Abstract
The “self-cleaving” 2A sequence of picornavirus, which mediates ribosome-skipping events, enables the generation of two or more separate peptide products from one mRNA containing one or more “self-cleaving” 2A sequences. In this study, we introduced a single 2A sequence of porcine teschovirus-1 (P2A) linked to two fluorescent protein genes, the enhanced yellow fluorescent protein (EYFP) gene and the red fluorescent protein (RFP) gene, in a single cassette into transgenic Eimeria tenella (EtER). As expected, we obtained two separated protein molecules rather than a fused protein, although the two molecules were translated from the same mRNA carrying a single “self-cleaving” 2A sequence. Importantly, RFP led by a secretion signal was secreted into parasitophorous vacuoles, while EYFP localized mainly to the nucleus of EtER. Our results demonstrate that the “self-cleaving” 2A sequence actively mediated cleavage of polyproteins in the apicomplexan parasite E. tenella.
Similar content being viewed by others
Introduction, methods and results
Eimeria tenella is an emerging model organism for studying the basic cell biology of protozoan parasites. Both transient and stable transfection systems in Eimeria species were developed successfully [1–4]. Transgenic parasites expressing fluorescent proteins, such as enhanced yellow fluorescent protein (EYFP), such as reporters, are particularly convenient to visually study the basic biology of Eimeria parasites [5–7]. The simultaneous expression of more than one reporter gene in a single construct is sometimes required, especially for studying multiple gene functions in different cellular compartments or organelles of organisms like the Eimeria parasite. So far, two open reading frames (ORF), each flanked with one set of regulatory sequences in a single plasmid (the so-called double cassette system) have been proven to be the only efficient way for co-expressing dual fluorescent protein genes in Eimeria parasites [4, 7, 8]. However, suppression of gene expression may occur in a single plasmid with multiple promoters due to the phenomenon called promoter interference [9].
This limitation has been overcome by a small “self-cleaving” peptide, F2A, first identified by Ryan et al. in the foot-and-mouth disease virus, a member of the picornavirus group [10]. Subsequently, “2A like” peptides from equine rhinitis A virus (E2A), porcine teschovirus-1 (P2A) and Thosea asigna virus (T2A) were identified, and their activities in proteolytic cleavage were shown in various in vitro and in vivo eukaryotic systems [10–13]. Here, we report a single cassette system which co-expresses two fluorescent proteins, EYFP and RFP (red fluorescent protein), and is cleaved by P2A in the apicomplexan parasite E. tenella.
Surface antigen 13 (SAG13) is a highly expressed sporozoite antigen of E. tenella. The expression level of SAG13 is nearly 5% of E. tenella total sporozoite soluble antigens [14]. We constructed a single cassette system in which TgDHFR/EYFP-P2A-ssRFP was flanked by the SAG13 promoter and its 3′ UTR (Figure 1A; Additional file 1). TgDHFR (pyrimethamine resistance gene) was used for transgenic parasite selection. P2A was a 66-base pair oligonucleotide from porcine teschovirus-1 [15]. RFP was preceded by GRA8, a signal sequence, and followed by a His6 tag [7]. Restriction enzyme-mediated integration (REMI) was adapted for the transfection of sporozoites as previously described [16, 17]. Five 3-day-old AA broilers were inoculated with 106 electroporated sporozoites via the cloacal route. Chicks were given a standard diet supplemented with 150 ppm pyrimethamine (Sigma, USA) 18 h after inoculation [18]. Oocysts from faeces excreted 6–10 days post-inoculation (dpi) were collected and sporulated for an additional four generations of in vivo passages as previously described [19].
Generation of stably transfected E. tenella line co-expressing EYFP and RFP. A Schematic of the transfected vector (pSDEP2ARS). 5′ UTR (1376 bp) and 3′ UTR (1002 bp) of E. tenella surface antigen 13 (SAG13) were amplified from genomic DNA with primers SAG13-5-F/SAG13-5-R and SAG13-3-F/SAG13-3-R (Additional file 1), respectively. The synthesized 66 bp nucleotides encoding porcine teschovirus 2A sequence (Gray background and Additional file 1) was fused with the RFP gene of pMIC-EYFP/ACTss-RFP [7] by three rounds of PCR (2A-F1/2A-F2/2A-F3 and 2A-R (Additional file 1). ss: T. gondii GRA8 signal sequence. B Both EYFP and RFP were expressed in sporulated oocysts whereas no fluorescence could be detected in unsporulated oocysts. C Genomic DNA and cDNA from EtER was amplified with the primers P1/P2 (giving a 564 bp product) or P3/P4 (giving a 586 bp product) to verify the recombination of EYFP and RFP genomic DNA and cDNA from wild type E. tenella were used as a control. M: marker. D Genomic DNA from EtER was amplified with arbitrary degenerate primers (AP 1, AP 2, AP 3 and AP 4) and specific primers (5-SP 1, 5-SP 2 and 5-SP 3/3-SP1, 3-SP2 and 3-SP3 (Additional file 1) from SAG13 5′ (upper) and 3′ (lower) UTR by thermal asymmetric interlaced PCR, and the products from the third-round PCR were cloned into pEasy-T1 vector for sequencing. M: marker. E One integration site (Eth_scaff16) was confirmed by BLAST from 58 clones in E. tenella GeneDB. F Genomic DNA from EtER was amplified with the primers P5/5-SP3 (giving a 607 bp product), 3-SP3/P6 (giving a 306 bp product) and P5/P6 (giving a 6523 bp product) to verify the integration site of transfected vector in Eth_scaff16 locus in the EtER genome. Genomic DNA from wild type E. tenella was used as a control. M: marker, NS: no specific band.
We then infected 30 two-day-old AA broilers with single transgenic parasite oocysts from the fifth generation and obtained a stable transgenic line, EtER (Additional file 2) [2, 20]. The stable line expressed EYFP and RFP in its sporulated oocyst stage but not in its unsporulated oocysts (Figure 1B), showing that the SAG13 promoter is a stage-specific regulatory sequence [21].
To further characterize the transgenic strain at the molecular level, we extracted DNA and RNA from EtER as previously described [20, 22]. cDNA was synthesized from the extracted RNA using random primers and a high capacity cDNA Reverse Transcription Kit (Applied Biosystems) [22]. PCR-based analysis using P1/P2 primers revealed that the transfection vector existed in the transgenic parasite genome (gDNA), and analysis using P3/P4 primers revealed that EYFP-RFP was translated by the same mRNA from cDNA (Figures 1A and C). We further analysed the integration sites of the 5′ and 3′ ends of the transfected vector in the EtER genome through genome walking (Figures 1D and E) [6, 17]. Specific primers (5-SP 1, 5-SP 2, 5-SP 3/3-SP1, 3-SP2 and 3-SP3; Additional file 1) were designed according to the SAG13 promoter and 3′ UTR sequence following the kit instructions. The flanking sequences to the 5′ and 3′ ends of the integrated vector were obtained after three rounds of thermal asymmetric interlaced PCR. We found that both the 5′ and 3′ ends of the exogenous plasmid were integrated in the Eth_scaff 16 locus (Figure 1E). To confirm that the exogenous vector was integrated into that specific site of the parasite genome, we designed specific primers, P5 and P6 (Figure 1E; Additional file 1), based on the 5′ and 3′ flanking sequences of the transfected vector in the Eth_scaff 16 locus. After amplification with various primer pairs (P5/5-SP3, P6/3-SP3 and P5/P6, Figure 1F; Additional file 1) from wild type E. tenella and EtER genomic DNA, we obtained the predicted specific bands from EtER genomic DNA (Figure 1F). All these data show that we obtained a stable transgenic E. tenella line (EtER).
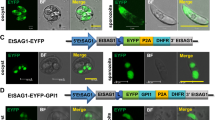
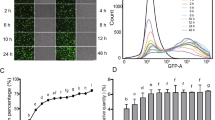
To assess if the “self-cleaving” 2A peptide could mediate proteolytic cleavage in transgenic E. tenella and to show that the introduction of a signal peptide to regulate RFP secretion did not interfere with EYFP and RFP distribution in EtER, (Figure 1A) [7, 23], we conducted an indirect immunofluorescence assay (IFA). In this assay, EtER sporozoites were treated with mouse anti-SAG13 polyclonal-antibody, followed by AMCA-conjugated goat anti-mouse IgG staining for confocal microscope imaging. We found that EYFP mainly localized to the nucleus, while RFP was found in the cytoplasm (Figure 2A). We also found that SAG13 was distributed in the cell surface, with some observed nuclear SAG signal interpreted as bleeding of EYFP (Figure 2A).
“Self-cleaving” 2A peptide cleaves EYFP and RFP in EtER. A EtER sporozoites were reacted with mouse anti-SAG13 polyclonal antibody, followed by the reaction with AMCA-conjugated goat anti-mouse IgG (H + L) (Proteintech, USA), revealed by a confocal laser scanning microscopy (SP5, Leica, Germany). EYFP mainly localized to the nucleus, while RFP was found in the cytoplasm. Bar 5 μm. B RFP was secreted into PV in trophozoites (24 hpi) and 1st-genernation schizont stages, while EYFP in the nucleus (48 and 72 hpi). Bar 5 μm. C Soluble proteins extracted from EtER and the wild-type E. tenella (WT) were resolved by SDS-PAGE and the immunoblot analysis was conducted following standard protocols. The primary antibody was the mouse anti-EYFP polyclonal antibody and mouse anti-His tag monoclonal antibody, the mouse anti-GAPDH polyclonal antibody served as the loading control, while the HRP-conjugated goat anti-mouse IgG was used as the secondary antibody. TgDHFR-EYFP was 95 kDa and RFP 28 kDa. M: marker.
To confirm that the function of the GRA8 signal peptide was not affected by P2A, we did an in vitro culture of primary chicken kidney cells (PCKC) inoculated with EtER sporozoites. As expected, EYFP localized mainly to the nucleus, while RFP was secreted into parasitophorous vacuoles (PV) in trophozoites [24 h post-inoculation (hpi)] and 1st-generation schizont stages (48 and 72 hpi) (Figure 2B) [24]. As a stronger promoter was used here, there are many red signals in the cytoplasm (Figure 2B).
To further test the self-cleavage efficiency of P2A in EtER, we used Western blotting. EtER sporozoite soluble antigens were subjected to SDS-PAGE, followed by reactions with mouse anti-EYFP polyclonal-antibody, mouse anti-His tag monoclonal antibody and HRP-conjugated goat anti-mouse IgG (Proteintech, USA) secondary antibody. Our results showed that aTgDHFR-EYFP (95 kDa) was efficiently cleaved from RFP (28 kDa) (Figure 2C).
The above results conclusively demonstrate that the “self-cleaving” 2A sequence of picornavirus works efficiently in the apicomplexan parasite E. tenella.
Discussion
Expression of multiple polypeptides from a single mRNA is made possible by inclusion of a short viral 2A peptide coding sequence between the polypeptide-encoding transgenes [11, 25]. In this study, we obtained a stably transfected E. tenella line (EtER) expressing separate EYFP and RFP from one mRNA mediated by P2A function. Our result encourages the use of P2A in cell biological studies of apicomplexan parasites that may require multi-reporter expression in a single cassette.
Interestingly, we found that SAG13 was a much stronger promoter than His4 as revealed by EYFP density via the confocal imaging measurement (data not shown), which indicates the usefulness of SAG13 for the study of gene overexpression without modifying the genome [26]. However, overexpression may also result in the appearance of unusual biological characteristics in transgenic parasites compared to wild-types [2].
Our previous study showed that transgenic E. tenella expressing EYFP in different cellular compartments elicit EYFP-specific, systemic and mucosal immune responses [22], and another group showed that E. tenella-delivered CjA stimulates protective immunity against Campylobacter jejuni infections in chickens [27]. The finding that P2A mediates separate expression of multiple genes in different compartments has great implications for the development of transgenic Eimeria parasites as vaccine vectors and beyond [2, 6, 7, 16, 17, 20, 22].
Abbreviations
- P2A:
-
2A sequences from porcine teschovirus-1
- EYFP:
-
enhanced yellow fluorescent protein
- RFP:
-
red fluorescent protein
- EtER:
-
transgenic Eimeria tenella line co-expressing EYFP and RFP
- SAG13:
-
Eimeria tenella sporozoites surface antigen 13
- GRA 8:
-
Toxoplasma gondii dense granule 8
References
Kelleher M, Tomley FM (1998) Transient expression of beta-galactosidase in differentiating sporozoites of Eimeria tenella. Mol Biochem Parasitol 97:21–31
Yan W, Liu X, Shi T, Hao L, Tomley FM, Suo X (2009) Stable transfection of Eimeria tenella: constitutive expression of the YFP-YFP molecule throughout the life cycle. Int J Parasitol 39:109–117
Kurth M, Entzeroth R (2009) Reporter gene expression in cell culture stages and oocysts of Eimeria nieschulzi (Coccidia, Apicomplexa). Parasitol Res 104:303–310
Hanig S, Entzeroth R, Kurth M (2012) Chimeric fluorescent reporter as a tool for generation of transgenic Eimeria (Apicomplexa, Coccidia) strains with stage specific reporter gene expression. Parasitol Int 61:391–398
Hao L, Liu X, Zhou X, Li J, Suo X (2007) Transient transfection of Eimeria tenella using yellow or red fluorescent protein as a marker. Mol Biochem Parasitol 153:213–215
Qin M, Liu X, Tang X, Suo J, Tao G, Suo X (2014) Transfection of Eimeria mitis with yellow fluorescent protein as reporter and the endogenous development of the transgenic parasite. PLoS One 9:e114188
Yin G, Liu X, Zou J, Huang X, Suo X (2011) Co-expression of reporter genes in the widespread pathogen Eimeria tenella using a double-cassette expression vector strategy. Int J Parasitol 41:813–816
Jonscher E, Erdbeer A, Günther M, Kurth M (2015) Two COWP-like cysteine rich proteins from Eimeria nieschulzi (coccidia, apicomplexa) are expressed during sporulation and involved in the sporocyst wall formation. Parasit Vectors 8:395
Emerman M, Temin HM (1984) Genes with promoters in retrovirus vectors can be independently suppressed by an epigenetic mechanism. Cell 39:449–467
Ryan MD, King AM, Thomas GP (1991) Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J Gen Virol 72:2727–2732
de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD (2006) E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol 24:68–75
Gauthier NP, Soufi B, Walkowicz WE, Pedicord VA, Mavrakis KJ, Macek B, Gin DY, Sander C, Miller ML (2013) Cell-selective labeling using amino acid precursors for proteomic studies of multicellular environments. Nat Methods 10:768–773
Zhang C, Xiao B, Jiang Y, Zhao Y, Li Z, Gao H, Ling Y, Wei J, Li S, Lu M, Su XZ, Cui H, Yuan J (2014) Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio 5:e1414
de Venevelles P, Chich JF, Faigle W, Loew D, Labbé M, Girard-Misguich F, Péry P (2004) Towards a reference map of Eimeria tenella sporozoite proteins by two-dimensional electrophoresis and mass spectrometry. Int J Parasitol 34:1321–1331
Doherty M, Todd D, McFerran N, Hoey EM (1999) Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J Gen Virol 80:1929–1941
Liu X, Shi T, Ren H, Su H, Yan W, Suo X (2008) Restriction enzyme-mediated transfection improved transfection efficiency in vitro in Apicomplexan parasite Eimeria tenella. Mol Biochem Parasitol 161:72–75
Li Z, Tang X, Suo J, Qin M, Yin G, Liu X, Suo X (2015) Transgenic Eimeria mitis expressing chicken interleukin 2 stimulated higher cellular immune response in chickens compared with the wild-type parasites. Front Microbiol 6:533
Clark JD, Billington K, Bumstead JM, Oakes RD, Soon PE, Sopp P, Tomley FM, Blake DP (2008) A toolbox facilitating stable transfection of Eimeria species. Mol Biochem Parasitol 162:77–86
Long PL, Millard BJ, Joyner LP, Norton CC (1976) A guide to laboratory techniques used in the study and diagnosis of avian coccidiosis. Folia Vet Lat 6:201–217
Liu X, Zou J, Yin G, Su H, Huang X, Li J, Xie L, Cao Y, Cui Y, Suo X (2013) Development of transgenic lines of Eimeria tenella expressing M2e-enhanced yellow fluorescent protein (M2e-EYFP). Vet Parasitol 193:1–7
Tabares E, Ferguson D, Clark J, Soon PE, Wan KL, Tomley F (2004) Eimeria tenella sporozoites and merozoites differentially express glycosylphosphatidylinositol-anchored variant surface proteins. Mol Biochem Parasitol 135:123–132
Huang X, Zou J, Xu H, Ding Y, Yin G, Liu X, Suo X (2011) Transgenic Eimeria tenella expressing enhanced yellow fluorescent protein targeted to different cellular compartments stimulated dichotomic immune responses in chickens. J Immunol 187:3595–3602
Zou J, Liu X, Shi T, Huang X, Wang H, Hao L, Yin G, Suo X (2009) Transfection of Eimeria and Toxoplasma using heterologous regulatory sequences. Int J Parasitol 39:1189–1193
Hofmann J, Raether W (1990) Improved techniques for the in vitro cultivation of Eimeria tenella in primary chick kidney cells. Parasitol Res 76:479–486
Daniels RW, Rossano AJ, Macleod GT, Ganetzky B (2014) Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS One 9:e100637
Pereira LM, Yatsuda AP (2014) The chloramphenicol acetyltransferase vector as a tool for stable tagging of Neospora caninum. Mol Biochem Parasitol 196:75–81
Clark JD, Oakes RD, Redhead K, Crouch CF, Francis MJ, Tomley FM, Blake DP (2012) Eimeria species parasites as novel vaccine delivery vectors: anti-Campylobacter jejuni protective immunity induced by Eimeria tenella-delivered CjaA. Vaccine 30:2683–2688
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XT and XS conceived and designed this study and analyzed the data. XT carried out the experiments and drafted the manuscripts. XL, GT, MQ, GY and JS contributed to help the statistical analysis and help to draft the manuscripts. XS and XL supervised the study implementation and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31330076 and 31472180).
Author information
Authors and Affiliations
Corresponding author
Additional files
13567_2016_351_MOESM2_ESM.docx
Additional file 2. Transgenic parasite (EtER) selection. The details of EtER selection including the inoculation dosage of each generation and the efficacy of drug selection are provided.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Tang, X., Liu, X., Tao, G. et al. “Self-cleaving” 2A peptide from porcine teschovirus-1 mediates cleavage of dual fluorescent proteins in transgenic Eimeria tenella . Vet Res 47, 68 (2016). https://doi.org/10.1186/s13567-016-0351-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-016-0351-z