Abstract
Nitrification inhibitors and urease inhibitors, such as nitrapyrin and N-(n-butyl) thiophosphoric triamide (NBPT), can improve the efficiencies of nitrogen fertilizers in cropland. However, their effects on ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) across different soil pH levels are still unclear. In the present work, vegetable soils at four pH levels were tested to determine the impacts of nitrification and urease inhibitors on the nitrification activities, abundances and diversities of ammonia oxidizers at different pHs by real-time PCR, terminal restriction fragment length polymorphism (T-RFLP) and clone sequence analysis. The analyses of the abundance of ammonia oxidizers and net nitrification rate suggested that AOA was the dominate ammonia oxidizer and the key driver of nitrification in acidic soil. The relationships between pH and ammonia oxidizer abundance indicated that soil pH dominantly controlled the abundance of AOA but not that of AOB. The T-RFLP results suggested that soil pH could significantly affect the AOA and AOB community structure. Nitrapyrin decreased the net nitrification rate and inhibited the abundance of bacterial amoA genes in this vegetable soil, but exhibited no effect on that of the archaeal amoA genes. In contrast, NBPT just lagged the hydrolysis of urea and kept low NH4 +-N levels in the soil at the early stage. It exhibited no or slight effects on the abundance and community structure of ammonia oxidizers. These results indicated that soil pH, rather than the application of urea, nitrapyrin and NBPT, was a critical factor influencing the abundance and community structure of AOA and AOB.
Similar content being viewed by others
Introduction
Nitrification is a necessary transition to convert ammonia to nitrate in soils, which can significantly affect the ecological system. Nitrogen losses by the leaching of nitrate and the emissions of N2O have become more and more severe in last several years (IPCC 2007). Although it occurs in the terrestrial environments at very low concentrations, the concentration of nitrous oxide has grown 18.5% since the preindustrial period (IPCC 2007). Among total global nitrous oxide emitted by various sources and human activities, soils account for about 62% (Thomson et al. 2012). In addition, the nitrate unabsorbed by plants in soils can be leached due to its mobility in soil. To reduce nitrogen loss and increase the nitrogen fertilizer use efficiency, nitrification inhibitors (NIs) and urease inhibitors (UIs) are usually applied to agricultural systems (Cui et al. 2013; Sanz-Cobena et al. 2011).
Among the nitrification inhibitors, nitrapyrin or N-serve (2-chloro-6-(trichloromethyl) pyridine) is an individual inhibitor of ammonium oxidation (Hughes and Welch 1970). It inhibits the first step of nitrification from ammonia to nitrite (Kangatharalingam and Priscu 1993) by targeting ammonia monooxygenase (AMO) that catalyzes the conversion of NH3 to NH2OH (Arp et al. 2002). N-(n-butyl) thiophosphoric triamide (NBPT) is one of the most effective urease inhibitor (Bremner and Chai 1986; Bronson et al. 1989). It can lower the hydrolysis rate and volatilization loss of urea as it is applied to soils at high concentrations (Antisari et al. 1996; Watson et al. 1994). NBPT is transformed into N-(butyl) phosphoric triamide (NBPTO) as it is directly applied to soil (Creason et al. 1990). Both NBPT and NBPTO inhibit urease activity by competing with urea molecules for the enzyme Ni receptor sites (Kolodziej 1994).
In general, ammonia-oxidizing Archaea (AOA) and ammonia-oxidizing Bacteria (AOB) are the key drivers of the ammonia oxidation in soil (Jin et al. 2010; Li and Gu 2013). AOA and AOB use the same substrate for energy metabolism, but differ in their biochemistries and physiologic properties, such as the molecular and cellular features (Lehtovirta-Morley et al. 2011; Kim et al. 2012). The differences of AOA and AOB membrane structures lead to different membrane permeability, and thus cause different nitrification activities (Shen et al. 2008; Schouten et al. 2000). In addition, they occupy different ecological niches due to their dissimilar sensitivity to soil properties, such as nitrogen concentration, pH, water content, and so on (Morimoto et al. 2011; Shen et al. 2008). By comparing the inhibitory effects of allylthiourea (ATU) and nitrapyrin on ammonia oxidizers, Jäntti et al. (2013) concluded that ATU was not a good nitrification inhibitor for the communities containing AOA and nitrapyrin exhibited good inhibitory effects in presences of both AOA and AOB. Lehtovirta-Morley et al. (2013) investigated the inhibitory effects of nitrapyrin at different concentrations on the growth of ammonia oxidizers in soil and liquid cultures at pH 4.5 and found that the abundance of amoA varied with the nitrapyrin concentration and culture environment. (Belser and Schmidt 1981) reported the inhibitory effects of nitrapyrin on seven strains of ammonia oxidizers. Although nitrapyrin has been well-studied for years, its effects on the diversity and richness of ammonia oxidizers across different soil pH levels have never been reported. In previous study real-time PCR was used regularly based on amoA gene copies, but there has been very little concern on the community shifts of ammonia oxidizers in the presence of NIs and UIs (Liu et al. 2015). For example, Shen et al. (2013) found that the inhibitory effect of nitrapyrin on Ca. Nitrososphaera was more effective than that on N. multiformis by cultivating two representative strains of AOA and AOB and calculating the effective concentration 50 (EC50). It was shown that nitrapyrin could increase the ammonium retention and decrease the gross nitrification at 40 °C, but had no effect on the abundances of the bacterial ammonia oxidizer genes (Fisk et al. 2015). Other studies on the effect of nitrapyrin on amoA gene copies, nitrous oxide emissions also have been reported (Regina et al. 1998).
Ammonia-oxidizing microorganisms are influenced by many environmental factors, like substrate concentration, land utilization, organic matter, temperature, pH, oxygen concentration, and so on (Di et al. 2009; Ying et al. 2010; Abell et al. 2011), among which the soil pH has a particularly important effect on the abundance and diversity of ammonia oxidizers (Liu et al. 2015; Nicol et al. 2008). An examination of 65 soil samples collected from different regions and ecosystems indicated that pH drove the distribution of ammonia oxidizers and the AOA/AOB ratio declined with the increase of soil pH (Hu et al. 2013). AOA exhibited a more competitive advantage than AOB in acidic soils. In addition, the diversity of AOA was mainly affected by pH at pHs below 3.5 and not significantly influenced by the soil type and land-use method (Stempfhuber et al. 2015). Nicol et al. (2008) confirmed that soil pH determined the phylotype distribution of bacterial and archaeal ammonia oxidizers. Li et al. (2015) also reported that the ammonia oxidizers community structure and nitrification activity were significantly affected by soil pH. The aim of our work was to investigate the short-term effects of nitrapyrin and NBPT on nitrification and the abundance and community structure of AOA and AOB in a vegetable soil across a pH-gradient. Four treatments at four pH levels in the range of 3.97–7.04 were conducted. Molecular biological technologies including quantitative PCR, terminal restriction fragment length polymorphism (T-RFLP) and clone libraries were used in our study.
Materials and methods
Sample collection and microcosm incubation
Soil samples (0–20 cm depth) were collected from a vegetable field in Ningbo (121°51′N, 29°54′E), Zhejiang Province in eastern China. The sampling site was planted with Chinese cabbages (Brassica campestris spp. Pekinensis) for over 10 years and fertilized with average 450 kg N ha−1 year−1. The soil was classified as red soil (equivalent to Ultisols in US soil taxonomy), and developed on quaternary red earth. Mean annual rainfall in this area is 1300–1500 mm and mean temperature is 16.6 °C. The vegetable field is a sandy loam soil composed of 13.97% clay, 21.75% silt and 64.28% sand with a pH of 3.97, total nitrogen (TN) content of 0.64%, microbial biomass carbon (MBC) of 382.12 mg kg−1, microbial biomass nitrogen (MBN) contents of 63.04 mg kg−1, and potential nitrification rate (PNR) of 0.44 mg NO3 −-N kg−1 h−1. The collected soil samples were ground to pass through a 2-mm sieve after air-dried. The water-holding capacity of the soil was determined to assure the consistent water content.
The pH of the soil sample was adjusted to 3.97, 4.82, 6.07 and 7.04 by CaCO3, separately. Four treatments including control, 200 mg kg−1 urea-N, 200 mg kg−1 urea-N + nitrapyrin (0.1% of urea-N) and 200 mg kg−1 urea-N + NBPT (2% of urea-N) were applied to the soil of each pH level in triplicates. Soil samples were thoroughly mixed with the composite of urea, nitrapyrin and NBPT, and put in a plastic jar, then kept the 50% water-holding capacity (WHC). The plastic jar was covered with a plastic lid with aeration holes to keep an aerobic environment. The soil samples were then incubated in the dark at 25 °C for 28 days in a chamber. The soil moisture was maintained at 50% WHC by weighing the soil sample once a day.
After the fertilizer, nitrapyrin and NBPT were supplied, the destructive sampling of ~20 g soil was conducted in triplicates at day 1, 3, 7, 14, 21 and 28. About 5.0 g of soil samples were stored at −80 °C for DNA extraction, the rest of the fresh soil samples was used for further chemical analysis.
Soil chemical analysis
The soil NO3 −-N and NH4 +-N were extracted from fresh soil samples using 2 mol l−1 KCl (soil/KCl, 1:5) and measured with a flow injection analyzer (FLA star 5000 Analyzer, Foss, Denmark). MBC and MBN were determined by the fumigation-extraction method (Wu et al. 1990). Briefly, the soil samples were fumigated with CHCl3 for 24 h at room temperature in the dark. The fumigated samples and samples without fumigated were extracted with 0.5 M K2SO4 for 30 min on a shaker and filtrated. The filtrates were measured on an automated TOC Analyzer (TOC-500, Japan). PNR was measured in triplicates by the shaken-slurry method (Yao et al. 2011). Fifteen grams of fresh soil samples were mixed with 7.5 ml of 0.2 M KH2PO4, 17.5 ml of 0.2 M K2HPO4 and 75 ml of 0.05 M (NH4)2SO4, respectively, and incubated in the dark at 25 °C for 24 h on a 180 rpm shaker. Suspension aliquots of 10 ml were sampled at 2, 4, 22 and 24 h incubation, respectively, and immediately analyzed on the continuous flow analyzer to determine their nitrate concentrations. The measurements of other soil properties were the same as described by Zhang et al. (2012).
The net nitrification rate (n) was calculated by the formula presented by Persson and Wirén (1995) as follows:
where (NO3 −-N) t2 and (NO3 −-N) t1 are the concentrations of NO3 −-N in the soil at time t2 and time t1 respectively, and t is the number of days between t2 and t1.
DNA extraction
DNA was extracted from 500 mg frozen soil using the FastDNA® SPIN Kit for Soil (Bio 101, Vista, CA) according to the manufacturer’s instruction, immediately diluted ten times and stored at −20 °C for molecular analyses. DNA concentration was measured on a NanoDrop ND-1000 UV–vis spectrophotometer (NanoDrop®, USA).
Real-time PCR assay of amoA genes
Quantitative PCR of amoA genes was conducted on a Light Cycler 480 real-time PCR detection system (Roche480, USA). Standard plasmids of AOA and AOB were constructed and diluted one- to nine-folds to construct the standard curve. Two different pairs of primers were used to target the AOA and AOB respectively (Additional file 1: Table S1). Each PCR reaction was performed in a 20-μl reaction mixtures consisting of 0.5 μM of each primer, 10 μl of SYBR® Premix, 1 μl of tenfold dilution DNA template, 0.5 μl of bovine serum albumin (BSA, 20 mg·ml−1), and the residual volume replenished by deionized water. For quantification of AOA and AOB, the amplification efficiencies were in the range from 90 to 96% and the correlation coefficient (r2) of the determination ranged from 0.95 to 0.99 for all of the standard curves.
T-RFLP of amoA genes for ammonia oxidizers
For analysis of the ammonia oxidizers community, T-RFLP was conducted from the soils of all treatments at day 28. Primers used in the qPCR with the forward primer marked with 6-carboxyfluorescein (FAM) (Additional file 1: Table S1) were used for the T-RFLP (Ying et al. 2010). The AOA and AOB samples were digested with restriction with HpyCH4V and MspI, respectively. The PCR products were purified using the concrete method presented by Yao et al. (2011). Fragments with sizes longer than 50 bp and percentages higher than 1% were kept for cluster analysis and the rest fragments were eliminated.
Cloning and sequencing
To identify the T-RFs, the AOA and AOB clone libraries from all the soil samples at day 28 were constructed with same primers CrenamoA23f/616r and amoA-1F/2R used in the qPCR analysis, but the different enzyme. One hundred clones were selected from these two clone libraries. The sequences displaying less than 2% nucleotide dissimilarities with each other were grouped into the different operational taxonomic unit (OTU). Representative sequences selected from each OTU were then used to build phylogenetic trees. Phylogenetic trees were constructed with Mega software (Tamura et al. 2013). The sequences of AOA and AOB were grouped into 6 and 16 OTUs, respectively. Eight representative sequences of AOA and 23 representative sequences of AOB were selected. Sequences that were analogous to the representative sequences most were selected from the GenBank to construct the phylogenetic tree.
Statistical analysis
To compare the amoA genes abundance of AOA and AOB among all treatments, data were analyzed using ANOVA with SPSS 19.0 software (IBM, USA). Pearson correlation analysis (P < 0.05) was also performed in SPSS 19.0 software. In order to anticipate the variations among the ammonia oxidizer community structures of different treatments and pHs, the T-RFLP data were analyzed in the CANOCO version 4.5. The construction of phylogenetic trees used MEGA version 6.0 software (Tamura et al. 2013). A 500 replicates bootstrap analysis was performed to evaluate the cluster stability.
Accession numbers of nucleotide sequences
The nucleotide sequences accession numbers of AOA and AOB were KX683109–KX683208 and KY073755–KY073854, respectively.
Results
Concentrations of inorganic nitrogen and net nitrification rate
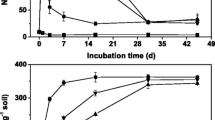
The temporal changes of NH4 +-N concentrations at four pH levels exhibited a similar trend. Compared with control soil, the soil treated with urea only contained a significantly higher NH4 +-N concentration (Fig. 1). The NH4 + concentration in urea treatment rapidly decreased after day 3 at all pH levels except for pH 7.04 and then remained higher than that in the control soil. Inhibitors exhibited different inhibiting effects on the soils of different pH levels. The NH4 +-N concentrations in urea + NBPT treatment were lower than in urea treatment at the early stage (Fig. 1). The NO3 −-N concentrations in soils of four pH levels gradually increased from day 1 to day 28. Control soil contained the lowest concentration of NO3 −-N and the highest concentration was found in soil of urea treatment. The NO3 −-N concentrations in the soils of four treatments remained relatively stable at pH 7.04 during the 28-day incubation (Fig. 2). The NO3 −-N concentrations in urea + nitrapyrin treatment at the four pH levels were ~8.2, ~5.2, ~1.1 and ~6.9%, respectively, lower than those in the soils of urea treatment at corresponding pH levels at day 28 (Fig. 2). The NO3 −-N concentrations in the soils of urea + NBPT treatment at the four pH levels were about ~14.5, ~2.7, ~7.9 and ~9.7%, also lower than those in soils of urea treatment at corresponding pH levels at day 28. Both inhibitors had a significant effect on nitrate concentration at pH 3.97 (Fig. 2).
Control soil exhibited the lowest net nitrification rates at all testing pHs. The application of urea increased the net nitrification rates 72.3, 134.8, 24.4, and 23.8%, respectively (Table 1). Net nitrification rates in urea + nitrapyrin and urea + NBPT treatments were higher than that in the control soil and lower than that in the soil of urea treatment at all testing pH levels except for pH 7.04. Net nitrification rate at pH 7.04 was the lowest for the soils of all treatments (Table 1). The correlation analysis suggested that net nitrification rate and soil pH in the tested soil existed a significant relationship (r = −0.736, P < 0.01) (Table 2).
amoA gene abundances
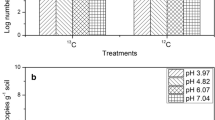
The variation in the AOA and AOB population sizes in the soils of different treatments was determined on day 28 (Fig. 3; Additional file 1: Figure S1). AOA amoA gene copy numbers were in the range from 9.49 × 104 to 3.43 × 109 g−1 dry soil. The highest AOA population (1.01 × 109 g−1dry soil) was found at soil pH 4.82 and the lowest (3.02 × 106 g−1dry soil) was observed at soil pH 7.04. No differences were found among the AOA amoA gene copy numbers of four treatments at the same pH level. The AOA abundance exhibited a positive correlation with the net nitrification rate (r = 0.790, P < 0.01) and a negative correlation with pH (r = −0.926, P < 0.01) (Table 2). The AOB amoA gene copy numbers ranged from 9.80 × 103 to 2.93 × 104 g−1 dry soil. The quantitative PCR analysis indicated that the AOA abundance was much higher than that of AOB abundance in the corresponding soil. No significant differences among the AOB abundances in the soils of all treatments except for the nitrapyrin treatment were observed (Fig. 3; Additional file 1: Figure S1).
Log number of AOA and AOB amoA copies in four different treatments (control; urea; urea + nitrapyrin; urea + NBPT) at different pH levels. Error bars indicate standard errors of three replicates, different small letters indicate the significant difference within the same treatment at different pH levels (P < 0.05)
AOA and AOB communities
The AOA and AOB communities in the vegetable soils of different treatments and at different pH levels were determined by T-RFLP (Figs. 4, 5). Seven AOA T-RFs were obtained, among which T-RF 166 bp (23.90–48.97%) and T-RF 217 bp (19.49–43.80%) accounted for the most proportion of the community. The relative abundance of AOA T-RF 166 bp was negatively correlated with soil pH (r = −0.963, P < 0.01) and the relative abundances of AOA T-RFs 217 bp and 205 bp exhibited extreme and positive correlations with soil pH (r = 0.905, P < 0.01; r = 0.778, P < 0.01, respectively) (Additional file 1: Table S2). PCA analyses further confirmed these community shifts among all treatments (Fig. 4b), and showed that two PCA axes explained 80.11% of the observed structure. A significant separation was found between treatments at pH 3.97 and 4.82 and those at pH 6.07 and 7.04 along PC1, and a distinct separation between treatments at pH 3.97 and those at pH 4.82 was observed along PC2. However, the applications of urea, nitrapyrin and NBPT exhibited no significant effect on the AOA T-RFLP pattern.
Abundance (a) and principle component analysis (b) of AOA T-RFs in vegetable soils treated with control, urea, urea + nitrapyrin, urea + NBPT at different pH. Error bars indicate standard errors of three replicates. Different pH represented by different colors, red, green, purple and black color indicate treatments at pH 3.97, pH 4.82, pH 6.07 and pH 7.04, respectively
Abundance (a) and principle component analysis (b) of AOB T-RFs in vegetable soils treated with control, urea, urea + nitrapyrin, urea + NBPT at different pH. Error bars indicate standard errors of three replicates. Different pH represented by different colors, red, green, purple and black color indicate treatments at pH 3.97, pH 4.82, pH 6.07 and pH 7.04, respectively
Four major AOB T-RFs of 56, 157, 235, and 256 bp were obtained. T-RF 157 bp accounted for about 62.73% of total T-RFs and its abundance had a significant and positive relationship with soil pH (r = 0.721, P < 0.01). The abundance of AOB T-RF 56 bp was negatively correlated to soil pH (r = −0.825, P < 0.01) (Additional file 1: Table S3). The two axes of PCA can explain 96.46% of the observed structures (Fig. 5b). The changes in the relative abundances of T-RFs 157 and 56 bp were the dominated alteration. The samples at pH 3.97 and 4.82 were separated from the samples at pH 6.07 and 7.04 along PC1 at a significant level. Similar to AOA, no significant separation was found between the four treatments at the same pH.
Genotypes of ammonia oxidizers and their phylogenetic tree analysis
The trees of amoA amino acid sequences and related GenBank sequences are shown in Figs. 6 and 7, respectively. There were 6 OTUs and 16 OTUs identified for AOA and AOB, respectively (Additional file 1: Table S4). Two representative OTUs (OTU 01 and OTU02) (Additional file 1: Table S4) and 8 representative sequences of AOA were included into three different clusters. Half of the representative sequences belonged to group Nitrososphaera and the rest were grouped into cluster I and cluster II (Fig. 6). The representative T-RFs 217 and 166 bp were related to OTU02 (KX683117) and OTU01 (KX683109) (Additional file 1: Table S4), respectively. One representative OTU (OTU05) of bacterial amoA genes was selected (Additional file 1: Table S4) and 23 AOB representative sequences were chosen and classified into five different clusters. Two of the clusters belonged to β-proteobacteria and the rest belonged to cluster I–III. Nine of the representative sequences belonged to β-proteobacteria and the rest 14 representative sequences were grouped into cluster I–III (Fig. 7). The representative T-RF 157 bp was most related to OTU05 (KY073756) (Additional file 1: Table S4).
Neighbor-joining phylogenetic tree of AOA amoA gene sequences retrieved from the vegetable soil. Sequences from this study are shown in bold and are described as clone name (accession number) T-RF size. Bootstrap values (>50%) are indicated at branch points. Reference sequences are described as clone name (environment, accession number). The number in bracket means clones. The scale bar represents 0.5% estimated sequence divergence
Neighbor-joining phylogenetic tree of AOB amoA gene sequences retrieved from the vegetable soil. Sequences from this study are shown in bold and are described as clone name (accession number) T-RF size. Bootstrap values (>50%) are indicated at branch points. Reference sequences are described as clone name (environment, accession number). The number in bracket means clones. The scale bar represents 1% estimated sequence divergence
The results of T-RFLP analyses and phylogenetic trees of AOA indicated that the dominant T-RFs 166 and 217 bp belonged to cluster II were the predominant AOA genotypes in the soils of all treatments (Additional file 1: Table S4, Fig. 6). T-RF 205 bp fell into both Nitrososphaera and Cluster II, but mainly into Nitrososphaera. The phylogenetic tree of AOB indicated that all amoA gene sequences of bacteria in this vegetable soil belonged into β-proteobacteria and cluster I–III (Fig. 7). T-RF 157 bp spread the whole clone library, but mostly belonged to cluster II. T-RF 56 bp fell into cluster I and β-proteobacteria.
Discussion
Abundances and activities of ammonia oxidizers
In the vegetable soil tested in the present work, the abundance of AOA was significantly higher than that of AOB, in agreement with previous findings (Chen et al. 2011, 2015; Gubry-Rangin et al. 2010). In addition, the abundance of AOA was negatively correlated to soil pH, indicating the preference of AOA to acidic soil. The AOA abundance sharply decreased at pH 7.04, indicating that AOA was inactivated in neutral soil. It might be explained by the competition between AOA and AOB for the limited energy source, especially the limited ammonia in the acidic soil (He et al. 2012). Zhang et al. (2012) found the important role of AOA in acidic soils was attributed to the low-pH-reduced availability of ammonia and high substrate affinity of AOA. In addition, the positive correlation between AOA abundance and net nitrification rate indicated that the ammonia oxidation was mainly attributed to AOA (Table 2) and AOA was the main driver of nitrification in the vegetable soil examined. The abundances of AOB in the soils of same fertilizer treatment at different pH levels showed no significant differences, indicating that the abundance of AOB was less affected by pH. The results were mainly due to the high AOA to AOB ratio in the original soil (Fig. 3).
AOA and AOB communities
Soil pH is a very important factor influencing the distribution of ammonia oxidizers. In the present work, the T-RFLP analysis suggested that the AOA and AOB community structure varied and were correlated with soil pH (Figs. 4, 5). It has been reported that soil pH exhibits similar effects on the AOA and AOB community structure in Chinese tea orchard soils (Yao et al. 2011). Nicol et al. (2008) also revealed that the community structure changed across a soil pH gradient with specific species in acidic and neutral soils. Studies have shown that the nitrogen fertilizer and nitrification inhibitors can affect the community composition of ammonia oxidizers (Yao et al. 2011, 2016; Mahmood and Prosser 2006). However, the T-RFLP and PCA analyses showed that urea, nitrapyrin and NBPT had less effects on the community composition of ammonia oxidizers than soil pH (Figs. 4, 5). Therefore, it can be concluded that the community structure of AOA and AOB are more impressionable to soil pH than to urea, nitrapyrin and NBPT. Shifts in the ammonia oxidizer community structure might be due to the variation of soil pH. The relative abundances of AOA T-RFs 217, 205 and 166 bp and AOB T-RFs 157 and 56 bp were related to soil pH. AOA T-RFs 217 and 205 bp and AOB T-RF 157 bp were more suitable to acidic soil and AOA T-RF 166 bp and AOB T-RF 56 bp were more suitable to neutral soil. This is similar to the findings in Chinese tea orchard soils where some T-RFs are correlated to soil pH and the relative abundances of AOA T-RF 166 bp and AOB T-RF 156 bp decrease and those of AOA T-RFs 205 bp and 217 bp increase with the increase of soil pH. It has also been reported in the National Soil Inventory of Scotland that AOA T-RF 217 bp is relevant to high pH (Yao et al. 2013). Our results are consistent with these findings that soil pH is the major driver of AOA and AOB community structure.
Effects of nitrapyrin on soil inorganic nitrogen availability and abundance of ammonia oxidizers
Nitrapyrin is the most well-studied highly effective nitrification inhibitor. It can keep nitrogen in the form of ammonia by chelating copper components of the cytochrome oxidase involved in ammonia oxidation (Subbarao et al. 2006). The concentrations of NO3 −-N in the soils of urea + nitrapyrin treatment across the pH gradient at day 28 were lower than those in the soils of urea treatment at corresponding pH levels, and the net nitrification rates in the soils of urea + nitrapyrin treatment across the pH gradient were also lower than those in the soils of urea treatment (Table 1), indicating that nitrapyrin inhibited nitrification and its inhibitory effect varied with soil pH. This is consistent with the finding reported previously (Hall 1984; Chancy and Kamprath 1982; Degenhardt et al. 2016; Sims and MacKown 1987; Touchton et al. 1979; Hendrickson and Keeney 1979). For example, (Hendrickson and Keeney 1979) found that the inhibitory efficiency of nitrapyrin increased with soil pH. However, the affecting mechanism of pH on the inhibitory efficiency of nitrapyrin is not clear. For the abundance of amoA genes, the abundance of AOB amoA genes decreased significantly in urea + nitrapyrin treatment in comparison with treatments without nitrapyrin (Additional file 1: Figure S1), indicating that nitrapyrin inhibited the growth of AOB. However, no significant effect of nitrapyrin on AOA abundance was observed in the vegetable soil. The inhibition of the abundance of ammonia oxidizers by nitrapyrin have been rarely studied. Cui et al. (2013) demonstrated that nitrapyrin reduced the AOB abundances in alluvial soil and paddy soil. Fisk et al. (2015) indicated that the abundance of bacterial amoA genes could be decreased by nitrapyrin at 20 °C. Our results further confirmed that nitrapyrin could significantly decrease the abundance of AOB, but not that of AOA. This can be justified by the different cell membrane compositions of AOA and AOB, which affects the permeability of membranes, and thus differ in their sensitivity to the NIs (Ruser and Schulz 2015).
Effects of NBPT on soil inorganic nitrogen availability and ammonia oxidizers
NBPT as a urease inhibitor can effectively delay the hydrolysis of urea (McCarty et al. 1989; Wang et al. 1991; Kawakami et al. 2012). Our results indicated that the addition of NBPT reduced the NO3 −-N concentration and net nitrification rate in the tested soil. However, the decreasing degree of the net nitrification rate varied with soil pH (Table 1), indicating that NBPT could inhibit the hydrolysis of urea and the inhibition efficiency was affected by soil pH. Hendrickson and Douglass (1993) revealed that pH was a key factor for NBPT to control the urea hydrolysis and both NBPT and BNPO (an oxon analog of NBPT) could inhibit the hydrolysis of urea more effectively in neutral soils than in acidic soils. However, Beyrouty et al. (1988) found that the effect of NBPT was rarely influenced by soil pH and NBPT could be applied to both acidic and neutral soils. Our results indicated that NBPT was a much more effective urease inhibitor at pH 3.97 than at other pHs. Therefore, the inhibition efficiency of NBPT might be also influenced by other environmental factors. Further study is essential to definite the transformation of NBPT under different environmental conditions. NBPT exhibited no significant effects on the abundance and community structure of ammonia oxidizers. The effects of NBPT on bacteria, fungi and actinomycetes have been well studied. For example, Zhao et al. (2007) found that high concentrations of NBPT could inhibit the growths of bacteria and actinomycete. Song and Sun (2006) reported that NBPT could promote the growth of soil bacteria, actinomycetes and fungi. However, NBPT has shown no effects on growth of ammonia oxidizers, consistent with our results.
In conclusion, our results indicated that nitrapyrin could inhibit the growth of AOB but not AOA. AOA were affected by pH more significantly than AOB. AOA was the dominate ammonia oxidizers and drove the nitrification in acidic soils. NBPT was able to inhibit the urea hydrolysis, but exhibited no significant effect on the abundance and community structure of ammonia oxidizers. Community populations of AOA and AOB were more susceptive to pH than to NIs and UIs. Future work will be focused on the roles of AOA and AOB in autotrophic nitrifying activity using DNA-SIP technologies.
Abbreviations
- NBPT:
-
N-(n-butyl) thiophosphoric triamide
- AOA:
-
ammonia-oxidizing archaea
- AOB:
-
ammonia-oxidizing bacteria
- T-RFLP:
-
terminal restriction fragment length polymorphism
- N2O:
-
nitrous oxide
- Nis:
-
nitrification inhibitors
- UIs:
-
urease inhibitors
- N-serve:
-
2-chloro-6-(trichloromethyl) pyridine
- AMO:
-
ammonia monooxygenase
- NBPTO:
-
N-(butyl) phosphoric triamide
- ATU:
-
allylthiourea
- EC50 :
-
effective concentration 50
- N:
-
nitrogen
- TN:
-
total nitrogen
- MBC:
-
microbial biomass carbon
- MBN:
-
microbial biomass nitrogen
- PNR:
-
potential nitrification rate
- WHC:
-
water-holding capacity
- BSA:
-
bovine serum albumin
- OTU:
-
operational taxonomic unit
- SIP:
-
stable isotope probing
References
Abell GC, Banks J, Ross DJ, Keane JP, Robert SS, Revill AT, Volkman JK (2011) Effects of estuarine sediment hypoxia on nitrogen fluxes and ammonia oxidizer gene transcription. FEMS Microbiol Ecol 75:111–122
Antisari LV, Marzadori C, Gioacchini P, Ricci S, Gessa C (1996) Effects of the urease inhibitor N-(n-butyl) phosphorothioic triamide in low concentrations on ammonia volatilization and evolution of mineral nitrogen. Biol Fert Soils 22:196–201
Arp DJ, Sayavedrasoto LA, Hommes NG (2002) Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol 178:250–255
Belser LW, Schmidt EL (1981) Inhibitory effect of nitrapyrin on three genera of ammonia-oxidizing nitrifiers. Appl Environ Microbiol 41:819–821
Beyrouty CA, Sommers LE, Nelson DW (1988) Ammonia volatilization from surface-applied urea as affected by several phosphoroamide compounds. Soil Sci Soc Am J 52:1173–1178
Bremner JM, Chai HS (1986) Evaluation of N-butyl phosphorothioic triamide for retardation of urea hydrolysis in soil. Commun Soil Sci Plant Anal 17:337–351
Bronson KF, Touchton JT, Hiltbold AE, Hendrickson LL (1989) Control of ammonia volatilization with N-(n-butyl) thiophosphoric triamide in loamy sands. Commun Soil Sci Plan 20:1439–1451
Chancy HF, Kamprath EJ (1982) Effect of nitrapyrin on N response of corn on sandy soils. Agron J 74:565–569
Chen X, Zhang LM, Shen JP, Wei WX, He JZ (2011) Abundance and community structure of ammonia-oxidizing archaea and bacteria in an acid paddy soil. Biol Fert Soils 47:323–331
Chen Q, Qi L, Bi Q, Dai P, Sun D, Sun C, Liu W, Lu L, Ni W, Lin X (2015) Comparative effects of 3,4-dimethylpyrazole phosphate (DMPP) and dicyandiamide (DCD) on ammonia-oxidizing bacteria and archaea in a vegetable soil. Appl Microbiol Biotechnol 99:477–487
Creason GL, Schmitt MR, Douglass EA, Hendrickson LL (1990) Urease inhibitory activity associated with N-(n-butyl) thiophosphoric triamide is due to formation of its oxon analog. Soil Biol Biochem 22:209–211
Cui P, Fan F, Yin C, Li Z, Song A, Wan Y, Liang Y (2013) Urea- and nitrapyrin-affected N2O emission is coupled mainly with ammonia oxidizing bacteria growth in microcosms of three typical Chinese arable soils. Soil Biol Biochem 66:214–221
Degenhardt RF, Juras LT, Smith LRA, Macrae AW, Ashigh J, Mcgregor WR (2016) Application of nitrapyrin with banded urea, urea ammonium nitrate, and ammonia delays nitrification and reduces nitrogen loss in Canadian soils. Crop Forage Turfgrass Manag 2:1–11
Di HJ, Cameron KC, Shen JP, Winefield CS, O’Callaghan M, Bowatte S, He JZ (2009) Nitrification driven by bacteria and not archaea in nitrogen-rich grassland soils. Nature Geosci 2:621–624
Fisk LM, Maccarone LD, Barton L, Murphy DV (2015) Nitrapyrin decreased nitrification of nitrogen released from soil organic matter but not amoA, gene abundance at high soil temperature. Soil Biol Biochem 88:214–223
Gubry-Rangin C, Nicol GW, Prosser JI (2010) Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol 74:566–574
Hall GH (1984) Measurement of nitrification rates in lake sediments: comparison of the nitrification inhibitors nitrapyrin and allylthiourea. Microb Ecol 10:25–36
He JZ, Hu HW, Zhang LM (2012) Current insights into the autotrophic thaumarchaeal ammonia oxidation in acidic soils. Soil Biol Biochem 55:146–154
Hendrickson LL, Douglass EA (1993) Metabolism of the urease inhibitor N-(n-butyl) thiophosphoric triamide (NBPT) in soils. Soil Biol Biochem 25:1613–1618
Hendrickson LL, Keeney DR (1979) A bioassay to determine the effect of organic matter and pH on the effectiveness of nitrapyrin (N-serve) as a nitrification inhibitor. Soil Biol Biochem 11:51–55
Hu HW, Zhang LM, Dai Y, Di HJ, He JZ (2013) pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J Soil Sediment 13:1439–1449
Hughes TD, Welch LF (1970) 2-chloro-6-(trichloromethyl) pyridine as a nitrification inhibitor for anhydrous ammonia applied in different seasons. Agron J 62:821–824
IPCC (2007) Mitigation of climate change. Contribution of working group III to the fourth assessment report of the intergovernmental panel on climate change. Cambridge University Press, UK
Jäntti H, Jokinen S, Hietanen S (2013) Effect of nitrification inhibitors on the baltic sea ammonia-oxidizing community and precision of the denitrifier method. Aquat Microb Ecol 70:181–186
Jin T, Zhang T, Yan Q (2010) Characterization and quantification of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in a nitrogen-removing reactor using T-RFLP and qPCR. Appl Microbiol Biotechnol 87:1167–1176
Kangatharalingam N, Priscu JC (1993) Nitrapyrin-ammonium combination induces rapid multiplication of mixed cultures of the stalked bacterium Nevskia ramosa, famintzin and other heterotrophic bacteria. Arch Microbiol 159:48–50
Kawakami EM, Snider JL, Oosterhuis DM, Mozaffari M (2012) Physiological and yield responses of field-grown cotton to application of urea with the urease inhibitor NBPT and the nitrification inhibitor DCD. Eur J Agron 43:147–154
Kim JG, Jung MY, Park SJ, Rijpstra WIC, Damsté JSS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK (2012) Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group l.1b from an agricultural soil. Environ Microbiol 14:1528–1543
Kolodziej AF (1994) Cheminform abstract: the chemistry of nickel-containing enzymes. ChemInform 41:493–597
Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW (2011) Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA 108:15892–15897
Lehtovirta-Morley LE, Verhamme DT, Nicol GW, Prosser JI (2013) Effect of nitrification inhibitors on the growth and activity of Nitrosotalea devanaterra, in culture and soil. Soil Biol Biochem 62:129–133
Li M, Gu JD (2013) Community structure and transcript responses of anammox bacteria, AOA, and AOB in mangrove sediment microcosms amended with ammonium and nitrite. Appl Microbiol Biotechnol 97:9859–9874
Li H, Weng BS, Huang FY, Su JQ, Yang XR (2015) pH regulates ammonia-oxidizing bacteria and archaea in paddy soils in southern china. Appl Microbiol Biotechnol 99:6113–6123
Liu R, Hayden H, Suter H, He J, Chen D (2015) The effect of nitrification inhibitors in reducing nitrification and the ammonia oxidizer population in three contrasting soils. J Soil Sediment 15:1113–1118
Mahmood S, Prosser JI (2006) The influence of synthetic sheep urine on ammonia oxidizing bacterial communities in grassland soil. FEMS Microbiol Ecol 56:444–454
McCarty GW, Bremner JM, Chai HS (1989) Effect of N-(n-butyl) thiophosphoric triamide on hydrolysis of urea by plant, microbial, and soil urease. Biol Fert Soils 8:123–127
Morimoto S, Hayatsu M, Takada HY, Nagaoka K, Yamazaki M, Karasawa T, Takenaka M, Akiyama H (2011) Quantitative analyses of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in fields with different soil types. Microb Environ 26:248–253
Nicol GW, Leininger S, Schleper C, Prosser JI (2008) The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10:2966–2978
Persson T, Wirén A (1995) Nitrogen mineralization and potential nitrification at different depths in acid forest soils. Plant Soil 169:55–65
Regina K, Silvola J, Martikainen PJ (1998) Mechanisms of N2O and NO production in the soil profile of a drained and forested peatland, as studied with acetylene, nitrapyrin and dimethyl ether. Biol Fert Soils 27:205–210
Ruser R, Schulz R (2015) The effect of nitrification inhibitors on the nitrous oxide (N2O) release from agricultural soils-a review. J Plant Nut Soil Sci 178:171–188
Sanz-Cobena A, Misselbrook T, Camp V, Vallejo A (2011) Effect of water addition and the urease inhibitor NBPT on the abatement of ammonia emission from surface applied urea. Atmo Environ 45:1517–1524
Schouten S, Hopmans EC, Pancost RD, Damsté JSS (2000) Widespread occurrence of structurally diverse tetraether membrane lipids: evidence for the ubiquitous presence of low-temperature relatives of hyperthermophiles. Proc Natl Acad Sci 97:14421–14426
Shen JP, Zhang LM, Zhu YG, Zhang JB, He JZ (2008) Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10:1601–1611
Shen TL, Stieglmeier M, Dai JL, Urich T, Schleper C (2013) Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol Lett 344:121–129
Sims JL, MacKown CT (1987) Rate of nitrapyrin and soil pH effects on nitrification of ammonium fertilizer and growth and composition of burley tobacco. Commun Soil Sci Plant Anal 18:1479–1500
Song ZY, Sun QY (2006) Effect of urease inhibitor NBPT on soil microorganisms. J Dalian Inst Light Ind 25:100–102
Stempfhuber B, Engel M, Fischer D, Neskovic-Prit G, Wubet T, Schöning I, Gubry-Rangin C, Kublik S, Schloter-Hai B, Rattei T, Welzl G, Nicol GW, Schrumpf M, Buscot F, Prosser JI, Schloter M (2015) pH as a driver for ammonia-oxidizing archaea in forest soils. Microb Ecol 69:879–883
Subbarao GV, Ito O, Sahrawat KL, Berry WL, Nakahara K, Ishikawa T, Watanabe T, Suenaga K, Rondon M, Rao IM (2006) Scope and strategies for regulation of nitrification in agricultural systems—challenges and opportunities. Crit Rev Plant Sci 25:303–335
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thomson AJ, Giannopoulos G, Pretty J, Baggs EM, Richardson DJ (2012) Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos Trans R Soc Lond 367:1157–1168
Touchton JT, Hoeft RG, Welch LF, Argyilan WL (1979) Loss of nitrapyrin from soils as affected by pH and temperature. Agron J 71:865–869
Wang Z, Cleemput OV, Demeyer P, Baert L (1991) Effect of urease inhibitors on urea hydrolysis and ammonia volatilization. Biol Fert Soils 11:43–47
Watson CJ, Miller H, Poland P, Kilpatrick DJ, Allen MDE, Garrett MK, Christianson CB (1994) Soil properties and the ability of the urease inhibitor N-(n-BUTYL) thiophosphoric triamide (nBTPT) to reduce ammonia volatilization from surface-applied urea. Soil Biol Biochem 26:1165–1171
Wu JS, Joergensen RG, Pommerening B, Chaussod R, Brookes PC (1990) Measurement of soil microbial biomass C-an automated procedure. Soil Biol Biochem 22:1167–1169
Yao H, Gao Y, Nicol GW, Campbell CD, Prosser JI, Zhang L, Han W, Singh BK (2011) Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl Environ Microbiol 77:4618–4625
Yao H, Campbell CD, Chapman SJ, Freitag TE, Nicol GW, Singh BK (2013) Multi-factorial drivers of ammonia oxidizer communities: evidence from a national soil survey. Environ Microbiol 15:2545–2556
Yao H, Sha H, Qiu Q, Li Y, Wu L, Mi W, Dai F (2016) Effects of different fertilizers on the abundance and community structure of ammonia oxidizers in a yellow clay soil. Appl Microbiol Biotechnol 100:6815–6826
Ying J, Zhang L, He J (2010) Putative ammonia-oxidizing bacteria and archaea in an acidic red soil with different land utilization patterns. Environ Microbiol R 2:304–312
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J 6:1032–1045
Zhao L, Sun QY, Yu YM, Ma GB (2007) Effect of urease inhibitor NBPT on soil urease activity and urease producing microorganisms. J Dalian Inst Light Ind 26:24–27
Authors’ contributions
HY and X-EL designed the experiments and revised the paper. RX performed the experiments and wrote the paper. RX, X-EL and SH analyzed the data. All authors read and approved the final manuscript.
Acknowledgements
None.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its Additional file 1.
Consent for publication
This article does not contain any individual person’s data.
Ethical approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This study was funded by the National Key Research Program of China (2016YFC0502704), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15020301), the National Natural Science Foundation of China (41471206, 41525002) and Ningbo Municipal Science and Technology Bureau (2015C1003).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional file
13568_2017_426_MOESM1_ESM.docx
Additional file 1: Table S1. Primers of AOA and AOB used for molecular analyses. Table S2. Pearson correlation between pH and the relative abundance of archaeal ammonia oxidizer TRFs. Table S3. Pearson correlation between pH and the relative abundance of bacterial ammonia oxidizer TRFs. Table S4. Genotype patterns based on the clone libraries of amoA genes. Figure S1. Log number of AOA and AOB amoA copies in four different treatments (control; urea; urea+nitrapyrin; urea+NBPT) at different pH levels. Error bars indicate standard errors of three replicates, different capital letters indicate the significant difference within different treatments at same pH level (P < 0.05).
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Xi, R., Long, XE., Huang, S. et al. pH rather than nitrification and urease inhibitors determines the community of ammonia oxidizers in a vegetable soil. AMB Expr 7, 129 (2017). https://doi.org/10.1186/s13568-017-0426-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-017-0426-x