Abstract
Background
Considering the limited accessibility of amyloid position emission tomography (PET) in patients with dementia, we proposed a deep learning (DL)-based amyloid PET positivity classification model from PET images with 2-deoxy-2-[fluorine-18]fluoro-D-glucose (2-[18F]FDG).
Methods
We used 2-[18F]FDG PET datasets from the Alzheimer's Disease Neuroimaging Initiative and Korean Brain Aging Study for the Early diagnosis and prediction of Alzheimer’s disease for model development. Moreover, we used an independent dataset from another hospital. A 2.5-D deep learning architecture was constructed using 291 submodules and three axes images as the input. We conducted the voxel-wise analysis to assess the regions with substantial differences in glucose metabolism between the amyloid PET-positive and PET-negative participants. This facilitated an understanding of the deep model classification. In addition, we compared these regions with the classification probability from the submodules.
Results
There were 686 out of 1433 (47.9%) and 50 out of 100 (50%) amyloid PET-positive participants in the training and internal validation datasets and the external validation datasets, respectively. With 50 times iterations of model training and validation, the model achieved an AUC of 0.811 (95% confidence interval (CI) of 0.803–0.819) and 0.798 (95% CI, 0.789–0.807) on the internal and external validation datasets, respectively. The area under the curve (AUC) was 0.860 when tested with the model with the highest value (0.864) on the external validation dataset. Moreover, it had 75.0% accuracy, 76.0% sensitivity, 74.0% specificity, and 75.0% F1-score. We found an overlap between the regions within the default mode network, thus generating high classification values.
Conclusion
The proposed model based on the 2-[18F]FDG PET imaging data and a DL framework might successfully classify amyloid PET positivity in clinical practice, without performing amyloid PET, which have limited accessibility.
Similar content being viewed by others
Background
Alzheimer’s disease (AD) is characterized by the accumulation of β-amyloid (Aβ) and tau proteins. Aβ can be measured in humans with specific position emission tomography (PET) tracers or an examination of the cerebrospinal fluid (CSF) [1,2,3]. Patients clinically diagnosed with mild cognitive impairment (MCI) or AD have been found Aβ-negative [4, 5], thus leading to a pathophysiology-based unbiased and descriptive amyloid, tau, and neurodegeneration classification [6, 7]. The aforementioned criterion considers the Alzheimer’s continuum only when the Aβ marker is positive. However, high cost and low availability make it difficult to conduct amyloid PET. Furthermore, the limitations of CSF examination can be attributed to its invasiveness, thus necessitating alternative ways to classification amyloid status.
2-deoxy-2-[fluorine-18]fluoro-D-glucose (2-[18F]FDG) is the most widely used PET tracer for measuring brain metabolism, which is related to neuronal activity [8]. Studies using 2-[18F]FDG PET images reported on lesser hypometabolism in the bilateral temporoparietal regions and hippocampus in Aβ-negative participants with MCI and AD, compared to their positive counterparts [4]. There had been several attempts to develop an amyloid status classification model using magnetic resonance imaging (MRI), neuropsychological, or laboratory tests [9,10,11]. However, the performance was unsatisfactory. No attempts have been made using 2-[18F]FDG PET images.
Deep learning (DL) is the state-of-the-art mathematical algorithms that enable computers to automatically find patterns in large datasets. DL is studied in the medical imaging field for classification (diagnosis) [12, 13], predict prognosis [14, 15], detection [16, 17], and segmentation [18, 19]. A convolutional neural network (CNN), a subset of DL [20], is a DL model mimicking visual recognition concept which can extract features that reflect spatial relationships by applying non-linear convolutional filters. These spatial features pass through artificial neurons whose weights in the layers are properly set during training. Through this process, the model automatically learns hidden representative features from images and labels. 2-D-based deep learning architectures for 3-D medical images had been proposed: grid method (single montage image made by 16 images [21]), surface projection method (volumetric information projected onto a surface [22, 23]), and 2.5-D model (three axes (axial, coronal, and sagittal) images of brain volume [24]).
We aimed to construct a model that classifies amyloid PET positivity using 2-[18F]FDG PET that reflects brain metabolism and CNN architecture. In addition, we intended to validate the model using an independent external dataset with various diseases and classification probability analysis of submodules.
Methods
Study participants and data collection
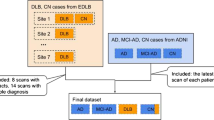
The following three datasets were used for model training, internal validation, and external validation: (i) Alzheimer’s Disease Neuroimaging Initiative (ADNI; adni.loni.usc.edu)-1, ADNI-GO (Grand Opportunities), ADNI-2, and ADNI-3 dataset [25], (ii) Korean Brain Aging Study for the Early diagnosis and prediction of Alzheimer’s disease (KBASE; kbase.kr) dataset [26], and (iii) dataset from the Severance Hospital. The inclusion criterion comprised individuals who underwent T1-weighted MRI, 2-[18F]FDG PET, and amyloid PET imaging. MRI and PET assessments were performed within 6 months. We excluded 11 and three participants from the ADNI and KBASE datasets, respectively, due to poor image quality. We eventually selected 963 and 470 participants from the ADNI and KBASE datasets, respectively (Fig. 1a). All data used in this study were from the baseline assessments. An additional 100 participants were recruited at the memory disorder clinic in the Department of Neurology at the Severance Hospital in Seoul, South Korea, between December 2017 and April 2019. The same criterion had been applied to ensure consistency (Fig. 1b). Table 1 summarizes the patient demographics and other information.
While 80% of the ADNI and KBASE datasets was used for model training, the remaining 20% was used for internal validation. The Severance Hospital dataset was used for externally validating the classification model.
Standard protocol approvals, registrations, and patient consent
The ADNI study protocol was approved by the institutional review board of each participating ADNI site (adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf). All participants provided written informed consent at the time of their enrollment in our study.
The use of the others datasets was approved by the Institutional Review Boards of the Seoul National University Hospital and the Severance Hospital. All participants provided their written informed consent.
Imaging acquisition and preprocessing
The detailed 2-[18F]FDG /18F-florbetapir PET imaging protocol for the ADNI dataset is described at adni.loni.usc.edu/method/documents/. All participants underwent simultaneous PET and MRI scans using a 3T Biograph mMR (PET-MR) scanner (Siemens, Washington DC, USA) for the KBASE dataset. The procedure was performed based on the guidelines approved by the manufacturer. 3-D T1-weighted images with 3-D T1-weighted magnetization-prepared rapid acquisition with gradient echo (MPRAGE) sequence were acquired in the sagittal orientation with the following acquisition parameters: repetition time (TR) = 1670 ms, echo time (TE) = 1.89 ms, field of view (FOV) = 250 mm, and a 256 × 256 matrix with a 1.0 mm slice thickness. For the [11C] Pittsburgh compound-B (PiB) PET, we obtained a 30-min emission, 40-min after the intravenous administration of 555 MBq 11C-PiB. In contrast, for the 2-[18F]FDG PET scans, we requested the participant to fast for at least 6 h, prior to receiving the intravenous administration of 3.7 MBq/kg 2-[18F]FDG. Following the intravenous injections, the participants rested for 40 min in a dimly lit waiting room, prior to scanning. We processed the images for routine corrections and reconstructed them into a 256 × 256 image matrix using iterative methods (six iterations with 21 subsets) [26].
The dataset of the Severance Hospital included all participants who underwent MRI scans using a 3T Achieva scanner (Philips Medical System, Best, The Netherlands). We acquired a 3-D T1-weighted MRI sequence with a 3-D T1-turbo field echo sequence in the axial orientation with the following acquisition parameters: TR = 73,421 ms, TE = 5.09 ms, FOV = 215 mm, and a 1024 × 1024 matrix with a 1.0 mm slice thickness. We used a Discovery 600 scanner (GEHealthcare, Milwaukee, WI, USA) for the 18F-florbetaben PET scan. Moreover, we obtained a 20-min emission scan after the intravenous administration of 300 MBq 18F-florbetaben. The images were processed for routine corrections and reconstructed into a 256 × 256 image matrix using iterative methods (four iterations with 32 subsets).
Data preprocessing
We conducted the preprocessing steps using SPM12 (Wellcome Trust Centre for Neuroimaging, University College London) and MATLAB R2019a (MathWork, Natick, MA). The amyloid and 2-[18F]FDG images were coregistered onto T1-weighted images and normalized into the Montreal Neurological Institute (MNI) template (McGill University, Montreal, Canada). Furthermore, we normalized each voxel of the 2-[18F]FDG PET image according to the mean intensity of the pons [27]. The pons were used as reference sites and extracted using an Automated Anatomical Labeling (AAL) template.
Decision of Aβ PET status
We downloaded the UC Berkeley 18F-florbetapir analysis data from the ADNI dataset. Moreover, we classified each participant as Aβ-positive PET scan on observing a global standardized uptake value ratio (SUVR) > 1.11 [28].
We extracted the mean regional 11C-PiB uptake values from the frontal, posterior cingulate-precuneus, and lateral temporal and lateral parietal cortices using the individual AAL atlas from T1-coregistered 11C-PiB PET images for the KBASE dataset [29, 30]. In addition, we calculated the SUVRs for each region of interest (ROI) by dividing the mean value for all voxels within each ROI by the mean cerebellar uptake value. This can be attributed to its relatively low Aβ deposition [31]. We classified each participant as Aβ-positive PET scan if the SUVR was > 1.4 in at least one of the four ROIs [32].
We eventually analyzed the Severance Hospital dataset using a method similar to the aforementioned ones. We extracted the mean regional 18F-florbetaben uptake values from the frontal, anterior/posterior cingulate, lateral parietal, and lateral temporal cortices using the individual AAL atlas from T1-coregistered 18F-florbetaben PET images. Moreover, we calculated the SUVRs for each ROI by dividing the mean value for all voxels within each ROI by the mean cerebellar uptake value. We classified each participant as Aβ-positive PET scan if the SUVR was > 1.478 in at least one of the four ROIs [33].
Deep learning architecture
Initially, we tried to create a 3-D model for considering the axial, sagittal, and coronal spatial relationships. This can be attributed to the importance of spatial relationships in imaging. We have constructed two versions of 3-D CNN model (Additional file 1: Fig. 1). Nonetheless, the datasets were not large enough for model training. Furthermore, the 3-D models could not be trained even after augmenting the small datasets and needed a high computing power. Thus, we eventually built a 2.5- model that used the axial, coronal, and sagittal images as inputs to incorporate the 3-D spatial relationships. We entered a total of 291 images (91 images each along the z-axis and x-axis and 109 images along the y-axis) as inputs from each participant to the DL architecture (Fig. 2). The 2-D CNN submodule consists of four convolutional layers. The number of filters used in each convolutional layer was 32, 64, 128, and 128, respectively. Each convolutional layer had a kernel size of 3 × 3 and a stride of 1. Following the convolutional layers, we added a batch normalization layer and a rectified linear unit (ReLU). While we used the ReLU for the nonlinear activation function, the batch normalization layer improved the training convergence speed. We then added a 2 × 2 max pooling layer with a stride of 2 for down-sampling of the feature maps. After the four convolutional layers, we used a completely connected layer to determine the Aβ PET status in each image with sigmoid activation. DL architecture consists of 291 submodules, and each submodule had been assigned a different weight value. We concatenated 291 prediction values as inputs to the aforementioned layer. In addition, we used sigmoid activation as a function for the final prediction. When the final prediction value was larger than 0.5, the model classified this participant Aβ-positive PET scan. We initialized the 2-D convolutional filters and the completely connected layers using the He-weight initialization [34] and the Xavier initialization, respectively. Our architecture was trained using a mini-batch size of four and an Adam optimizer with a 0.0001 learning rate for a maximum of 30 epochs. We monitored the internal validation loss after every epoch. Moreover, we saved the model weights upon encountering the lowest internal validation loss. We used the DL model with the lowest internal validation loss to classify the amyloid PET positivity on the external validation dataset. All model was made in open-source package PyTorch on NVIDIA GTX 2080Ti.
Convolutional neural network architecture in 2.5 dimensions (2.5-D). A total of 291 2-D images were used as inputs for the model. 291 number of prediction values obtained from 291 submodules pass through the fully connected layer for the final prediction. Abbreviation: Conv = convolution layer; MaxPool = Max pooling layer; FC = fully connected layer
Performance evaluation
First, our model was trained on the ADNI and KBASE datasets. These datasets were randomly divided into two groups for training and internal validation. Moreover, our model was externally validated on the Severance Hospital dataset. This process was repeated 50 times.
We computed the receiver operating characteristic (ROC) curve, the area under curve (AUC) values, accuracy, sensitivity, and specificity of model classifications on all datasets to evaluate the performance of our model. In addition, we computed the F1-score, the harmonized mean of the positive predictive value and sensitivity.
The F1-score is defined as follows:
In addition, performance of our model was evaluated within each subgroup, such as CU, MCI, and demented participants.
Second, to evaluate the model performance between two distinct datasets with different diagnoses, composition, and ethnicity, our model was trained and internally validated on the ADNI dataset, and externally validated on the KBASE dataset.
Voxel-wise analysis
We conducted a two-sample t test to find substantial differences in glucose metabolism between the Aβ-positive and Aβ-negative PET scan participants. In addition, we conducted the vowel-wise analysis using SPM12 [35]. The threshold for statistical mapping was at FWE-corrected p < 0.05 or uncorrected p < 0.001. We applied the spatial clustering of regions with statistically relevant voxels using a clustering threshold of k > 50 voxels for eliminated voxel clusters with smaller sizes.
Classification basis for understanding model decision
We commonly used a class activation map (CAM) to confirm the classification basis. Nonetheless, we could not apply it as our model was built using a 2.5-D CNN [36]. Thus, we suggested a method to approximately understand the decision basis of our model. Figure 3 illustrates the way of analysis of classification probability from the submodules. This facilitates the verification of the ROIs that contribute to our classification. First, we extracted the prediction values from the submodules. Second, we performed a two-sample t test based on the final classification between the Aβ-positive and Aβ-negative PET scan participants (p value below 0.05, two-tailed) and obtained a substantial slice number. Third, we considered the (x, y, z) point as statistically significant when the ‘x’th, ‘y’th, and ‘z’th planes of the sagittal, the coronal, and axial axis showed substantial differences. We eventually plotted the aforementioned points in the MNI space. If ×0, ×1 in the sagittal; y0, y1 in the coronal; and z0, z1 in the axial planes were significant slices, a total of 8 points could be plotted in the MNI space. Assuming that (× 0, y0, z0) and (× 1, y1, z1) are significant points among the plotted points, the unrelated six points ((× 0,y0,z1), (× 0,y1,z0), (× 0,y1,z1), (× 1,y0,z0), (× 1,y0,z1), (× 1,y1,z0)) may also be included in the MNI space. Therefore, to exclude these unrelated points, we finally extracted clusters with more than 50 points (same threshold as voxel-wise analysis) from the AAL atlas. Moreover, we compared the aforementioned regions with the results of our classification-based voxel-wise analysis.
Analysis of prediction values from submodules to understand the model’s decision. a Acquiring prediction values from submodules along the 3 axes. b The Aβ-positive and Aβ-negative PET scan participants are divided based on the final classification, and a two-sample t test is performed to determine substantial slice numbers. c) Points are plotted for each slice number in the MNI space and correlated labels are extracted from the AAL atlas. In the example, if a significant slice number is x = 46, 51, y = 34, 64, 66, and z = 37, 45, a total of 12 points are plotted in the MNI space. However, actual significant points are (46,34,37), (51,54,45), and (51,66,45). d) To exclude unrelated points, ROIs in the AAL atlas with more than 50 points are used for comparison with the result of the voxel-wise analysis. Abbreviation: Aβ = β-amyloid; MNI = Montreal Neurological Institute; AAL = Automated Anatomical Labeling; ROI = region of interest
Results
Participant demographics
Table 1 shows the demographic data of the participants in the three datasets. There were 686 out of 1433 (47.9%) and 50 out of 100 (50%) Aβ-positive PET scan participants in the ADNI/KABSE and Severance Hospital datasets, respectively.
Performance of deep learning model
The internal validation dataset included 129 and 138 Aβ-negative and Aβ-positive PET scan participants (20% of ADNI/KBASE datasets), respectively. In contrast, the external validation dataset included 50 participants each in the abovementioned categories. The model achieved an AUC of 0.811 (95% confidence interval (CI) of 0.803–0.819) and 0.798 (95% CI, 0.789–0.807) on the internal and external validation datasets, respectively. Table 2 summarizes the accuracy, sensitivity, specificity, and F1 score. When we performed external validation with the model shown best AUC value (0.864) on the internal validation dataset, the AUC, accuracy, sensitivity, specificity, and F1 score of 0.860, 0.770, 0.800, 0.740, and 0.780, respectively. Figure 4 shows the ROC curves and confusion matrix of the aforementioned model for classifying Aβ PET positivity in the internal and external validation datasets.
Internal validation model performance by the subgroups (CU, MCI, and demented (AD)) were shown in the Additional file 1: Fig. 2. The model achieved an AUC of 0.717 (95% CI, 0.705–0.729), 0.757 (95% CI, 0.746–0.768) and 0.816 (95% CI, 0.789–0.843) on CU (n = 105), MCI (n = 141) and AD (n = 41) participants, respectively. When AUC value was 0.864 on internal validation dataset, the AUC were 0.738, 0.839 and 0.887 on CU, MCI, and AD, respectively. Moreover, external validation model performance by the subgroups were shown in the Additional file 1: Fig. 3. The model achieved an AUC of 0.798 (95% CI, 0.789–0.807) and 0.787 (95% CI, 0.774–0.800) on MCI (n = 75) and demented (n = 24) participants. When we performed external validation with the model that showed best AUC value (0.864) on the internal validation dataset, the AUC were 0.865 and 0.832 on MCI and demented participants, respectively. Table 2 summarizes the accuracy, sensitivity, specificity, and F1 scores.
Additional file 1: Table 1 shows model performance by all datasets and subgroups in the internal validation (part of the ADNI dataset) and external validation (KBASE dataset). The model achieved an AUC of 0.769 (95% CI, 0.762–0.776) and 0.806 (95% CI, 0.803–0.809) on the internal and external validation datasets, respectively. When we performed external validation with the model, it indicated a good AUC value (0.800) on the internal validation, the AUC previously was 0.822. The accuracy, sensitivity, specificity, and F1 score and values on subgroups are summarized in the Additional file 1: Table 1.
Voxel-wise analysis
In the ADNI and KBASE datasets, Aβ-negative PET scan participants showed higher glucose metabolism than Aβ-positive PET scan participants at bilateral angular, supramarginal, precuneus, middle and posterior cingulate, inferior and middle temporal, parahippocampal, and fusiform gyri (Fig. 5a). In contrast, Aβ-positive PET scan participants showed higher glucose metabolism in the bilateral paracentral lobules, pre- and postcentral gyri (Fig. 5b).
Result of voxel-wise 2-[18F]FDG PET analysis performed in the ADNI and KBASE datasets. a) Areas of higher glucose metabolism in Aβ-negative PET scan participants compared to Aβ-positive PET scan participants. b) Areas of higher glucose metabolism in Aβ-positive PET scan participants compared to Aβ-positive PET scan participants. All results are presented with a threshold of p < 0.05, FWE-corrected, and 50 voxels. Abbreviation: ADNI = Alzheimer’s Disease Neuroimaging Initiative; KBASE = Korean Brain Aging Study for the Early diagnosis and prediction of Alzheimer’s disease; FDG = fluorodeoxyglucose; PET = positron emission tomography; Aβ = β-amyloid
Classification basis for understanding model decision
Analyzing of the prediction values from the submodules derived the following nine ROIs from the external validation dataset: bilateral postcentral gyri, superior parietal gyri and precuneus, left superior frontal gyrus, middle frontal gyrus, and calcarine gyrus (Fig. 6b). The voxel-wise analysis of 2-[18F]FDG PET showed differences in the left inferior temporal, left middle temporal gyrus, right angular, right inferior parietal, right superior parietal, right middle occipital gyri, and right cuneus (Fig. 6a).
Result of voxel-wise analysis and analyzing prediction values from submodules in the external validation dataset. a Areas of higher glucose metabolism in Aβ-negative PET scan participants compared to Aβ-positive PET scan participants. The results are presented with a threshold of p < 0.001, uncorrected, and 50 voxels. b Analysis result of prediction values from submodules. Abbreviation: AAL = Automated Anatomical Labeling; Aβ = β-amyloid
Discussion
A sound understanding of amyloid pathology is critical for the accurate diagnosis and prognosis prediction in the Alzheimer’s disease continuum or in patients with dementia. The following two methods are currently available to examine amyloid pathology: amyloid PET and CSF analysis through lumbar puncture. However, amyloid PET is expensive and not universally available. Moreover, lumbar puncture is relatively invasive and poses potential side effects. Thus, the classification of amyloid pathology using long-established and widely distributed imaging techniques, such as MRI or 2-[18F]FDG PET, appears promising. Several studies have attempted to classify amyloid pathology by analyzing the cortical thickness from structural MRI (sMRI) [9, 10]. Nonetheless, sMRI has its limitations in early classification. This can be attributed to the occurrence of cortical thinning primarily at the late stage of the disease. Moreover, it does not provide any functional information. Glucose metabolism changes before a change in the cortical thickness in AD [37]. Hence, we assumed that 2-[18F]FDG PET would better classify amyloid pathology. However, the use of 2-[18F]FDG PET images for amyloid classification is rare. Thus, we developed a DL model that determines amyloid pathology from 2-[18F]FDG PET imaging data.
We found a lower glucose metabolism in the Aβ-PET-positive participants in the bilateral angular, supramarginal, precuneus, middle and posterior cingulate, inferior and middle temporal, parahippocampal, and fusiform gyri, consistent with previous reports (Fig. 4a) [4]. However, we recorded a higher glucose metabolism in the bilateral paracentral lobules and pre- and postcentral gyri in the Aβ-positive participants. According to Braak staging, the aforementioned regions are reportedly preserved during the progress of Aβ pathology [38]. These findings support the possibility of its classification power and form the basis of model interpretation.
Despite these possibilities, the datasets were extremely small for building a 3-D DL model. However, the model could extract more spatial relationships while considering the coronal and sagittal images, in addition to the axial images. Therefore, we designed a 2.5-D DL model for completely considering the 3-D spatial relationships. The CNN DL model can be usually explained using CAM for visualization. However, the last layer of each submodule gets replaced with a global average pooling layer during CAM. Moreover, the values from the submodules passed the fully connected layer for the final classification. This in turn leads to the disappearance of the location. Thus, the analysis would provide limited spatial information in 2.5-D. Therefore, we developed a 2.5-D method that analyzed the prediction values from the submodules to understand the basis of this decision.
We trained and internally validated the combined internal datasets. The internal validation results were impressive, with AUC values, accuracy, sensitivity, specificity, and F1 score of 0.811, 0.733, 0.678, 0.785, and 0.709, respectively. We encounter several diseases characterized by memory impairment in clinical practice. In contrast, the ANDI and KBASE datasets only contained the cognitive unimpaired and Alzheimer’s disease continuum. Thus, we externally validated our proposed model using a dataset, including various neurological diseases that cause memory impairment. The external validation dataset included clinically diagnosed AD, Lewy body dementia, epileptic cognitive disorder, normal pressure hydrocephalus, frontotemporal dementia, and vascular dementia. In addition, it included various clinical stages, such as one, 75, and 24 patients with cognitive unimpaired, MCI, and dementia, similar to the clinical situation. Moreover, the application of the proposed model to the external validation dataset also resulted in a high AUC value, accuracy, sensitivity, specificity, and F1 score of 0.798, 0.690, 0.768, 0.612, and 0.712, respectively. Furthermore, when we performed external validation with the model shown the best AUC value (0.864) on the internal validation dataset, the AUC value, accuracy, sensitivity, specificity, and F1 score was 0.860, 0.770, 0.800, 0.740, and 0.780, respectively. The performance of our model is comparable to that of the model based on longitudinal sMRI (AUC value was 0.86) [9]. However, it is better than that of the model which used patient demographics and sMRI (AUC value was 0.79) [10]. This enhanced performance could be attributed to the changes in glucose metabolism before cortical atrophy. Thus, our model that only used one-time 2-[18F]FDG PET images could be effectively used for classifying amyloid PET positivity in memory disorder clinics.
The performances by subgroups in the internal validation resulted in a high AUC of 0.717, 0.757, and 0.816 in the CU, MCI, and AD participants, respectively. Our model outperformed conventional models by producing comparatively better AUC in AD subgroup that indicated many significant differences within the group on voxel-wise analysis. Additionally, it resulted in a good AUC in CU subgroup that indicated a few significant differences within the group on voxel-wise analysis. Moreover, the application of the proposed model to the external validation dataset also resulted in a high AUC of 0.798 and 0.787 in the MCI and demented participants, respectively. These results were achieved because the external validation dataset contained many MCI and demented participants data. Additionally, the result of the internal validation in the Alzheimer’s Continuum was impressive, but the results for when this model was applied on the external validation dataset with various clinical diagnosis were impressive as well. This reflects the applicability in clinical practice.
When our model was trained and internally validated in the ADNI dataset and externally validated in the KBASE dataset, the model achieved the AUC values, accuracy, sensitivity, specificity, and F1 score were 0.769, 0.698, 0.702, 0.694, and 0.719, respectively. These results suggested that the performance was comparatively decent than when we used the ADNI and KBASE dataset for training and internal validation (Additional file 1: Fig. 4 and Additional file 1: Table 1). The reasons of the differences were suggested as follows: First, the ADNI and KBASE had different patients’ composition. Within the ADNI dataset, MCI participants were the most common, and 55% of the participants had amyloid PET-positive scan, while, within the KBASE, CU participants were the most common, and 34% of the participants had amyloid PET-positive scan. Second, the two datasets were obtained under different ethnicities, different amyloid tracers, and various PET manufacturers. Although we did spatial normalization with ethnicity specific template in the preprocessing pipeline, there may be remained differences. However, from the other perspective, the above results were also affirmative. The model with the best AUC value (0.800) in the internal validation showed good AUC values of 0.745 and 0.984 in MCI and AD, respectively, which also showed good AUC values of 0.837 and 0.721 in MCI and AD in the external validation, respectively. These results were encouraging since one could classify amyloid PET positivity status from the dataset obtained regardless of the ethnicities, tracers, and MRI manufacturers.
On internal and external validation, 10–15% of false negative and false positive cases were found. According to Alzheimer’s disease biomarker curve [39], amyloid deposition preceded the hypometabolism, which could explain the false negative cases. There are two speculation for comprehend false positive cases. First, approximately 10% of false negative cases are due to sensitivity of amyloid PET [40, 41]. Second, other cause of dementia, including Lewy body dementia, could be shown similar hypometabolism pattern [42].
The common region between the analysis of prediction value from the submodules and voxel-wise analysis included the right superior parietal gyrus in the external validation dataset (Fig. 6). However, we observed additional regions with different metabolism in the training and internal validation datasets that only included AD (Fig. 5). The above-mentioned difference could be attributed to the characteristics of the patient population, including only AD or other diseases. Therefore, our model might have extracted these hidden features and used them for the classification.
Our study had some limitations. First, we used several datasets that used different protocols and amyloid tracers for data collection. Moreover, these datasets included patients of various ethnicities. However, their characteristics suggest the general applicability of our model regardless of the aforementioned variabilities. Second, amyloid pathology could be detected by CSF Aβ42 or amyloid PET studies; most cases show concordant results in the Alzheimer’s disease continuum, whereas 6–21% show discordant results [43]. Both results should be considered for more accurate amyloid pathology; however, CSF Aβ42 was not available in all datasets. Especially in the ADNI dataset, only 66 participants underwent sMRI, 2-[18F]FDG PET, amyloid PET, and CSF Aβ42. We chose amyloid PET only because deep learning requires a large amount of data. Third, the DL framework had a black box limitation. Thus, the model visualization method, such as CAM, can further explain our classification model. We could not apply CAM because of the limitation of available datasets. In contrast, we used three axes images (2.5-D) as the inputs to improve the classification accuracy. The development of a 3-D DL model from larger datasets would facilitate the interpretation through CAM. Fourth, we suggested a novel method for the brief understanding of our model’s decision. After the t test between the amyloid PET-positive and PET-negative participants on SPSS, we obtained 20 slices in the sagittal plane, 27 slices in the coronal plane, and 24 slices in the coronal plane. We considered the (x, y, z) point as statistically significant when the ‘x’th, ‘y’th, and ‘z’th planes of the sagittal, coronal, and axial axes, respectively, showed substantial differences; a total of 12,960 (20×27×24) points were plotted in the MNI space. However, our method suffers from a limitation due to the following assumption. If the (x,y,z) point is significant, the ‘x’ th plane in the sagittal, ‘y’th plane in the coronal, and ‘z’th plane in the axial could be significant; however, the opposite was not assumed. Therefore, many unrelated points were plotted in the MNI space; ROIs clustered with more than 50 points were extracted to remove these unrelated points. Further investigation is required to accurately understand the model’s decision. Fifth, we did not include other biomarkers or neuropsychological test results. An incorporation of these features might have improved the performance. However, we tried to build a classification model by only using images, which can be applied more generally.
In conclusion, we proposed a DL model that can classify the amyloid PET positivity from 2-[18F]FDG PET imaging and demonstrated its high performance across an external test dataset. A large-scale external validation of multi-institutional data, model calibration, and optimization of sensitivity needs to be incorporated into the clinical workflow. The aforementioned model can serve as an important decision supporting tool to aid clinicians while classifying amyloid PET positivity.
Conclusion
The proposed model based on the 2-[18F]FDG PET imaging data and a DL framework might successfully classify amyloid PET positivity in clinical practice, without performing amyloid PET, which have limited accessibility.
Availability of data and materials
All imaging, and demographics data used in this article from ADNI are available and were downloaded from the ADNI website (adni.loni.usc.edu). Upon request, we will provide a list of ADNI participants for replication purposes. The data for this analysis are owned by the KBASE research group. Requests for data access can be submitted to the administrative coordinator of the group by e-mail (kbasecohort@gmail.com).
Code availability
Request for code access can be submitted to the author by e-mail (drshkim@kaist.ac.kr).
References
Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, et al. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–19. https://doi.org/10.1002/ana.20009.
Xia CF, Arteaga J, Chen G, Gangadharmath U, Gomez LF, Kasi D, et al. [F-18]T807, a novel tau positron emission tomography imaging agent for Alzheimer’s disease. Alzheimers Dement. 2013;9:666–76. https://doi.org/10.1016/j.jalz.2012.11.008.
Tamaoka A, Sawamura N, Fukushima T, Shoji S, Matsubara E, Shoji M, et al. Amyloid beta protein 42(43) in cerebrospinal fluid of patients with Alzheimer’s disease. J Neurol Sci. 1997;148:41–5. https://doi.org/10.1016/S0022-510x(96)00314-0.
Landau SM, Horng A, Fero A, Jagust WJ, Alzheimer's Disease Neuroimaging I. Amyloid negativity in patients with clinically diagnosed Alzheimer disease and MCI. Neurology. 2016;86:1377–85. doi:https://doi.org/10.1212/WNL.0000000000002576.
Ossenkoppele R, Jansen WJ, Rabinovici GD, Knol DL, van der Flier WM, van Berckel BNM, et al. Prevalence of amyloid PET positivity in dementia syndromes a meta-analysis. Jama-J Am Med Assoc. 2015;313:1939–49. https://doi.org/10.1001/jama.2015.4669.
Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Feldman HH, Frisoni GB, et al. A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. 2016;87:539–47. https://doi.org/10.1212/WNL.0000000000002923.
Jack CR, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–62. https://doi.org/10.1016/j.jalz.2018.02.018.
Kim EJ, Cho SS, Jeong Y, Park KC, Kang SJ, Kang E, et al. Glucose metabolism in early onset versus late onset Alzheimer’s disease: an SPM analysis of 120 patients. Brain. 2005;128:1790–801. https://doi.org/10.1093/brain/awh539.
Petrone PM, Casamitjana A, Falcon C, Artigues M, Operto G, Cacciaglia R, et al. Prediction of amyloid pathology in cognitively unimpaired individuals using voxel-wise analysis of longitudinal structural brain MRI. Alzheimers Res Ther. 2019;11:72. https://doi.org/10.1186/s13195-019-0526-8.
ten Kate M, Redolfi A, Peira E, Bos I, Vos SJ, Vandenberghe R, et al. MRI predictors of amyloid pathology: results from the EMIF-AD Multimodal biomarker discovery study. Alzheimers Res Ther. 2018;10. doi:https://doi.org/10.1186/s13195-018-0428-1.
Kim SE, Woo S, Kim SW, Chin J, Kim HJ, Lee BI, et al. A nomogram for predicting amyloid PET positivity in amnestic mild cognitive impairment. J Alzheimers Dis. 2018;66:681–91. https://doi.org/10.3233/Jad-180048.
Lee H, Yune S, Mansouri M, Kim M, Tajmir SH, Guerrier CE, et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng. 2019;3:173–82. https://doi.org/10.1038/s41551-018-0324-9.
Tang Z, Chuang KV, DeCarli C, Jin LW, Beckett L, Keiser MJ, et al. Interpretable classification of Alzheimer’s disease pathologies with a convolutional neural network pipeline. Nat Commun. 2019;10:2173. https://doi.org/10.1038/s41467-019-10212-1.
van der Burgh HK, Schmidt R, Westeneng HJ, de Reus MA, van den Berg LH, van den Heuvel MP. Deep learning predictions of survival based on MRI in amyotrophic lateral sclerosis. Neuroimage Clin. 2017;13:361–9. https://doi.org/10.1016/j.nicl.2016.10.008.
Lee G, Nho K, Kang B, Sohn KA, Kim D, Weiner MW, et al. Predicting Alzheimer's disease progression using multi-modal deep learning approach. Sci Rep-Uk. 2019;9. doi: https://doi.org/10.1038/s41598-018-37769-z.
Jiang H, Ma H, Qian W, Gao M, Li Y, Hongyang J, et al. An automatic detection system of lung nodule based on multigroup patch-based deep learning network. IEEE J Biomed Health Inform. 2018;22:1227–37. https://doi.org/10.1109/JBHI.2017.2725903.
Vente C, Vos P, Hosseinzadeh M, Pluim J, Veta M. Deep learning regression for prostate cancer detection and grading in bi-parametric MRI. IEEE Trans Biomed Eng. 2021;68:374–83. https://doi.org/10.1109/TBME.2020.2993528.
Henschel L, Conjeti S, Estrada S, Diers K, Fischl B, Reuter M. FastSurfer: a fast and accurate deep learning based neuroimaging pipeline. Neuroimage. 2020;219: 117012. https://doi.org/10.1016/j.neuroimage.2020.117012.
Tang X, Jafargholi Rangraz E, Coudyzer W, Bertels J, Robben D, Schramm G, et al. Whole liver segmentation based on deep learning and manual adjustment for clinical use in SIRT. Eur J Nucl Med Mol Imaging. 2020;47:2742–52. https://doi.org/10.1007/s00259-020-04800-3.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. https://doi.org/10.1038/nature14539.
Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, et al. A deep learning model to predict a Diagnosis of Alzheimer disease by using (18)F-FDG PET of the Brain. Radiology. 2019;290:456–64. https://doi.org/10.1148/radiol.2018180958.
Lee P, Kim HR, Jeong Y, Alzheimer's Disease Neuroimaging I. Detection of gray matter microstructural changes in Alzheimer's disease continuum using fiber orientation. BMC Neurol. 2020;20:362. doi:https://doi.org/10.1186/s12883-020-01939-2.
Iizuka T, Fukasawa M, Kameyama M. Deep-learning-based imaging-classification identified cingulate island sign in dementia with Lewy bodies. Sci Rep. 2019;9:8944. https://doi.org/10.1038/s41598-019-45415-5.
Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang SJ, et al. A New 2.5D Representation for lymph node detection using random sets of deep convolutional neural network observations. Lect Notes Comput Sc. 2014;8673:520–7.
Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, et al. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15(869–77):xi–xii. https://doi.org/10.1016/j.nic.2005.09.008.
Byun MS, Yi D, Lee JH, Choe YM, Sohn BK, Lee JY, et al. Korean brain aging study for the early diagnosis and prediction of Alzheimer’s disease: methodology and baseline sample characteristics. Psychiat Invest. 2017;14:851. https://doi.org/10.4306/pi.2017.14.6.851.
Nugent S, Croteau E, Potvin O, Castellano CA, Dieumegarde L, Cunnane SC, et al. Selection of the optimal intensity normalization region for FDG-PET studies of normal aging and Alzheimer’s disease. Sci Rep. 2020;10:9261. https://doi.org/10.1038/s41598-020-65957-3.
Joshi AD, Pontecorvo MJ, Clark CM, Carpenter AP, Jennings DL, Sadowsky CH, et al. Performance characteristics of amyloid PET with florbetapir F 18 in patients with alzheimer’s disease and cognitively normal subjects. J Nucl Med. 2012;53:378–84. https://doi.org/10.2967/jnumed.111.090340.
Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106:6820–5. https://doi.org/10.1073/pnas.0900345106.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15:273–89. https://doi.org/10.1006/nimg.2001.0978.
Lopresti BJ, Klunk WE, Mathis CA, Hoge JA, Ziolko SK, Lu XL, et al. Simplified quantification of Pittsburgh compound B amyloid imaging PET studies: a comparative analysis. J Nucl Med. 2005;46:1959–72.
Jack CR Jr, Wiste HJ, Weigand SD, Knopman DS, Lowe V, Vemuri P, et al. Amyloid-first and neurodegeneration-first profiles characterize incident amyloid PET positivity. Neurology. 2013;81:1732–40. https://doi.org/10.1212/01.wnl.0000435556.21319.e4.
Sabri O, Sabbagh MN, Seibyl J, Barthel H, Akatsu H, Ouchi Y, et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer’s disease: phase 3 study. Alzheimers Dement. 2015;11:964–74. https://doi.org/10.1016/j.jalz.2015.02.004.
He KM, Zhang XY, Ren SQ, Sun J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. IEEE I Conf Comp Vis. 2015:1026–34. doi:https://doi.org/10.1109/Iccv.2015.123.
Friston KJ. Commentary and opinion: II. Statistical parametric mapping: ontology and current issues. J Cereb Blood Flow Metab. 1995;15:361–70. doi:https://doi.org/10.1038/jcbfm.1995.45.
Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning Deep Features for Discriminative Localization. Proc Cvpr Ieee. 2016:2921–9. doi:https://doi.org/10.1109/Cvpr.2016.319.
Gordon BA, Blazey TM, Su Y, Hari-Raj A, Dincer A, Flores S, et al. Spatial patterns of neuroimaging biomarker change in individuals from families with autosomal dominant Alzheimer’s disease: a longitudinal study. Lancet Neurol. 2018;17:241–50. https://doi.org/10.1016/S1474-4422(18)30028-0.
Braak H, Alafuzoff I, Arzberger T, Kretzschmar H, Del Tredici K. Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. 2006;112:389–404. https://doi.org/10.1007/s00401-006-0127-z.
Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–16. https://doi.org/10.1016/S1474-4422(12)70291-0.
Clark CM, Pontecorvo MJ, Beach TG, Bedell BJ, Coleman RE, Doraiswamy PM, et al. Cerebral PET with florbetapir compared with neuropathology at autopsy for detection of neuritic amyloid-beta plaques: a prospective cohort study. Lancet Neurol. 2012;11:669–78. https://doi.org/10.1016/S1474-4422(12)70142-4.
Jack CR Jr, Barrio JR, Kepe V. Cerebral amyloid PET imaging in Alzheimer’s disease. Acta Neuropathol. 2013;126:643–57. https://doi.org/10.1007/s00401-013-1185-7.
Brown RK, Bohnen NI, Wong KK, Minoshima S, Frey KA. Brain PET in suspected dementia: patterns of altered FDG metabolism. Radiographics. 2014;34:684–701. https://doi.org/10.1148/rg.343135065.
Mattsson N, Insel PS, Donohue M, Landau S, Jagust WJ, Shaw LM, et al. Independent information from cerebrospinal fluid amyloid-beta and florbetapir imaging in Alzheimer’s disease. Brain. 2015;138:772–83. https://doi.org/10.1093/brain/awu367.
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report.
A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
The coinvestigators of the KBASE Research Group are listed elsewhere (http://kbase.kr).
Funding
This research was supported by the Bio & Medical Technology Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2016941946). This research was supported by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2016M3C7A1913844). This research was supported by the Ministry of Science, ICT, and Future Planning, Republic of Korea (NRF-2014M3C7A1046042). This research was supported by the Basic Research Lab (BRL) Program ( NRF-2020R1A4A1018714 ) funded by the Korea Government (MSIP) through the National Research Foundation (NRF).
Author information
Authors and Affiliations
Consortia
Contributions
Suhong Kim and Yong Jeong contributed to conceptualization and methodology; Kyeong Taek Oh, Min Soo Byun, Dahyun Yi, Jun Ho Lee, Yu Kyeong Kim, Byoung Seok Ye, Mi Jin Yun and Dong Young Lee performed data curation; Suhong Kim, Peter Lee, Kyeong Taek Oh, Yu Kyeong Kim and Mi Jin Yun carried out formal analysis; Yong Jeong, Dong Young Lee and Mi Jin Yun performed supervision and writing—review and editing; Suhong Kim done writing—original draft. All authors read and approved the final manuscripts.
Corresponding authors
Ethics declarations
Ethics approvals and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was approved by the Institutional Review Boards of the Seoul National University Hospital and the Severance Hospital. All participants provided their written informed consent.
Consent for publication
Informed consent was obtained from all individual participants included in the study.
Compering of interests
Suhong Kim, Kyeong Taek Oh, Min Soo Byun, Dahyun Yi, Jun Ho Lee, Yu Kyeong Kim, Byoung Seok Ye, Mi Jin Yun, Dong Young Lee, and Yong Jeong report no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Supplementary Fig. 1. Convolutional neural network architecture in 3 dimensions. (3-D) a) Custom ResNet with 15 layers and 4 residual layers. b) A structure that simply changed our 2.5-D architecture to 3-D. This consist of 4 convolution layers. Abbreviation: Conv = convolution layer; BN = batch normalization; ReLU = rectified linear unit; @ = number of channels; MaxPool = Max pooling layer; AvgPool = average pooling; FCN = fully connected layer. Supplementary Fig. 2. ROC curve and confusion matrix by the subgroups (CU, MCI, and AD participants) in the interval validation. (a, b) CU participants, (c, d) MCI participants and (e, f) AD participants. Abbreviation: ROC = receiver operating characteristic; AUC = area under curve, CU = cognitively unimpaired, MCI = mild cognitive impairment, AD = Alzheimer’s dementia. Supplementary Fig. 3. ROC curve and confusion matrix by the subgroups (MCI and demented participants) in the external validation. (a, b) MCI participants and (c, d) demented participants. Abbreviation: ROC = receiver operating characteristic; AUC = area under curve, MCI = mild cognitive impairment. Supplementary Fig. 4. (a-d) ROC curve and confusion matrix by all datasets and subgroups in the internal validation (part of the ADNI dataset). a) all participants, b) CU participants, c) MCI participants, and d) AD participants. (e-h) ROC curve and confusion matrix by all datasets and subgroups in the external validation (the KBASE dataset). e) all participants, f) CU participants, g) MCI participants, and h) AD participants. Abbreviation: ROC = receiver operating characteristic; AUC = area under curve, CU = cognitively unimpaired, MCI = mild cognitive impairment, AD = Alzheimer’s dementia. Supplementary table 1. Classification performance for Aβ PET positivity on ADNI and KBASE datasets.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, S., Lee, P., Oh, K.T. et al. Deep learning-based amyloid PET positivity classification model in the Alzheimer’s disease continuum by using 2-[18F]FDG PET. EJNMMI Res 11, 56 (2021). https://doi.org/10.1186/s13550-021-00798-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13550-021-00798-3