx2Dox cells were treated with Dox for the times indicated and subjected to real-time qPCR analysis for p65 mRNA (A) and western blot analysis with anti-p65 antibody to detect p65 (B). Similar results were obtained in three independent experiments.
13578_2016_73_MOESM9_ESM.tif
Additional file 9: Figure S9. IL-6 is upregulated by overexpression of p65 in C2C12 cells. The empty vector (pcDNA3.1) or p65 expression vector (pcDNA3.1-p65) was transfected into C2C12/Runx2Dox cells. The transfected cells were cultured for 3 days in the absence of Dox, and then subjected to real-time qPCR analysis for IL-6 mRNA (left panel). Data are presented as mean ± SD (n = 3). *P < 0.05 compared with pcDNA3.1-transfected cells. Western blot was performed to assess the overexpression level of p65 (right panel).
13578_2016_73_MOESM10_ESM.tif
Additional file 10: Figure S10. p65 exerts its negative effect on the expression of OC partially through IL-6 upregulation. (A) C2C12/Runx2Dox cells were transfected with si-NC (50 nM) and si-IL-6 (50 nM), respectively, and then cultured for 3 days in the absence of Dox. The mRNA level of IL-6 was examined by real-time qPCR. *P < 0.05 compared with si-NC sample. (B) si-NC (50 nM) or si-IL-6 (50 nM) was co-transfected with p65 expression vector (pcDNA3.1-p65) into C2C12/Runx2Dox cells, and then the transfected cells were treated with Dox for 3 days. The mRNA level of Alp and OC was examined by real-time qPCR. *P < 0.05 compared with si-NC sample.
13578_2016_73_MOESM11_ESM.pdf
Additional file 11: Table S1–S7. Table S1. Nucleotide sequence of 3′UTR primer for p65 WT reporter plasmid. Table S2. Nucleotide sequence of primers used for RT-PCR. Table S3. Nucleotide sequence of miRNA RT primers. Table S4. Nucleotide sequence of primers used for miRNA detection. Table S5. Nucleotide sequence of primers used for real-time qPCR analysis. Table S6. Nucleotide sequence of the oligonucleotides (probes) used in EMSA. Table S7. Nucleotide sequence of primers used for ChIP assay.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, S., Geng, Q., Pan, Q. et al. MiR-690, a Runx2-targeted miRNA, regulates osteogenic differentiation of C2C12 myogenic progenitor cells by targeting NF-kappaB p65. Cell Biosci 6, 10 (2016). https://doi.org/10.1186/s13578-016-0073-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13578-016-0073-y