Abstract
Vegetative insecticidal proteins 3A (Vip3A) were important insecticidal proteins for control of lepidopteran pests. Previous study demonstrated that Vip3Aa and Vip3Ad showed significant difference in insecticidal activities against Spodoptera exigua, while the molecular mechanism remained ambiguous. Here we demonstrated that the difference in insecticidal activities between Vip3Aa and Vip3Ad might be caused by the difference in stability of Vip3Aa and Vip3Ad in S. exigua midgut protease. Vip3Aa was quite stable while Vip3Ad could be further degraded. Molecular dynamics simulation revealed that Vip3Aa was more stable than Vip3Ad, with smaller RMSD and RMSF value. Amino acid sequence alignment indicated that three were three extra prolines (P591, P605 and P779) located on Vip3Aa. We further identified that residue P591 played a crucial role on stability and insecticidal activity of Vip3Aa. Taken together, our study demonstrated that the stability was essential for the insecticidal activity of Vip3A toxins, which might provide new insight into the action mode of Vip3A toxins and contribute to the design Vip3A variants with improved stability and insecticidal activity.
Similar content being viewed by others
Introduction
Vegetative insecticidal proteins 3A (Vip3A), secreted from Bacillus thuringiensis (Bt) during their vegetative growth stage, are important insecticidal proteins that show toxicity against a wide variety of lepidopteran pests (Chakrabarty et al. 2020; Chakroun et al. 2016a, b; Gupta et al. 2021). It was worth mentioning that Vip3A proteins displayed synergistic effect when coupling with Cry toxins for the control of agricultural pests (Wang et al. 2020; Lemes et al. 2014). It was also reported that Vip3A proteins were very active against insect species of Spodoptera, which showed low susceptibility to Cry proteins (Zhu et al. 2006; Baranek et al. 2015; Song et al. 2016). Furthermore, because Vip3 proteins did not share the binding sites with Cry toxins, it was not easy for target pests to develop cross-resistance between Vip3A and Cry toxins (Walsh et al. 2014; Jackson et al. 2007). These properties made Vip3A toxins suitable candidates to complement Cry toxins in Bt crops to broaden the insecticidal spectrum and for resistance management purposes.
Up to date, there are 10 Vip3A subtypes, including Vip3Aa to Vip3Aj, have been identified and reported (Crickmore et al. 2022). The different Vip3A proteins exhibited significant differences in toxicity and insecticidal spectrum against target insects (Boukedi et al. 2018; de Escudero et al. 2014; Hernández-Martínez et al. 2013). For example, Vip3Aa, Vip3Ae and Vip3Af were toxic to Spodoptera littoralis, with LC50 of 228.42 ng/cm2, 65.71 ng/cm2, and 388.90 ng/cm2, respectively. By contrast, Vip3Ad showed little insecticidal activity against this species (Boukedi et al. 2018). Vip3Aa exhibited toxicity against considerable number of lepidopteran species, while the insecticidal spectrum of Vip3Ab was relatively narrow (de Escudero et al. 2014; Hernández-Martínez et al. 2013). Similar to Cry toxins, Vip3A toxins were produced in protoxin form and could be cleaved into activated-toxin in target insect midgut (Sellami et al. 2015; Kunthic et al. 2017; Chakroun et al. 2016a, b). Some researches revealed that Vip3A activated-toxins were more susceptible to the action of proteases when compared with Cry toxins (Caccia et al. 2014; Abdelkefi-Mesrati et al. 2011). These findings suggested that excessive degradation might also affect the activities of Vip3A toxins. The binding ability of Vip3A toxin was also reported involving in the action of Vip3A toxins (Jiang et al. 2018; Liu et al. 2011). The analysis of different Vip3A toxins with different activity could contribute to better understanding of the insecticidal mechanism of Vip3A toxin and designing of Vip3A toxins with improved activity.
Previous study demonstrated that Vip3Aa and Vip3Ad showed significant difference in insecticidal activities against lepidopteran pest Spodoptera exigua, while the molecular mechanism was still unclear (Pan et al. 2017). Here we found that both of Vip3Aa and Vip3Ad could be cleaved into activated-toxins by Spodoptera exigua midgut protease and their cleavage sites were the same. However, Vip3Aa activated-toxin was stable when incubation with S. exigua midgut protease while Vip3Ad activated-toxin was less stable and could be further degraded. Molecular dynamics simulation also revealed that Vip3Aa was more stable than Vip3Ad. Amino acid sequence alignment displayed that three were three extra prolines (P591, P605 and P779) located on Vip3Aa and the mutation of residue P591 could lead to the decline of stability and insecticidal activity of Vip3Aa. Taken together, our study demonstrated that the stability of Vip3A toxin might play a key role for its insecticidal activity.
Materials and methods
Insects
A laboratory population of S. exigua larvae was purchased from Henan Jiyuan Baiyun Industry Co., Ltd, China. S. exigua larvae were fed with artificial diet provided by Hubei Biopesticide Engineering Research Center under the conditions of 26 ± 2 °C, 70% humidity and photoperiod of 14: 10 h (light: dark).
The S. exigua midgut proteases were prepared as previously described (Xu et al., 2016). 5 midguts were extracted from third instar larvae of S. exigua and followed by two washes with cold sodium chloride (128 mM). Midgut tissues were homogenized with 200 μL of phosphate-buffered saline (PBS) buffer (50 mM, pH 7.4) and centrifuged at 25,000 g for 30 min at 4 °C. The supernatants, which contained midgut proteases, were quantified by the BCA Protein Assay Kit (Beyotime, China) according to the instructions of the manufacturers.
Expression and purification of Vip3Aa and Vip3Ad
E. coli BL21 (DE3) cells harboring pET30a-vip3Aa and pET30a-vip3Ad (the DNA sequences of vip3Aa and vip3Ad were listed in Additional file 1: Table S1) were grown in LB medium containing 35 μg/ mL kanamycin. The expression of Vip3Aa and Vip3Ad were induced overnight at 20 °C with 0.1 mM isopropyl-B-D-thiogalactopyranoside (IPTG) after OD600 nm reached 0.4–0.6. Cells were pelleted at 6000 g at 4 °C and were resuspended with Tris–HCl buffer (100 mM Tris–HCl, 200 mM NaCl, 25 mM imidazole, pH 7.0). Cells were lysed by ultrasonication and the soluble protein was purified by a Ni-IDA Prepacked Column (Sangon, China) according to the manufacturer’s instructions. The purified Vip3Aa and Vip3Ad were exchanged into Sodium carbonate buffer (50 mM, pH 9.5) by a PD-10 desalination column (GE Healthcare, USA). Protein concentration was determined using a BCA Protein Assay Kit (Beyotime, China). Trypsin or S. exigua midgut juice was used to produce Vip3Aa and Vip3Ad activated-toxins as previous described (Pan et al. 2017).
Bioassays
The insecticidal activities of Vip3Aa and Vip3Ad against first-instar larvae of S. exigua were estimated according to Pan et al. (2017). Six concentrations of toxins were set up, and sodium carbonate buffer (50 mM, pH 9.5) was served as a negative control. Twenty S. exigua larvae were treated with each concentration and the bioassays were repeated two times. Mortalities were recorded after 7 days. The LC50 values were analysed by SPSS 17.0 (Statistical Product and Service Solutions) using PROBIT analysis (Finney 1971).
Transmission electron microscopy
Second-instar larvae of S. exigua were starved for 12 h and fed an artificial diet covered with Vip3Aa and Vip3Ad protoxins (4000 ng/cm2) for 48 h. After that, the midgut tissues of S. exigua were isolated, immediately fixed in 2.5% glutaraldehyde, and postfixed in 1% OsO4. The fixed midgut tissues were immerged into Epon for embedding. Ultrathin sections were sliced using a Leica ultramicrotome (EM UC7), stained with uranyl acetate and then lead citrate. The ultrastructure of the midgut epithelium was examined using a JEOL JEM-2100HC transmission electron microscope (Shao et al. 2016).
Edman degradation sequencing analysis
N-terminal sequencing was conducted to determine the cleavage site of Vip3Ad processed by S. exigua midgut protease as previous described. Specially, 300 μg Vip3Ad activated-toxin was separated by SDS-PAGE electrophoresis, electro-transferred onto a PVDF membrane and submitted for amino acid sequencing using SHIMADZU automated protein/peptide sequencer (PPSQ-333A, JAPAN) (Xu et al. 2016).
Proteolysis assay
Proteolysis assay was performed to evaluate the stability of Vip3Aa and Vip3Ad activated-toxins under S. exigua midgut juice. 20 μg Vip3Aa and Vip3Ad protoxin were mixed with S. exigua midgut juice (0.2 μg) in 50 μL final volume of sodium carbonate buffer (50 mM, pH 9.5). The mixtures were incubated at 30 °C for different incubation times, with constant shaking (about 40 rpm). The proteolytic reactions were stopped by boiling for 10 min and SDS-PAGE electrophoresis was used to assess the stability of Vip3Aa and Vip3Ad activated-toxins under S. exigua midgut juice.
Site-directed mutagenesis
Site-directed mutagenesis was carried out to create Vip3Aa variants P591A, P605A and P779A by polymerase chain reaction (PCR) using PrimeSTAR Max DNA Polymerase (TaKaRa, Japan). Plasmid pET30a-vip3Aa was used as template. Primers used for mutagenesis were listed in Additional file 1: Table S2. The PCR program was as follows: 98 °C for 10 min for pre-denaturation, (98 °C for 15 s, 60 °C for 15 s, 72 °C for 2 min) × 25 cycles, and a final elongation step at 72 °C for 5 min. After reaction, PCR product (10 μL) were digested with the Dpn I (1U) at 37 °C for 1 h and further transformed into E. coli BL21 (DE3) competent to obtain the relevant mutants.
Homology modeling and molecular dynamic simulation
The structure of Vip3Aa (PDB: 6TFJ) was obtained from PDB database (https://www.rcsb.org/). The homology model of and Vip3Ad was constructed by Swiss-Model (https://swissmodel.expasy.org/) with the crystal structure of Vip3Aa protoxin structure (PDB: 6TFJ) as the template (86.1% identity), and evaluated in SAVES (https://servicesn.mbi.ucla.edu/SAVES/).
The pretreatment of Vip3Aa and Vip3Ad for molecular dynamic (MD) were performed as previous study with AmberTools18. The system was heated from 0 to 300 K at constant volume in 50 ps with the protein restricted, equilibrated at constant pressure in 50 ps with the protein restricted and equilibrated for 500 ps without restriction of the protein. After equilibration, normal temperature and pressure (NPT) simulation was conducted for 20 ns to produce trajectories of MD simulation The root mean square deviation (RMSD) and the root mean square fluctuation (RMSF) were calculated for the protein backbone atoms using least-square fitting derived from the MD trajectories (Xu et al. 2020).
Results
Insecticidal activities of Vip3Aa and Vip3Ad against S. exigua
It was reported that Vip3Aa was toxic to S. exigua while Vip3Ad showed little insecticidal activity against S. exigua, although they shared more than 85% identity in amino acid sequence. To further investigate the molecular mechanism of the difference in insecticidal activities between Vip3Aa and Vip3Ad, we over-expressed Vip3Aa and Vip3Ad protoxin using E. coli Bl21 (DE3) expression system. SDS-PAGE indicated that Vip3Aa and Vip3Ad protoxins were both 89 kDa in molecular weight and could be cleaved into 65 kDa activated-toxins by S. exigua midgut juice (Fig. 1A). In fact, the 65 kDa activated-toxins were not a sole protein but composed of two attached fragments with molecular weight of 65 and 19 kDa, respectively (Jiang et al. 2020; Nuñez-Ramirez et al. 2020). We than evaluated the insecticidal activities of Vip3Aa and Vip3Ad against first instar larvae S. exigua larvae. As shown in Additional file 1: Fig S1, high concentration of Vip3Aa (400–1000 ng/cm2) could kill S. exigua larvae and low concentration of Vip3Aa (50–200 ng/cm2) could inhibit the growth of larvae. By contrast, it did not have any insecticidal or inhibitory activity under low concentration range of Vip3Ad (50–400 ng/cm2), inhibition effects were observed when the concentration of Vip3Ad increased to 1000 or 4000 ng/cm2 (Additional file 1: Fig S1). The LC50 value of Vip3Aa against S. exigua larvae was determined as 142 ng/cm2, much lower than that of Vip3Ad (> 4000 ng/cm2) (Fig. 1B). The disruptions of S. exigua midgut epithelium by Vip3Aa and Vip3Ad were also assessed by transmission electron microscopy. It demonstrated that serious pathological changes of the midgut epithelium and complete disintegration of the epithelial microvilli were clearly observed in Vip3Aa treatment group. While in Vip3Ad treatment group, the S. exigua midgut epithelial cells were integral, and the epithelial microvilli were neatly arranged, consistent with its nontoxicity against S. exigua larvae (Fig. 1C). The above results clearly confirmed the great distinction in insecticidal activities among Vip3Aa and Vip3Ad.
Studies on the activities of Vip3Aa and Vip3Ad against S. exigua. A Expression and purification of Vip3Aa and Vip3Ad. lane 1: Vip3Aa protoxin, lane 2: Vip3Aa activated-toxin, lane 3: Vip3Ad protoxin, lane 4: Vip3Ad activated-toxin. B Insecticidal activities of Vip3Aa and Vip3Ad against S. exigua. C The ultrastructure of S. exigua midgut epithelial cell treated with sodium carbonate buffer, Vip3Aa group and Vip3Ad group were observed using transmission electron microscope. EC: epithelial cell, M: microvilli
Vip3Aa was more stable than Vip3Ad in S. exigua midgut juice
Vip3A toxins were produced in protoxin form and activated by insect midgut protease. Our previous study showed that the activation of Vip3Aa by S. exigua midgut protease was cleaved at K198, while the cleavage site of Vip3Ad was unknown (Zhang et al., 2018). In present study, Edman degradation sequencing was performed and showed that the N-terminal sequence of Vip3Ad activated-toxin was “D-X-P-P-A” (Additional file 1: Fig. S2), which matched the amino acid sequence of “199DSPPA203” in Vip3Ad. These results indicated that proteolysis of Vip3Aa and Vip3Ad were both cleaved at K198 and produced the relevant activated-toxins.
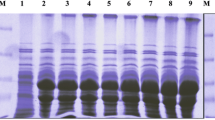
For Cry toxins that also produced by Bacillus thuringiensis, it was reported that the stability of activated-toxin was associative with its insecticidal activity. We therefore estimated the time course degradation of Vip3Aa and Vip3Ad under 10% S. exigua midgut juice. As shown in Fig. 2A, Vip3Aa protoxin was cleaved into 70 kDa activated-toxin by S. exigua midgut protease and the activated-toxin was not further degraded, suggesting that it was stable when incubation with S. exigua midgut protease. For Vip3Ad protoxin, we also observed that the protoxin was gradually cleaved into 70 kDa activated-toxin. However, the Vip3Ad activated-toxin could be further degraded and completely disappeared when the incubation time was over 24 h (Fig. 2B). These results clearly demonstrated that Vip3Aa activated-toxin was more stable than Vip3Ad activated-toxin.
To precisely interpret the differences in stability of Vip3Aa and Vip3Ad, these two toxins were subjected for 20 ns MD simulations at 303 K, respectively. The value of Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) were calculated. As shown in Fig. 3A, it demonstrated that both of Vip3Aa and Vip3Ad gradually became equilibrated at about 6 ns, however, the RMSD value of Vip3Ad fluctuated more than that of Vip3Aa, indicating that the overall structures of Vip3Aa was more stable than Vip3Ad. The RMSF results indicated that higher volatilities were clearly observed on residues 0–25, 370–382, 423–441 and 667–786 in Vip3Aa and Vip3Ad, also demonstrating that Vip3Aa was more stable than Vip3Ad (Fig. 3B).
Residue P591 was essential for stability and insecticidal activity of Vip3Aa
To explore the mechanism for the different stability of Vip3Aa and Vip3Ad activated-toixn, we first compared the amino acid sequence of Vip3Aa and Vip3Ad toxin since they are about 15% differentiation in amino acid sequence. We found that three were three proline (P591, P605 and P779) located on Vip3Aa, while the corresponding residues on Vip3Ad were S, A and T, respectively (Additional file 1: Fig S3). As far as we know, proline had only one rotatable angle compared to the other 19 natural amino acids, resulting a relatively rigid structure and losing less entropy (Qu et al. 2022). This founding let us speculate that the more proline on Vip3Aa might contribute to the stability of Vip3Aa during the processing of midgut juice.
To evaluate the residues P591, P605 and P779 on insecticidal activity of Vip3Aa, these three residues were mutated to alanine, constructing three mutants P591A, P605A and P779A. Time course degradation of wild-type and mutants Vip3Aa were performed. As shown in Fig. 4, wild-type and mutants Vip3Aa could be cleaved into 70 kDa activated-toxin. Observation after 24 h revealed that wild-type Vip3Aa, P605A and P779A activated-toxins were still existed and not further degraded, suggesting that they are stable under S. exigua midgut protease. By contrast, variant P591A was less stable in the S. exigua midgut protease and could be total degraded after 24 h. This result suggested that residue P591 was essential for the stability of Vip3Aa and its mutation could lead to more easily degradation of Vip3Aa when incubated with S. exigua midgut protease.
Bioassays were performed to further evaluate the insecticidal activities of wild-type, P591A, P605A and P779A Vip3Aa. As expected, the LC50 value of mutant P591A was 391 μg/cm2, higher when compared to wild-type, P605A, and P779A Vip3Aa, with LC50 values of 124, 195, and 135 ng/cm2, respectively (Table 1). These results indicated that residue P591A was essential for the stability and insecticidal activities of Vip3Aa.
Discussion
Previous study indicated that both Vip3Aa and Vip3Ad could bind to S. exigua BBMV, while the binding amount of Vip3Aa was more than Vip3Ad, which might be one reason for the difference in insecticidal activities between Vip3Aa and Vip3Ad (Pan et al. 2017). However, considering that Vip3Ad was nontoxic at all to S. exigua, we believed there were other factors resulting in the total inactivity of Vip3Ad.
The present study indicated that the cleavage site of Vip3Ad was at K198. This result demonstrated that the proteolysis activation of Vip3Aa and Vip3Ad were the same since previous study showed that the activation of Vip3Aa by S. exigua midgut protease was also cleaved at K198 (Zhang et al. 2018). The 65 kDa C-terminal fragments of Vip3A used to be considered as the toxic core. However, recent studies indicated that the N-terminal fragment (about 19 kDa) and the C-terminal fragment of Vip3A still bind together after digestion, and the N-terminus is required for the stability and toxicity of Vip3A (Jiang et al. 2020; Nuñez-Ramirez et al. 2020). We speculated that the stabilities of Vip3Aa and Vip3Ad were different and showed that Vip3Aa was more stable than Vip3Ad gut protease. Molecular dynamics simulation also revealed that Vip3Aa was more stable than Vip3Ad, with smaller RMSD and RMSF value. Interestingly, three were three extra prolines (P591, P605 and P779) located on Vip3Aa. To the best of our knowledge, the introduction of proline into proteins had been studied numerous times, generally with the aim of increasing stability (Qu et al. 2022). For example, the proline substitution in chitinase from Paenibacillus pasadenensis CS0611 resulted in a 26.3-fold increase in half-life at 50 °C and a 7.9 °C increase in half-inactivation temperature relative to wild-type enzyme (Xu et al. 2020). It also reported that the frequency of proline residues in thermophilic enzymes is higher than in mesophilic enzymes (Land et al. 2019). This founding let us speculate that the more proline on Vip3Aa might contribute to the stability of Vip3Aa during the processing of midgut juice. We further constructed three Vip3Aa mutants (P591A, P605A and P779A) and evaluated their stability and insecticidal activities compared with wild-type Vip3Aa. The result indicated that residue P591 played a crucial role on activity of Vip3Aa.
Taken together, our study demonstrated that the stability was essential for the insecticidal activity of Vip3A toxins, which might provide new insight into the action mode of Vip3A toxins and contribute to the design Vip3A variants with improved stability and insecticidal activity.
Availability of data and materials
The additional files are available online.
References
Abdelkefi-Mesrati L, Boukedi H, Chakroun M, Kamoun F, Azzouz H, Tounsi S, Jaoua S (2011) Investigation of the steps involved in the difference of susceptibility of Ephestia kuehniella and Spodoptera littoralis to the Bacillus thuringiensis Vip3Aa16 toxin. J Invertebr Pathol 107:198–201
Baranek J, Kaznowski A, Konecka E, Naimov S (2015) Activity of vegetative insecticidal proteins Vip3Aa58 and Vip3Aa59 of Bacillus thuringiensis against lepidopteran pests. J Invertebr Pathol 130:72–81
Boukedi H, Khedher SB, Abdelkefi-Mesrati L, Van Rie J, Tounsi S (2018) Comparative analysis of the susceptibility/tolerance of Spodoptera littoralis to Vip3Aa, Vip3Ae, Vip3Ad and Vip3Af toxins of Bacillus thuringiensis. J Invertebr Pathol 152:30–34
Caccia S, Chakroun M, Vinokurov K, Ferre J (2014) Proteolytic processing of Bacillus thuringiensis Vip3A proteins by two Spodoptera species. J Insect Physiol 67:76–84
Chakrabarty S, Jin M, Wu C, Chakraborty P, Xiao Y (2020) Bacillus thuringiensis vegetative insecticidal protein family Vip3A and mode of action against pest Lepidoptera. Pest Manag Sci 76:1612–1617
Chakroun M, Banyuls N, Bel Y, Escriche B, Ferré J (2016a) Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol Mol Biol Rev 80:329–350
Chakroun M, Banyuls N, Walsh T, Downes S, James B, Ferré J (2016b) Characterization of the resistance to Vip3Aa in Helicoverpa armigera from Australia and the role of midgut processing and receptor binding. Sci Rep 6:1–11
Crickmore N, Baum J, Bravo A, Lereclus D, Narva K, Sampson K, Schnepf E, Sun M, Zeigler DR (2022) “Bacillus thuringiensis toxin nomenclature”. http://www.lifesci.sussex.ac.uk/home/Neil_Crickmore/Bt/. Accessed 11 Jan 2022
de Escudero IR, Banyuls N, Bel Y, Maeztu M, Escriche B, Muñoz D, Ferré J (2014) A screening of five Bacillus thuringiensis Vip3A proteins for their activity against lepidopteran pests. JInvertebr Pathol 117:51–55
Finney DJ (1971) Probit analysis, 3rd edn. Cambridge University Press, London
Gupta M, Kumar H, Kaur S (2021) Vegetative insecticidal protein (Vip): a potential contender from Bacillus thuringiensis for efficient management of various detrimental agricultural pests. Front Microbiol 12:1139
Hernández-Martínez P, Hernández-Rodríguez CS, Van Rie J, Escriche B, Ferré J (2013) Insecticidal activity of Vip3Aa, Vip3Ad, Vip3Ae, and Vip3Af from Bacillus thuringiensis against lepidopteran corn pests. J Invertebr Pathol 113:78–81
Jackson RE, Marcus MA, Gould F, Bradley JJR, Van Duyn JW (2007) Cross-resistance responses of Cry1Ac-selected Heliothis virescens (Lepidoptera: Noctuidae) to the Bacillus thuringiensis protein Vip3A. J Econ Entomol 100:180–186
Jiang K, Hou X, Han L, Tan T, Cao Z, Cai J (2018) Fibroblast growth factor receptor, a novel receptor for vegetative insecticidal protein Vip3Aa. Toxins 10:546
Jiang K, Zhang Y, Chen Z, Wu D, Cai J, Gao X (2020) Structural and functional insights into the C-terminal fragment of insecticidal Vip3A toxin of Bacillus thuringiensis. Toxins 12:438
Kunthic T, Surya W, Promdonkoy B, Torres J, Boonserm P (2017) Conditions for homogeneous preparation of stable monomeric and oligomeric forms of activated Vip3A toxin from Bacillus thuringiensis. Eur Biophys J 46:257–264
Land H, Campillo-Brocal JC, Svedendahl Humble M, Berglund P (2019) B-factor guided proline substitutions in Chromobacterium violaceum amine transaminase: evaluation of the proline rule as a method for enzyme stabilization. ChemBioChem 20:1297–1304
Lemes ARN, Davolos CC, Legori PCBC, Fernandes OA, Ferre J, Lemos MVF, Desiderio JA (2014) Synergism and antagonism between Bacillus thuringiensis Vip3A and Cry1 proteins in Heliothis virescens, Diatraea saccharalis and Spodoptera frugiperda. PLoS ONE 9:e107196
Liu JG, Yang AZ, Shen XH, Hua BG, Shi GL (2011) Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J Invertebr Pathol 108:92–97
Núñez-Ramírez R, Huesa J, Bel Y, Ferré J, Casino P, Arias-Palomo E (2020) Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis. Nat Commun 11:1–9
Pan ZZ, Xu L, Zhang J, Liu B, Chen QX, Gao HJ, Guan X (2017) Screening of Vip3Aa60 and Vip3Ad5 and characterization of their binding to Spodoptera exigua midguts. Process Biochem 61:189–194
Qu G, Bi Y, Liu B, Li J, Han X, Liu W, Sun Z (2022) Unlocking the stereoselectivity and substrate acceptance of enzymes: proline-induced loop engineering test. Angew Chem 134:e202110793
Sellami S, Cherif M, Abdelkefi-Mesrati L, Tounsi S, Jamoussi K (2015) Toxicity, activation process, and histopathological effect of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 on Tuta absoluta. Appl Biochem Biotechnol 175:1992–1999
Shao E, Lin L, Chen C, Chen H, Zhuang H, Wu S, Huang Z (2016) Loop replacements with gut-binding peptides in Cry1Ab domain II enhanced toxicity against the brown planthopper, Nilaparvata lugens (Stål). Sci Rep 6:20106
Song F, Lin Y, Chen C, Shao E, Guan X, Huang Z (2016) Insecticidal activity and histopathological effects of Vip3Aa protein from Bacillus thuringiensis on Spodoptera litura. J Microbiol Biotechnol 26:1774–1780
Walsh TK, Downes SJ, Gascoyne J, James W, Parker T, Armstrong J, Mahon RJ (2014) Dual Cry2Ab and Vip3A resistant strains of Helicoverpa armigera and Helicoverpa punctigera (Lepidoptera: Noctuidae); testing linkage between loci and monitoring of allele frequencies. J Econ Entomol 107:1610–1617
Wang Z, Wang K, Bravo A, Soberón M, Cai J, Shu C, Zhang J (2020) Coexistence of cry9 with the vip3a gene in an identical plasmid of Bacillus thuringiensis indicates their synergistic insecticidal toxicity. J Agric Food Chem 68:14081–14090
Xu L, Pan ZZ, Zhang J, Liu B, Zhu YJ, Chen QX (2016) Proteolytic activation of Bacillus thuringiensis Cry2Ab through a belt-and-braces approach. J Agric Food Chem 64:7195–7200
Xu P, Ni ZF, Zong MH, Ou XY, Yang JG, Lou WY (2020) Improving the thermostability and activity of Paenibacillus pasadenensis chitinase through semi-rational design. Int J Biol Macromol 150:9–15
Zhang J, Pan ZZ, Xu L, Liu B, Chen Z, Li J, Niu LY, Chen QX (2018) Proteolytic activation of Bacillus thuringiensis Vip3Aa protein by Spodoptera exigua midgut protease. Int J Biol Macromol 107:1220–1226
Zhu C, Ruan L, Peng D, Yu Z, Sun M (2006) Vegetative insecticidal protein enhancing the toxicity of Bacillus thuringiensis subsp kurstaki against Spodoptera exigua. Lett Appl Microbiol 42:109–114
Acknowledgements
Not applicable.
Funding
The present investigation was supported by the National Natural Science Foundation of China (31972335), the Extended Project of National Natural Science Foundation of Fujian Academy of Agricultural Sciences (GJYS2019003).
Author information
Authors and Affiliations
Contributions
YZ and YS provided conceptual framework and technical oversight on all experiments; BF and LX conducted the experiments and analyzed the experimental data. YZ, BF and LX wrote the manuscript; QC and MZ revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
All authors have provided consent for this publication.
Competing interests
All the authors declare that they do not have any conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Primer sequences used for the generation of Vip3Aa mutants. Table S2. DNA sequences of Vip3Aa and Vip3Ad. Figure S1. The insecticidal activities of Vip3Aa and Vip3Ad against first-instar larvae of S. exigua. Figure S2. N-terminal sequencing identified cleavage-sites of Vip3Ad processed by Spodoptera exigua midgut juice. (A) Spectrum of 19 PTH-amino acids standards; (B)-(F) N-terminal amino acid identification of Vip3Aa activated-toxin. Figure S3. Amino acid sequence alignment of Vip3Aa and Vip3Ad. The sequences in blue box represented the 65 kDa activated-toxins of Vip3Aa and Vip3Ad. The positions of three extra prolines (P591, P605 and P779) was indicated as (●).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fu, BW., Xu, L., Zheng, MX. et al. Stability is essential for insecticidal activity of Vip3Aa toxin against Spodoptera exigua. AMB Expr 12, 92 (2022). https://doi.org/10.1186/s13568-022-01430-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-022-01430-w