Abstract
Background
To investigate the value of 18F-FDG PET/CT molecular radiomics combined with a clinical model in predicting thoracic lymph node metastasis (LNM) in invasive lung adenocarcinoma (≤ 3 cm).
Methods
A total of 528 lung adenocarcinoma patients were enrolled in this retrospective study. Five models were developed for the prediction of thoracic LNM, including PET radiomics, CT radiomics, PET/CT radiomics, clinical and integrated PET/CT radiomics-clinical models. Ten PET/CT radiomics features and two clinical characteristics were selected for the construction of the integrated PET/CT radiomics-clinical model. The predictive performance of all models was examined by receiver operating characteristic (ROC) curve analysis, and clinical utility was validated by nomogram analysis and decision curve analysis (DCA).
Results
According to ROC curve analysis, the integrated PET/CT molecular radiomics-clinical model outperformed the clinical model and the three other radiomics models, and the area under the curve (AUC) values of the integrated model were 0.95 (95% CI: 0.93–0.97) in the training group and 0.94 (95% CI: 0.89–0.97) in the test group. The nomogram analysis and DCA confirmed the clinical application value of this integrated model in predicting thoracic LNM.
Conclusions
The integrated PET/CT molecular radiomics-clinical model proposed in this study can ensure a higher level of accuracy in predicting the thoracic LNM of clinical invasive lung adenocarcinoma (≤ 3 cm) compared with the radiomics model or clinical model alone.
Similar content being viewed by others
Background
Lung adenocarcinoma accounts for over 30% of all lung cancers. Furthermore, approximately 20% of patients with invasive lung adenocarcinoma (≤ 3 cm) already have thoracic lymph node metastasis(LNM)at the time of diagnosis [1, 2]. Identifying the presence of thoracic LNM before surgery can indicate the necessity of intraoperative mediastinal lymph node dissection and subsequent radical resection in some invasive lung adenocarcinomas (≤ 3 cm) [2]. In addition, determining LNM status in advance is also important for the selection of the target range of radiotherapy [3]. Unfortunately, conventional approaches to thoracic LNM detection, thoracoscopy and transbronchial biopsies can lead to complications, including haemorrhage, infection and pneumothorax, which increases the cost of treatment despite high accuracy [4]. In addition, positron emission tomography/computed tomography (PET/CT), considered the most accurate thoracic LNM staging method, is subject to low diagnostic sensitivity due to several factors, such as lymph node hypertrophy, false positives caused by infection and inflammation and limited spatial resolution [5,6,7,8,9]. For example, Liu et al. showed that the sensitivity and specificity of PET/CT in the preoperative diagnosis of mediastinal LNM in non-small-cell lung cancer (NSCLC) patients were 65% and 96.8%, respectively [8]. On the one hand, the micrometastasis of certain lymph nodes with a diameter smaller than the spatial resolution range of PET/CT may cause false negatives [10]. On the other hand, long-term smoking and lung infections can lead to false positives [11]. In addition, PET/CT imaging reading is dependent on visual assessment and semiautomated measurements, which in turn rely on different interpretations by different observers. The accuracy of PET/CT in predicting the incidence of occult LNM in NSCLC is only approximately 14–19% [12, 13].
As a supplement to the abovementioned approaches, radiomics can help improve the diagnostic efficiency of lung cancer LNM [14]. Coroller et al. reported 35 CT radiomics features of lung adenocarcinoma for predicting distant metastasis [15]. Yang et al. demonstrated the value of CT radiomics in predicting lung adenocarcinoma LNM [16]. Cong et al. used the random forest method to establish models to predict the LNM of lung adenocarcinoma [17]. Although the CT-based evaluation of radiomics features has been shown to be a promising predictor of lung adenocarcinoma LNM, PET/CT molecular radiomics-clinical models, including PET/CT molecular radiomics features and clinical factors, have not been investigated for their potential in predicting the thoracic LNM of lung adenocarcinoma. To address the literature gap, we conducted a comparative study of five models, including integrated PET/CT molecular radiomics-clinical, PET/CT radiomics, PET radiomics, CT radiomics, and clinical models, for predicting the thoracic LNM of lung adenocarcinoma. Among them, for PET/CT radiomics, the primary lesions on PET and CT images were delineated before extracting and analysing radiomics features [18]. The current study explored the diagnostic efficacy of PET/CT, CT, and PET radiomics models in predicting the thoracic LNM of lung adenocarcinoma by analysing the features of PET/CT images of lung adenocarcinoma patients. The study aimed to develop an integrated PET/CT molecular radiomics-clinical model for predicting the thoracic LNM of invasive lung adenocarcinoma (≤ 3 cm).
Methods
Patient selection and pathological evaluation
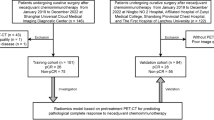
A retrospective analysis of 802 patients with lung adenocarcinoma (diameter ≤ 3 cm) was performed in Shanghai Chest Hospital from February 2016 to January 2021. The inclusion and exclusion criteria are illustrated in Additional file 1: Figure S1. A total of 528 patients with invasive lung adenocarcinoma were enrolled, including 379 patients with lung adenocarcinoma without thoracic LNM, accounting for 71.78%, and 149 patients with lung adenocarcinoma with thoracic LNM, accounting for 28.22%. Thoracic LNM was defined as lung cancer with N1 or N2 LNM. All patients underwent surgeries after diagnosis, including N1 and N2 resection. Two pathologists evaluated the tumour histology of these patients by following the 2015 WHO classification of lung adenocarcinoma. Lymph node staging was defined according to the eighth version of the TNM staging method. The study protocol was approved by the institutional ethics review committee of Shanghai Chest Hospital. Informed consent was not required because of the retrospective nature of the study. All the patient data were anonymized in this paper.
Clinical information of the selected lung adenocarcinoma patients
The clinical information of the patients included age, sex, smoking history, and the tumour marker carcinoembryonic antigen (CEA). The CT features included lung tumour location, lobulation sign, burr sign, pleural traction, and solid component size. The PET parameter included the maximum standardized uptake value (SUVmax). The clinical information of the enrolled patients is summarized in Table 1. The size of the solid component of pulmonary nodules, including mixed ground-glass nodules, refers to the average value of the longest cross-section length and the vertical diameter length of the solid component on the pulmonary window [19].
PET/CT scan procedures
All patients were examined under the same scanning conditions on the same device (Siemens Biograph MCT-S PET/CT). A 64-slice spiral CT was used. 18F-fluorodeoxyglucose (18F-FDG) was provided by Shanghai Atom Kexing Pharmaceuticals Co., Ltd. All patients had withheld from eating and drinking for more than 6 h before the PET/CT procedure, thus keeping the blood glucose level below 150 mg/dL. All patients were injected with 18F-FDG at 5 MBq/kg ± 10% of body weight and then rested for 60 min. The PET scan was divided into 5 or 6 beds, and each bed was checked for approximately 2 min. The CT data were used to attenuate corrected PET images, and Truex + TOF was used to reconstruct PET images. The PET and CT scan thickness of all patients was 5 mm. The matrix size of all PET reconstructions was 200 × 200, and the anisotropic voxel was 4.07 × 4.07 × 3.0 mm3. After regular PET and CT scans, a 1 mm breath-hold lung CT scan was added. CT was reconstructed by a conventional algorithm, while PET was reconstructed by an iterative method.
Lesion segmentation
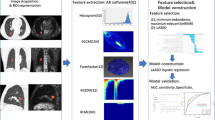
The 5-mm slice thickness PET images and 1-mm slice thickness CT images of all patients were exported from the PACS workstation in DICOM format and then imported into ITK-SNAP software (version 3.8.0-beta, www.itksnap.org) to outline lung lesions in 3D mode. The entire delineation procedure was performed by two radiologists with over 10 years of work experience, and neither of the radiologists was informed of the patients’ pathological results. For CT image delineation, the lesion was observed on the lung window (window width 1600 HU, window level -600 HU). The two radiologists delineated the primary tumour on PET images using a 40% SUVmax threshold to characterize the volume of interest (VOI) [20,21,22,23,24]. To avoid including the physiologic uptake in the VOI, a combined CT and PET scan reading was performed [22, 24]. An example of VOI delineation is shown in Fig. 1.
Workflow for developing a radiomics model based on PET/CT images to predict the thoracic LNM of lung adenocarcinoma. GLCM, gray level co-occurrence matrix; GLSZM, grey level size zone matrix; RLM, run length matrix; mRMR, maximum relevance minimum redundancy; LASSO, least absolute shrinkage and selection operator; ROC, receiver operating characteristic
Image preprocessing
The original 5 mm PET, 1 mm breath-holding thin-layer CT (DICOM format), and outlined VOI of each lung tumour were imported into IBSI-compatible Artificial Intelligence Kit software (AK analysis kit, GE healthcare, 3.2.2) for image preprocessing [23,24,25,26,27]. The μ ± 3σ method was used to remove data with a brightness greater than 3σ to normalize image brightness [23, 24, 28]. The images were resampled to 1 × 1 × 1 mm3 by using linear interpolation to improve the image resolution. The preprocessed images were then imported into ITK-SNAP to delineate the VOI.
Feature extraction and selection
The inter- and intraclass correlation coefficients (ICCs) were evaluated, in which 50 cases were randomly selected from the enrolled study cases. Two observers (Observers A and B) with more than 10 years of working experience in PET and CT applications delineated the VOIs. Observer A delineated the VOIs of CT and PET images twice at an interval of 4 weeks, and the intraobserver correlation coefficients of the extracted features were evaluated between the two delineations of Observer A. Observer B delineated the VOIs independently once, and the interobserver correlation coefficients between the radiomics features extracted by Observers A (the first delineation) and B were evaluated. ICC > 0.75 indicates good agreement. Observer A then finished the remaining delineation work. Based on the VOIs of lung tumours outlined by Observer A on CT and PET images, 402 radiomics features were extracted from every image by using AK software, including 42 histograms features, 154 grey level co-occurrence matrix (GLCM) features, 15 formfactor features describing the shape of the VOI, 180 run length matrix (RLM) features and 11 grey level size zone matrix (GLSZM) features. The bin width was set to 25 during feature extraction.
Model construction and validation
The patients were randomly assigned into training (371 patients) and test groups (157 patients) at a ratio of 7 to 3 using a stratified sampling method to ensure the balance of positive and negative samples in both groups [20, 23, 28]. Bootstrapping was used to split the data into training and validation groups. For the bootstrap samples, simple random sampling was used. To improve the representativeness of the minority group in the training group, the synthetic minority oversampling technique (SMOTE) was used to generate samples of the minority group from the joint weighting of optimal features. In the training group, the maximum relevance minimum redundancy (mRMR) and least absolute shrinkage and selection operator (LASSO) methods were applied to select the most valuable radiomics features (ICC > 0.75) for predicting lung adenocarcinoma LNM. Three multivariate logistic regression models based on PET/CT, CT, and PET were then established in the training group.
The radiomic score of each patient was calculated based on the combination of the retained features weighted by their LASSO logistic regression coefficients (Additional file 6: Methods). The area under the curve (AUC) was used to evaluate the diagnostic efficacy of the three radiomics models in predicting the thoracic LNM of lung adenocarcinoma. The efficacy of predicting the thoracic LNM of lung adenocarcinoma was evaluated in the test group. The DeLong test was employed to compare the performance of the three different models based on PET/CT, CT, and PET to determine the most powerful predictive model. To verify the reliability of the model, a cross-validation test was performed 100 times. The workflow of radiomics analysis is shown in Fig. 1.
Construction of the radiomics nomogram
The clinical factors (p < 0.1) were analysed using univariate logistic regression to identify whether the features were discriminative (p < 0.05). Then, multivariate logistic regression was applied to these discriminative clinical features to construct a clinical model, and the clinical features, as well as the radiomics score, were integrated to establish a predictive nomogram. Moreover, the variance inflation factor (VIF) was used for collinearity analysis, and factors with VIF > 10 were eliminated. All the models were constructed in the training group and then validated in the test group.
Statistical analysis
In this study, the programming language R (software version 3.5.1) was used for statistical analysis. For clinical data, the chi-square test was applied to features with a normal distribution, which were presented as the mean ± SD, while the Wilcoxon test was applied to features with a nonnormal distribution, which were presented as the median (lower and upper quartiles). In this study, the ModelGood package of R was used to construct the calibration curve. Decision curve analysis (DCA) was used to evaluate the clinical value of the PET/CT molecular radiomics-clinical model in predicting lung adenocarcinoma LNM in the test group.
Results
Radiomics feature extraction and selection
For the radiomics features extracted twice by Observer A, the intra-ICC ranges in the CT group and PET group were 0.06–1 and 0.32–1, respectively. For the features extracted by Observer A (for the first time) and Observer B, the inter-ICC ranges in the CT groups and PET group were 0.15–1 and 0.3–1, respectively, and the features with ICC > 0.75 in both the intragroup and intergroup comparisons were retained for further analysis (Additional file 5: Table S1). The PET/CT, CT, and PET data sets were further analysed by the mRMR algorithm and LASSO regression model. After feature extraction and selection, 10 PET/CT (6 CT and 4 PET), 12 CT, and 10 PET radiomics features were retained. These features and their corresponding coefficients are shown in Additional file 2: Figures S2–4.
Evaluation of the performance of the three radiomics models
The results showed that all three models (PET/CT, CT, and PET) could predict LNM with cut-off values of -0.66, -0.75, and -0.13, respectively (Additional file 2: Figures S2C, S3C and S4C). The cut-off values were obtained based on Youden's J statistic, and the optimal cut-off was defined as the threshold that maximizes the distance to the identity (diagonal) line. The value can be abbreviated as “y.” The optimum criterion is the max sensitivity + specificity. To determine the stability of the models, 100 rounds of repeated cross-validation were performed (Additional file 1: Figure S2D). The AUC values of the PET/CT, CT and PET radiomics models in the training group were 0.92 (95% CI: 0.89–0.95), 0.87 (95% CI: 0.83–0.90), and 0.83 (95% CI: 0.78–0.86), respectively. The AUC values of these three models in the test group were 0.91 (95% CI: 0.86–0.95), 0.87 (95% CI: 0.80–0.92), and 0.80 (95% CI: 0.73–0.86), respectively (Fig. 2). The sensitivity, specificity, and accuracy of the PET/CT, CT, and PET radiomics models for predicting the thoracic LNM of lung adenocarcinoma in the training and test groups are shown in Table 2. The DeLong test showed that the PET/CT model outperformed the PET and CT models (p < 0.05) in the training group (Table 3).
Development of a clinical model for predicting the thoracic LNM of lung adenocarcinoma
Further steps were taken to establish a clinical model for predicting the thoracic LNM of lung adenocarcinoma. After the screening of clinical models using univariate logistic analysis of clinical features, it was found that clinical characteristics, including pleural traction, size of the solid component, and location, were statistically significant in predicting thoracic LNM in the training group (Table 4). According to the results of multivariate logistic regression analysis, size of the solid component and location of the lesion were independent predictors (p < 0.05) of the LNM of lung adenocarcinoma, as shown in Table 5. The AUC values of the clinical model in the training and test groups were 0.93 (95% CI: 0.90–0.95) and 0.91 (95% CI: 0.85–0.95), respectively (Table 2).
Construction of an integrated PET/CT radiomics-clinical model for predicting the thoracic LNM of lung adenocarcinoma
The integrated PET/CT molecular radiomics-clinical logistic regression model was constructed using the radiomics score and two independent clinical risk factors, and the results are shown in the nomogram in Fig. 3A. In both groups, the PET/CT molecular radiomics-clinical model showed satisfactory performance in predicting LNM (Table 2). The AUC values of the training and test groups were 0.95 (95% CI: 0.93–0.97) and 0.94 (95% CI: 0.89–0.97), respectively. The AUC values of the PET/CT radiomics-clinical, PET/CT radiomics, and clinical models were compared using the DeLong test. It was found that the PET/CT radiomics-clinical model significantly outperformed the PET/CT radiomics model and the clinical model alone (Table 3). The calibration curve of the nomogram, shown in Fig. 3B, displays good calibration of the nomogram. Finally, the clinical usefulness of these models was compared using DCA. When the threshold probability of predicting lung adenocarcinoma LNM is between 1 and 70%, the application of the PET/CT molecular radiomics-clinical model in predicting the thoracic LNM of lung adenocarcinoma has greater advantages than the clinical model (Fig. 3C).
Evaluation of the performance of the integrated PET/CT molecular radiomics-clinical model. A The nomogram was developed by combining the PET/CT radiomic score and the clinical features of solid composition and location/body part (1, 2, 3, 4, 5 represent the upper lobe, middle lobe, and lower lobe of the right lung and the upper lobe and lower lobe of the left lung, respectively). B Calibration curve with the Hosmer–Lemeshow test of the nomogram in the training cohort (left panel) and test cohort (right panel). The calibration curve shows the calibration of the model in terms of the consistency between the predicted risk of thoracic LNM and the real observed thoracic LNM status. The x-axis represents the predicted risk of thoracic LNM, and the y-axis represents the real thoracic LNM status. C Decision curve analysis of the nomograms. The y-axis measures the standardized net benefit. The dark line represents the PET/CT molecular radiomics-clinical nomogram model, the red line represents the clinical features nomogram, the grey line represents the assumption that all patients are negative for thoracic LNM, and the blue line represents the assumption that all patients are positive for thoracic LNM
Discussion
Patients with occult thoracic LNM of lung adenocarcinoma tend to have short disease-free survival and overall survival. Although CT has been routinely used in the diagnosis of LNM of lung cancer, it shows tumour lesions based on only morphological characteristics and thus suffers from significant limitations. In contrast, PET/CT provides a high accuracy rate in diagnosing local LNM, as it relies on the metabolic characteristics of a tissue, and metabolic changes often occur ahead of morphologic changes [5]. Upon the intake of an imaging reagent, metastatic lesions of lung adenocarcinoma can be judged from the perspective of molecular metabolism [5, 29]. Importantly, PET/CT radiomics has been investigated in lung cancers [30,31,32]. For example, Mu et al. reported that deep learning of PET/CT images could predict PD-L1 status and immunotherapy response in NSCLC [33]. Du et al. reported that a PET/CT radiomics nomogram showed potential for the individualized differential diagnosis of solid active pulmonary tuberculosis and lung cancer [34]. In our previous studies [23, 24], a PET/CT radiomics model was developed to predict epidermal growth factor receptor (EGFR) mutation and anaplastic lymphoma kinase (ALK) rearrangement status in lung adenocarcinoma. This study aimed to predict the thoracic LNM of lung adenocarcinoma by PET/CT radiomics.
Multiple studies have demonstrated the value of CT radiomics in predicting the LNM of lung cancer [16, 17, 35]. For example, Yang et al. developed a nomogram with 14 CT radiomics features to predict LNM in solid lung adenocarcinoma, and the results showed that the AUC values for the training and validation cohorts were 0.871 and 0.856, respectively [16]. Moreover, the LASSO algorithm was used to choose the best set of CT radiomics features and develop a predictive LNM model in IA NSCLC patients based on radiomics and clinical features. The predictive performance for LNM of the combined model was further improved (the AUC values for training and testing were 0.911 and 0.860, respectively) [17]. In addition, PET/CT has also been used to predict LNM in NSCLC [8, 9]. However, only one PET/CT parameter was used alone to predict the LNM of NSCLC in these studies, which yielded high specificity but low sensitivity of diagnosis [8, 9]. In this study, 10 parameters of PET/CT images were extracted using the LASSO algorithm and were used to construct a predictive radiomics model, thus showing more sensitivity in predicting the thoracic LNM of lung adenocarcinoma. The AUC value of the ROC curve of the radiomics model was 0.92 in the training group and 0.91 in the test group. In addition, the clinical model (the visual assessment) was constructed by the subjective sign of the solid component and location of lung adenocarcinoma after feature selection, and the AUC value reached 0.93 in the training group and 0.91 in the test group. It was found that both the clinical model and radiomics model achieved similarly good performance outcomes in predicting LNM. Furthermore, compared with the PET/CT clinical model, the integrated PET/CT molecular radiomics-clinical model shows more power for predicting the local LNM of lung adenocarcinoma. The AUC value increased from 0.93 to 0.95 in the training group and from 0.91 to 0.94 in the test group.
The present results are significant in several major aspects as follows. First, the location of lung cancer is closely related to LNM [36, 37]. Ketchedjian et al. showed that the incidence of LNM increased as the size of peripheral T1 tumours of lung adenocarcinoma increased, whereas central T1 tumours demonstrated a 50% incidence of lymph node involvement irrespective of tumour size [36]. This study found that the location of lung adenocarcinoma was an important factor in predicting the LNM of lung adenocarcinoma. Second, it has been reported that there is a strong correlation between the diameter of solid components on CT images and the invasive components revealed by pathology, and the size of the solid component is an important factor affecting prognosis [19, 38, 39]. In this study, the solid component size was shown to serve as an effective predictor of thoracic LNM. Third, the nomogram provides a quantitative and intuitive method for clinicians to predict the LNM of lung adenocarcinoma. Finally, our study addresses the limitation of the CT radiomics model where the LNM of lung adenocarcinoma is predicted from CT morphology alone, while incorporating PET radiomics to indicate the level of tumour molecular metabolism. The overall objective of the current study is to further confirm the application value of PET radiomics. To date, there is no literature report comparing PET/CT and CT radiomics models in the prediction of the LNM of lung adenocarcinoma. This study found that PET/CT radiomics was superior to CT radiomics in the training group, indicating that PET radiomics has certain diagnostic value in predicting the LNM of lung adenocarcinoma.
Despite the promising statistical results, several limitations of this study need to be acknowledged. First, in view of the single-centre retrospective nature of the current study, a multicentre study with a larger sample size should be conducted for further verification. Second, it is time-consuming for radiologists to semi-manually delineate the segmentation of lung adenocarcinoma lesions by means of ITK software. It is expected that with the development of artificial intelligence software such as deep learning, fully automatic computer segmentation can be realized [20, 40, 41]. Third, this study lacks external validation to refine the proposed model. Finally, due to limited time after surgery and incomplete follow-ups, a predictive model for survival rate has yet to be established.
Conclusion
In summary, the PET/CT molecular radiomics-clinical model demonstrated its high diagnostic value in predicting the thoracic LNM of lung adenocarcinoma. A PET/CT molecular radiomics-clinical nomogram model was developed as a visualization tool to help predict thoracic LNM in newly diagnosed lung adenocarcinoma patients.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- LNM:
-
Lymph node metastasis
- NSCLC:
-
Non-small-cell lung cancer
- DCA:
-
Decision curve analysis
- PET/CT:
-
Positron emission tomography/computed tomography
- SUVmax:
-
Maximum standardized uptake value
- CEA:
-
Carcinoembryonic antigen
- VOI:
-
Volume of interest
- ICCs:
-
Inter- and intraclass correlation coefficients
- GLCM:
-
Grey level co-occurrence matrix
- RLM:
-
Run length matrix
- GLSZM:
-
Grey level size zone matrix
- mRMR:
-
Maximum relevance minimum redundancy
- LASSO:
-
Least absolute shrinkage and selection operator
- AUC:
-
Area under the curve
- VIF:
-
Variance inflation factor
- SMOTE:
-
Synthetic minority oversampling technique
- EGFR:
-
Epidermal growth factor receptor
- ALK:
-
Anaplastic lymphoma kinase
References
Travis W, Brambilla E, Noguchi M, Nicholson A, Geisinger K, Yatabe Y, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–85. https://doi.org/10.1097/JTO.0b013e318206a221.
Postmus PE, Kerr KM, Oudkerk M, Senan S, Waller DA, Vansteenkiste J, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017; 28(suppl_4): iv1–iv21. https://doi.org/10.1093/annonc/mdx222.
Ramnath N, Dilling TJ, Harris LJ, Kim LJ, Michaud GC, Balekian AA, et al. Treatment of stage III non-small cell lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e314S – e340. https://doi.org/10.1378/chest.12-2360.
Czarnecka-Kujawa K, Yasufuku K. The role of endobronchial ultrasound versus mediastinoscopy for non-small cell lung cancer. J Thorac Dis. 2017; 9(Suppl 2): S83–S97. https://doi.org/10.21037/jtd.2017.03.102.
Birim O, Kappetein AP, Stijnen T, Bogers AJ. Meta-analysis of positron emission tomographic and computed tomographic imaging in detecting mediastinal lymph node metastases in nonsmall cell lung cancer. Ann Thorac Surg. 2005;79(1):375–82. https://doi.org/10.1016/j.athoracsur.2004.06.041.
Gould MK, Kuschner WG, Rydzak CE, Maclean CC, Demas AN, Shigemitsu H, et al. Test performance of positron emission tomography and computed tomography for mediastinal staging in patients with non-small-cell lung cancer: a meta-analysis. Ann Intern Med. 2003;139(11):879–92. https://doi.org/10.7326/0003-4819-139-11-200311180-00013.
Groheux D, Quere G, Blanc E, Lemarignier C, Margerie-Mellon C, Merlet P, et al. FDG PET-CT for solitary pulmonary nodule and lung cancer: literature review. Diagn Interv Imaging. 2016;97(10):1003–17. https://doi.org/10.1016/j.diii.2016.06.020.
Liu B, Dong J, Xu C, Zuo C, Le J, Guan Y, et al. Accuracy of 18F-FDG PET/CT for lymph node staging in non-small-cell lung cancers. Chin Med J (Engl). 2009;122(15):1749–54.
Lee J, Kim E, Kim D, Lee JH, Kang W, Lee JD, et al. The diagnostic ability of 18F-FDG PET/CT for mediastinal lymph node staging using 18F-FDG uptake and volumetric CT histogram analysis in non-small cell lung cancer. Eur Radiol. 2016;26(12):4515–23. https://doi.org/10.1007/s00330-016-4292-8.
Liu Y. Role of FDG PET-CT in evaluation of locoregional nodal disease for initial staging of breast cancer. World J Clin Oncol. 2014;5(5):982–9. https://doi.org/10.5306/wjco.v5.i5.982.
Capitanio S, Nordin AJ, Noraini AR, Rossetti C. PET/CT in nononcological lung diseases: current applications and future perspectives. Eur Respir Rev. 2016;25(141):247–58. https://doi.org/10.1183/16000617.0051-2016.
Shimada Y, Tsuboi M, Saji H, Miyajima K, Usuda J, Uchida O, et al. The prognostic impact of main bronchial lymph node involvement in non-small cell lung carcinoma: suggestions for a modification of the staging system. Ann Thorac Surg. 2009;88(5):1583–8. https://doi.org/10.1016/j.athoracsur.2009.04.065.
El-Sherief AH, Lau CT, Carter BW, Wu CC. Staging lung cancer: regional lymph node classification. Radiol Clin North Am. 2018;56(3):399–409. https://doi.org/10.1016/j.rcl.2018.01.008.
Zhu X, Dong D, Chen Z, Fang M, Zhang L, Song J, et al. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur Radiol. 2018;28(7):2772–8. https://doi.org/10.1007/s00330-017-5221-1.
Coroller TP, Grossmann P, Hou Y, Velazquez ER, Leijenaar RT, Hermann G, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114(3):345–50. https://doi.org/10.1016/j.radonc.2015.02.015.
Yang X, Pan X, Liu H, Gao D, He J, Liang W, et al. A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J Thorac Dis. 2018; 10(Suppl 7):S807–S19. https://doi.org/10.21037/jtd.2018.03.126.
Cong M, Feng H, Ren JL, Xu Q, Cong L, Hou Z, et al. Development of a predictive radiomics model for lymph node metastases in pre-surgical CT-based stage IA non-small cell lung cancer. Lung Cancer. 2020;139:73–9. https://doi.org/10.1016/j.lungcan.2019.11.003.
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, Jong EEC, Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–62. https://doi.org/10.1038/nrclinonc.2017.141.
Chang C, Sun X, Zhao W, Wang R, Qian X, Lei B, et al. Minor components of micropapillary and solid subtypes in lung invasive adenocarcinoma (</= 3 cm): PET/CT findings and correlations with lymph node metastasis. Radiol Med. 2020;125(3):257–64. https://doi.org/10.1007/s11547-019-01112-x.
Bashir U, Azad G, Siddique MM, Dhillon S, Patel N, Bassett P, et al. The effects of segmentation algorithms on the measurement of (18)F-FDG PET texture parameters in non-small cell lung cancer. EJNMMI Res. 2017;7(1):60. https://doi.org/10.1186/s13550-017-0310-3.
Cook GJR, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and promises of PET radiomics. Int J Radiat Oncol Biol Phys. 2018;102(4):1083–9. https://doi.org/10.1016/j.ijrobp.2017.12.268.
Ren C, Zhang J, Qi M, Zhang J, Song S, Sun Y, et al. Machine learning based on clinico-biological features integrated (18)F-FDG PET/CT radiomics for distinguishing squamous cell carcinoma from adenocarcinoma of lung. Eur J Nucl Med Mol Imaging. 2021;48(5):1538–49. https://doi.org/10.1007/s00259-020-05065-6.
Chang C, Zhou S, Yu H, Zhao W, Ge Y, Duan S, et al. A clinically practical radiomics-clinical combined model based on PET/CT data and nomogram predicts EGFR mutation in lung adenocarcinoma. Eur Radiol. 2021;31(8):6259–68. https://doi.org/10.1007/s00330-020-07676-x.
Chang C, Sun X, Wang G, Yu H, Zhao W, Ge Y, et al. A Machine learning model based on PET/CT radiomics and clinical characteristics predicts ALK rearrangement status in lung adenocarcinoma. Front Oncol. 2021;11: 603882. https://doi.org/10.3389/fonc.2021.603882.
Liu H, Zhang C, Wang L, Luo R, Li J, Zheng H, et al. MRI radiomics analysis for predicting preoperative synchronous distant metastasis in patients with rectal cancer. Eur Radiol. 2019;29(8):4418–26. https://doi.org/10.1007/s00330-018-5802-7.
Shu Z, Shao Y, Xu Y, Ye Q, Cui S, Mao D, et al. Radiomics nomogram based on MRI for predicting white matter hyperintensity progression in elderly adults. J Magn Reson Imaging. 2020;51(2):535–46. https://doi.org/10.1002/jmri.26813.
Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med. 2003;50(1):92–8. https://doi.org/10.1002/mrm.10496.
Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging. 2004;22(1):81–91. https://doi.org/10.1016/j.mri.2003.09.001.
Chung HH, Lee I, Kim HS, Kim JW, Park NH, Song YS, et al. Prognostic value of preoperative metabolic tumor volume measured by (1)(8)F-FDG PET/CT and MRI in patients with endometrial cancer. Gynecol Oncol. 2013;130(3):446–51. https://doi.org/10.1016/j.ygyno.2013.06.021.
Avanzo M, Stancanello J, Pirrone G, Sartor G. Radiomics and deep learning in lung cancer. Strahlenther Onkol. 2020;196(10):879–87. https://doi.org/10.1007/s00066-020-01625-9.
Chetan MR, Gleeson FV. Radiomics in predicting treatment response in non-small-cell lung cancer: current status, challenges and future perspectives. Eur Radiol. 2021;31(2):1049–58. https://doi.org/10.1007/s00330-020-07141-9.
Manafi-Farid R, Karamzade-Ziarati N, Vali R, Mottaghy FM, Beheshti M. 2-[(18)F]FDG PET/CT radiomics in lung cancer: an overview of the technical aspect and its emerging role in management of the disease. Methods. 2020. https://doi.org/10.1016/j.ymeth.2020.05.023.
Mu W, Jiang L, Shi Y, Tunali I, Gray JE, Katsoulakis E, et al. Non-invasive measurement of PD-L1 status and prediction of immunotherapy response using deep learning of PET/CT images. J Immunother Cancer. 2021;9(6). https://doi.org/10.1136/jitc-2020-002118.
Du D, Gu J, Chen X, Lv W, Feng Q, Rahmim A, et al. Integration of PET/CT radiomics and semantic features for differentiation between active pulmonary tuberculosis and lung cancer. Mol Imaging Biol. 2021;23(2):287–98. https://doi.org/10.1007/s11307-020-01550-4.
Mimae T, Miyata Y, Tsutani Y, Mimura T, Nakayama H, Okumura S, et al. What are the radiologic findings predictive of indolent lung adenocarcinoma? Jpn J Clin Oncol. 2015;45(4):367–72. https://doi.org/10.1093/jjco/hyv005.
Ketchedjian A, Daly B, Fernando H, Florin L, Hunter C, Morelli D, et al. Location as an important predictor of lymph node involvement for pulmonary adenocarcinoma. J Thorac Cardiovasc Surg. 2006;132(3):544–8. https://doi.org/10.1016/j.jtcvs.2006.05.023.
Watanabe S, Suzuki K, Asamura H, et al. Superior and basal segment lung cancers in the lower lobe have different lymph node metastatic pathways and prognosis. Ann Thorac Surg. 2008;85(3):1026–31. https://doi.org/10.1016/j.athoracsur.2007.10.076.
Lee KH, Goo JM, Park SJ, Wi JY, Chung DH, Go H, et al. Correlation between the size of the solid component on thin-section CT and the invasive component on pathology in small lung adenocarcinomas manifesting as ground-glass nodules. J Thorac Oncol. 2014;9(1):74–82. https://doi.org/10.1097/JTO.0000000000000019.
Tsutani Y, Miyata Y, Nakayama H, Okumura S, Adachi S, Yoshimura M, et al. Prognostic significance of using solid versus whole tumor size on high-resolution computed tomography for predicting pathologic malignant grade of tumors in clinical stage IA lung adenocarcinoma: a multicenter study. J Thorac Cardiovasc Surg. 2012;143(3):607–12. https://doi.org/10.1016/j.jtcvs.2011.10.037.
Shiri I, Rahmim A, Ghaffarian P, Geramifar P, Abdollahi H, Bitarafan-Rajabi A. The impact of image reconstruction settings on 18F-FDG PET radiomic features: multi-scanner phantom and patient studies. Eur Radiol. 2017;27(11):4498–509. https://doi.org/10.1007/s00330-017-4859-z.
van Velden FH, Kramer GM, Frings V, Nissen IA, Mulder ER, Langen AJ, et al. Repeatability of radiomic features in non-small-cell lung cancer [(18)F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol. 2016;18(5):788–95. https://doi.org/10.1007/s11307-016-0940-2.
Acknowledgements
This study was financially supported by the Special Project of Integrated Traditional Chinese and Western Medicine in the General Hospital of Shanghai Health Committee (No. ZHYY-ZXYJHZX-202023); the Nurture Projects for Basic Research of Shanghai Chest Hospital (Nos. 2021YNJCM09, 2020YNJCM07, 2020YNJCQ11, 2020YNJCQ06); the National Natural Science Foundation of China (Nos. 82071873, 81871353); the Natural Science Foundation of Shanghai (No. 21ZR1458900); the Youth Medical Talents–Medical Imaging Practitioner Program (No. SHWRS(2020)_087); the Shanghai Sailing Program (Nos. 19YF1445400, 20YF1444500, 21YF1442700); and the Shanghai University of Traditional Chinese Medicine Budget Project (No. 18LK055).
Author information
Authors and Affiliations
Contributions
CC, MMR, BL, JF, and WHX contributed to the data analysis and interpretation, manuscript drafting, critical review and revision; LHW, CYL, and LL contributed to the data collection; CC, MMR, HYu, WJT, QFW, HYan, LL, and WHX contributed to the study design and funding acquisition; YQG, SFD, and XHQ contributed to the software and data statistics; and WLZ, WJT, and QFW contributed to the study design and data analysis and interpretation. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol and methods were approved by the institutional ethics review committee of Shanghai Chest Hospital in accordance with the Declaration of Helsinki (Project Number KS2035). Patient consent was waived by the institutional review board due to the retrospective and anonymized nature of the data analysis.
Consent for publication
Not applicable.
Competing interests
Two of the authors of this manuscript (YQG, SFD) are employees of GE Healthcare. The remaining authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Figure S1
: Flowchart of lung adenocarcinoma patient selection.
Additional file 2. Figure S2
: Construction of a PET/CT radiomics model based on PET/CT images. (A) A total of 10 radiomics features were identified by mRMR and LASSO logistic regression based on PET and CT features. (B) List of 10 radiomics features chosen to construct the PET/CT radiomics model. (C) Representative results of the PET/CT radiomics model for predicting thoracic LNM in the training (left) and test (right) groups of lung adenocarcinoma patients. 0, negative thoracic LNM; 1, positive thoracic LNM. (D) Cross-validation analysis showed that the PET/CT radiomics model has good reliability for predicting thoracic LNM in the training (left) and test (right) groups of lung adenocarcinoma patients
Additional file 3. Figure S3
: Construction of a CT radiomics model based on CT images. (A) A total of 12 radiomics features were identified by mRMR and LASSO logistic regression based on CT features. (B) List of 12 radiomics features chosen to construct the CT radiomics model. (C) Representative results of the CT radiomics model for predicting thoracic LNM in the training (left) and test (right) groups of lung adenocarcinoma patients. 0, negative thoracic LNM; 1, positive thoracic LNM
Additional file 4. Figure S4
: Construction of a PET radiomics model based on PET images. (A) A total of 10 radiomic features were identified by mRMR and LASSO logistic regression based on PET features. (B) List of 10 radiomisc features chosen to construct the PET radiomics model. (C) Representative results of the PET radiomics model for predicting thoracic LNM in the training (left) and test (right) groups of lung adenocarcinoma patients. 0, negative thoracic LNM; 1, positive thoracic LNM
Additional file 5. Table S1
: The intra-and interobserver ICCs of the consistency of lesion segmentation
Additional file 6. Methods
: The formula for the calculation of PET/CT, CT, PET radiomics scores
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chang, C., Ruan, M., Lei, B. et al. Development of a PET/CT molecular radiomics-clinical model to predict thoracic lymph node metastasis of invasive lung adenocarcinoma ≤ 3 cm in diameter. EJNMMI Res 12, 23 (2022). https://doi.org/10.1186/s13550-022-00895-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13550-022-00895-x