Abstract
Chemogenomics data generally refers to the activity data of chemical compounds on an array of protein targets and represents an important source of information for building in silico target prediction models. The increasing volume of chemogenomics data offers exciting opportunities to build models based on Big Data. Preparing a high quality data set is a vital step in realizing this goal and this work aims to compile such a comprehensive chemogenomics dataset. This dataset comprises over 70 million SAR data points from publicly available databases (PubChem and ChEMBL) including structure, target information and activity annotations. Our aspiration is to create a useful chemogenomics resource reflecting industry-scale data not only for building predictive models of in silico polypharmacology and off-target effects but also for the validation of cheminformatics approaches in general.
Similar content being viewed by others
Background
In pharmacology, “Big Data” on protein activity and gene expression perturbations has grown rapidly over the past decade thanks to the tremendous development of proteomics and genome sequencing technology [1, 2]. Similarly there has also been a remarkable increase in the amount of available compound structure and activity relation (SAR) data, contributed mainly by the development of high throughput screening (HTS) technologies and combinatorial chemistry for compound synthesis [3]. These SAR data points represent an important resource for chemogenomics modelling, a computational strategy in drug discovery that investigates an interaction of a large set of compounds (one or more libraries) against families of functionally related proteins [4].
Frequently, the “Big Data” in chemogenomics refers to large databases recording the bioactivity annotation of chemical compounds against different protein targets. Databases such as PubChem [5], BindingDB [6], and ChEMBL [7] are examples of large public domain repositories of this kind of information. PubChem is a well-known public repository for storing small molecules and their biological activity data [5, 8]. It was originally started as a central repository of HTS experiments for the National Institute of Health (USA) Molecular Libraries Program, but nowadays also incorporates data from other sources. ChEMBL contains data that was manually extracted from numerous peer reviewed journal articles, as do WOMBAT [9], BindingDB [6], and CARLSBAD [10]. Similarly, commercial databases, such as SciFinder [11], GOSTAR [12] and Reaxys [13] have accumulated a large amount of data from publications as well as patents. Besides these sources, large pharmaceutical companies maintain their own data collections originating from in-house HTS screening campaigns and drug discovery projects.
This data serves as a valuable source for building in silico models for predicting polypharmacology and off-target effects, and benchmarking the prediction performance and computation speed of machine-learning algorithms. The aforementioned publicly available databases have been widely used in numerous cheminformatics studies [14–16]. However, the curated data are quite heterogeneous [17] and lack a standard way for annotating biological endpoints, mode of action and target identifier. There is an urgent need to create an integrated data source with a standardized form for chemical structure, activity annotation and target identifier, covering as large a chemical and target space as possible. There are also irregularities within databases: the public screening data in PubChem, especially the inactive data points, are spread in different assay entries uploaded by data providers from around world and cannot be directly compared without processing. This makes curating SAR data for quantitative structure–activity relationship (QSAR) modeling very tedious. An example of work to synthesize the curated and uncurated data is Mervin et al. [15], where a dataset with ChEMBL active compounds and Pubchem inactive compounds was constructed, including inactive compounds for homologous proteins. However, the dataset can only be accessed as a plain text file, not as a searchable database.
In this work, by combining active and inactive compounds from both PubChem and ChEMBL, we created an integrated dataset for cheminformatics modeling purposes to be used in the ExCAPE [18] (Exascale Compound Activity Prediction Engine) Horizon 2020 project. ExCAPE-DB, a searchable open access database, was established for sharing the dataset. It will serve as a data hub for giving researchers around world easy access to a publicly available standardized chemogenomics dataset, with the data and accompanying software available under open licenses.
Dataset curation
The standardized ChEMBL20 data from an in-house database ChemistryConnect [3] was extracted and PubChem data was downloaded in January 2016 from the PubChem website (https://pubchem.ncbi.nlm.nih.gov/) using the REST API. Both data sources are heterogeneous. Data cleaning and standardisation procedures were applied in preparing both chemical structures and bioactivity data.
Chemical structure standardisation
Standardisation of PubChem and ChEMBL chemical structures was performed with ambitcli version 3.0.2. The ambitcli tool is part of the AMBIT cheminformatics platform [19–21] and relies on The Chemistry Development Kit library 1.5 [22, 23]. It includes a number of chemical structure processing options (fragment splitting, isotope removal, handling implicit hydrogens, stereochemistry, InChI [24] generation, SMILES [25] generation and structure transformation via SMIRKS [26], tautomer generation and neutralisation etc.). The details of the structure processing procedure can be found in Additional file 1. All standardisation rules were aligned between Janssen Pharmaceutica, AstraZeneca and IDEAConsult to reflect industry standards and implemented in open source software (https://doi.org/10.5281/zenodo.173560).
Bioactivity data standardisation
The processing protocol for extracting and standardizing bioactivity data is shown in Fig. 1. First, bioassays were restricted to only those comprising a single target; the black box (target unknown) or multi-target assays were excluded. 58,235 and 92,147 single targets containing concentration response (CR) type assays (confirmatory type in PubChem) remained in PubChem and ChEMBL, respectively. The assay target was further limited to human, rat and mouse species, and data points missing a compound identifier (CID) were removed. For those filtered assays, active compounds whose dose–response value was equal to or lower than 10 μM were kept as active entries and others were removed. Inactive compounds in CR assays were kept as inactive entries. Compounds that were labelled as inactive in PubChem screening assays (assays run with a single concentration) were also kept as inactive records.
The chemical structure identifiers (InChI, InChIKey and SMILES) generated from the standardized compound structures (as explained above) were joined with the compounds obtained after the filtering procedure.
The compound set was further filtered by the following physicochemical properties: organic filters (compounds without metal atoms), molecular weight (MW) <1000 Da, and a number of heavy atoms (HEV) >12. This was done to remove small or inorganic compounds not representative for modelling the chemical space relevant for a normal drug discovery project. This is a much more generous rule than the Lipinski rule-of-five [27], but the aim was to keep as much useful chemical information as possible while still removing some non-drug like compounds. Finally, fingerprint descriptors were generated for all remaining compounds. So far JCompoundMapper (JCM) [28], CDK circular fingerprint descriptors and signature descriptors [29] were generated respectively. For circular fingerprint and signature calculation, the maximum topological radius for fragment generation was set to 3.
From each data source, various attributes were read and converted into controlled vocabularies. The most important of these are target (Entrez ID), activity value, mode of action, assay type and assay technology etc. The underlying data sources contain activity data with various result types; the results were unified as best possible to make them comparable across tests (and data sources) irrespective of the original result type. The selected compatible dose–response result types are listed in Additional file 2: Table S1. Generally, the end-point name of a concentration related assay (e.g. IC50, units in µM) should match one of the keywords in this list. In the case when a compound has multiple activity data records for the same target, the records are aggregated so that one compound only has one record per target and the best (maximal) potency was chosen as the final aggregated value for a compound–target pair. The AMBIT generated InChIKey from the standardisation procedure was used as the molecular identifier to identify duplicate structures in the data aggregation. Finally, targets which have <20 active compounds were removed from the final dataset.
Entrez ID [30], gene symbol [31–33] and gene orthologue were collected as information for the target. The gene symbol was converted from Entrez ID with the gene2accession table [34] provided by National Center for Biotechnology Information (NCBI). Gene orthologues was included from the orthologue table [34] from NCBI.
Database and web interface
The ExCAPE-DB is built based on the AMBIT database and web application [19], enhanced with a free text search engine (Apache Solr [35]). An instance of the AMBIT web application (ambit2.war) was installed and the chemical structures were imported. This enables chemistry-aware search (similarity, substructure) and depiction, all exposed via a REST API and the web interface provided by the web application itself. The bioactivity data, consisting of compound related information (e.g. target activity label and InChIKey) and target related information (e.g. Entrez IDs and official gene symbols), is imported into an Apache Solr collection (http://lucene.apache.org/solr/) and exposed through the Solr REST API. The open source JavaScript client library jToxKit (https://github.com/ideaconsult/jToxKit) is used to interact with the AMBIT REST API and the Solr REST API. A dedicated JavaScript web interface was developed for ExCAPE-DB, integrating the chemical search, as well as the free text and faceted search functionality for biological activities.
The ExCAPE-DB is available online (https://solr.ideaconsult.net/search/excape/) and a screenshot of the web browser interface is shown in Fig. 2a. The dataset can be searched both by target name and CID. For target based searches, the Entrez ID, gene symbol, gene orthologous group and target species can be used for subsetting datasets. For compound searches, a user can choose to input the InChIKey or specify a CID (SMILES, InChI or IUPAC chemical name) for doing free-text search or use the embedded structure editor for doing substructure or similarity search (Fig. 2b). It is also possible to follow a link to the original ChEMBL or PubChem page of the specific compound from the search result. The download tab on the web page provides several download options. The “Filtered entries” download option allows the downloading of all of the current search result. For downloading specific entries, it is possible to include “Add to selection” links and compile a subset of selected entries, which will be available for download as “Selected entries”. A static link for downloading the entire ExCAPE-DB dataset is available at the download tab. The dataset is also uploaded to the Zenodo.org repository and available for download from there as doi:10.5281/zenodo.173258.
Discussion
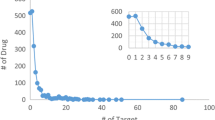
The dataset composition is described in Table 1. In total there are 998,131 unique compounds and 70,850,163 SAR data points. These SAR data points cover 1667 targets (Additional file 3: Table S2). It constitutes a curated large scale chemogenomics set freely available in the public domain under the Creative Commons Attribution Share-Alike 4.0 license. The dataset is useful for building QSAR models for predicting activity against one or more specific targets for novel compounds and will also serve as a benchmark dataset for evaluating the performance of various machine-learning algorithms, especially multi-target learning algorithms. The distribution of active compounds of ExCAPE-DB and ChEMBL themselves are shown in Fig. 3. Overall, most targets have far fewer inactive compounds than active compounds, which means that the chemogenomics dataset is highly imbalanced in both the ChEMBL and ExCAPE-DB datasets.
Composition of active compounds in the dataset. The distribution of active compounds among the targets in a ExCAPE-DB, b ChEMBL part of ExCAPE-DB and c the fraction span of actives in both datasets. We note that the ChEMBL dataset is shown here before the filtering and aggregation process and contains only single-target assays. Active compounds should have a pXC50 no less than 5 and only targets with at least 20 active compounds were considered
By adding inactive compounds from PubChem, the ExCAPE-DB has many more targets where the fraction of active compounds is <10% of the total number of compounds. Inclusion of inactive compounds from PubChem better mimics chemogenomics datasets available in the pharmaceutical industry, and it has been shown that inclusion of true inactive compounds results in better models than using random compounds as inactive compounds [15]. A low ratio between active and inactive compounds also reflects better the results of high-throughput screening where the hit rate is usually around 1%.
A clustering analysis was carried out for ChEMBL, PubChem and ExCAPE-DB compounds (as shown in Table 1) using an in-house program Flush [36] with a default Tanimoto similarity threshold of 0.7 that was calculated based on Foyfi fingerprints [37]. The distribution of cluster size for active compounds and inactive compounds is shown in Fig. 4. Here the singletons and small clusters whose size is <4 are excluded to give a better comparison. It can be seen that the cluster sizes of ChEMBL active and inactive compounds are very similar, while Pubchem active compounds tend to have a larger cluster size than the inactive compounds and hence they are less diverse than the inactive compounds. This is probably due to the fact that ChEMBL is composed of a series of analogue compounds, while the inactive compounds from screening campaigns in PubChem are more likely to be structurally diverse compounds. The SAR data is provided as is, but the underlying differences on structural diversity between active and inactive compounds should be considered when using ExCAPE-DB data for modelling.
The target class distribution across the dataset was also examined. The results are described in Fig. 5 for several major target families. The most common target class is enzymes followed by membrane receptors and then ion channels and transcription factors. The physicochemical property distribution of the dataset is shown in Fig. 6. Figure 6a–d are for MW, ClogP [38] representing calculated lipophilicity, polar surface area (PSA) which represent compounds polarity, and fraction of sp3 carbon atoms (Fsp3) in the compound which is a measure of the “flatness” of a compound [39], respectively. The MW of most compounds is between 220 and 540 Da. ClogP is mainly between 1 and 8. Most compounds have a PSA <150 and Fsp3 <0.7. In general, these distributions show that most compounds in the dataset fulfil the Lipinski rule-of-five [27] and are considered to be drug like compounds.
As an example of the utility of the generated dataset, 18 targets which have imbalance level varying from 1:10 to 1:1000 (ratio of active/inactive) were chosen for building support vector machine (SVM) models using LIBSVM [40]. Signature descriptors were used as input features. The performance of binary classification is given in Table 2 and model metrics shown are sensitivity, precision, specificity and Cohen’s κ value [41]. The results show that performance as expected varies from case to case and reasonable SVM models can be built even for some severely imbalanced datasets. This validates that the generated data set can be useful for predicting activity for novel compounds and for benchmarking studies.
Conclusion
ExCAPE-DB is a large public chemogenomics dataset based on the PubChem and ChEMBL databases, and large scale standardisation (including tautomerization) of chemical structures using open source cheminformatics software was performed in data curation. Comprehensive compound related information such as target activity label, fingerprint based descriptors and InChIKey, and target related information such as Entrez IDs and official gene symbols were collected and are easily accessible in the publicly available database. The active labels were determined based on their dose–response data to make sure the data quality is as high as possible. This ‘Big Data’ set covers large number of targets reported in the literature and can be used for building holistic multi-target QSAR models for target prediction. The data set will be used as a comprehensive benchmark set to evaluate the performance of various machine-learning algorithms in the ExCAPE project. To the best of our knowledge, this is first attempt to build such a large scale and searchable open access database for QSAR modelling.
References
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A et al (2015) Proteomics. Tissue-based map of the human proteome. Science 347:1260419
Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA et al (2013) The cancer genome atlas pan-cancer analysis project. Nat Genet 45:1113–1120
Muresan S, Petrov P, Southan C, Kjellberg MJ, Kogej T, Tyrchan C et al (2011) Making every SAR point count: the development of chemistry connect for the large-scale integration of structure and bioactivity data. Drug Discov Today 16:1019–1030
Bredel M, Jacoby E (2004) Chemogenomics: an emerging strategy for rapid target and drug discovery. Nat Rev Genet 5:262–275
Wang Y, Suzek T, Zhang J, Wang J, He S, Cheng T et al (2014) PubChem BioAssay: 2014 update. Nucleic Acids Res 42:D1075–D1082
Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J (2016) BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44:D1045–D1053
Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M et al (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res 42:D1083–D1090
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A et al (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213
Olah M, Rad R, Ostopovici L, Bora A, Hadaruga N, Hadaruga D et al (2007) WOMBAT and WOMBAT-PK: bioactivity databases for lead and drug discovery. In: Schreiber SL, Kapoor TM, Wess G (eds) Chemical biology: from small molecules to systems biology and drug design. Wiley-VCH, pp 760–786
Mathias SL, Hines-Kay J, Yang JJ, Zahoransky-Kohalmi G, Bologa CG, Ursu O et al (2013) The CARLSBAD database: a confederated database of chemical bioactivities. Database 2013:bat044
Williams J (1995) SCiFinder: information at the desktop for scientists. Online. ETATS-UNIS, Wilton, CT, pp 60–66
GOSTAR database release 2016. http://www.gostardb.com/. Accessed 1 Oct 2016
Reaxys database. http://www.reaxys.com. Accessed 1 Oct 2016
Lusci A, Browning M, Fooshee D, Swamidass J, Baldi P (2015) Accurate and efficient target prediction using a potency-sensitive influence-relevance voter. J Cheminform 7:63
Mervin LH, Afzal AM, Drakakis G, Lewis R, Engkvist O, Bender A (2015) Target prediction utilising negative bioactivity data covering large chemical space. J Cheminform 7:51
Helal KY, Maciejewski M, Gregori-Puigjane E, Glick M, Wassermann AM (2016) Public domain HTS fingerprints: design and evaluation of compound bioactivity profiles from PubChem’s bioassay repository. J Chem Inf Model 56:390–398
Fourches D, Muratov E, Tropsha A (2015) Curation of chemogenomics data. Nat Chem Biol 11:535
ExCAPE project website. http://www.excape-h2020.eu. Accessed 1 Oct 2016
Jeliazkova N, Jeliazkov V (2011) AMBIT RESTful web services: an implementation of the OpenTox application programming interface. J Cheminform 3:18
Kochev NT, Paskaleva VH, Jeliazkova N (2013) Ambit-Tautomer: an open source tool for tautomer generation. Mol Inform 32:481–504
Jeliazkova N, Kochev N (2011) AMBIT-SMARTS: efficient searching of chemical structures and fragments. Mol Inform 30:707–720
Steinbeck C, Han Y, Kuhn S, Horlacher O, Luttmann E, Willighagen E (2003) The chemistry development kit (CDK): an open-source java library for chemo- and bio-informatics. J Chem Inf Comput Sci 43:493–500
Steinbeck C, Hoppe C, Kuhn S, Floris M, Guha R, Willighagen EL (2006) Recent developments of the chemistry development kit (CDK)—an open-source java library for chemo- and bio-informatics. Curr Pharm Des 12:2111–2120
Heller SR, Mcnaught A, Pletnev I, Stein S, Tchekhovskoi D (2015) InChI, the IUPAC international chemical identifier. J Cheminform 7:23
Weininger D (1988) SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J Chem Inf Comput Sci 28:31–36
SMIRKS web site. http://www.daylight.com/dayhtml/doc/theory/theory.smirks.html. Accessed 1 Oct 2016
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Hinselmann G, Rosenbaum L, Jahn A, Fechner N, Zell A (2011) jCompoundMapper: an open source java library and command-line tool for chemical fingerprints. J Cheminform 3:3
Carbonell P, Carlsson L, Faulon J-L (2013) Stereo signature molecular descriptor. J Chem Inf Model 53:887–897
Maglott D, Ostell J, Pruitt KD, Tatusova T (2005) Entrez gene: gene-centered information at NCBI. Nucleic Acids Res 33:D54–D58
Gray KA, Yates B, Seal RL, Wright MW, Bruford EA (2015) Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 43:D1079–D1085
Shimoyama M, De Pons J, Hayman GT, Laulederkind SJ, Liu W, Nigam R et al (2015) The rat genome database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res 43:D743–D750
Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Mouse Genome Database Group (2015) The mouse genome database (MGD): facilitating mouse as a model for human biology and disease. Nucleic Acids Res 43:D726–D736
NCBI Gene. https://www.ncbi.nlm.nih.gov/gene. Accessed 12 Jan 2016
Apache Solr. https://lucene.apache.org/solr. Accessed 1 Oct 2016
Flush program. https://github.com/OpenEye-Contrib/Flush. Accessed 1 Oct 2016
Blomberg N, Cosgrove DA, Kenny PW, Kolmodin K (2009) Design of compound libraries for fragment screening. J Comput Aided Mol Des 23:513–525
ClogP version 4.3. http://www.biobyte.com/. Accessed 1 Apr 2016
Lovering F, Bikker J, Humblet C (2009) Escape from flatland: increasing saturation as an approach to improving clinical success. J Med Chem 52:6752–6756
Chang C-C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2:1–27
Cohen J (1960) A coefficient of agreement for nominal scales. Educ Psychol Meas 20:37–46
Authors’ contributions
JS performed data collection, clean and analysis. NJ, VJ performed the chemical structure standardisation and setup the database. IG customized the web interface. JS and HC drafted manuscript. HC, NJ and VC aligned the chemical standardisation rules. NK updated the standardisation software and implemented the agreed rules. JS and JG calculated the chemical descriptors. JS, HC, NJ, VC and TA contributed data interpretation. All authors contributed writing and/or editing the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank all ExCAPE partners for helpful discussions on data curation.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Interactive access as well as download links of user selected subsets or the entire dataset are available at https://solr.ideaconsult.net/search/excape/. The whole dataset is also available at https://zenodo.org/record/173258.
Funding
This project has received funding from the European Union’s Horizon 2020 Research and Innovation programme under Grant Agreement No. 671555.
Author information
Authors and Affiliations
Corresponding authors
Additional information
The original version of this article was revised. After publication of this work [1] it was noticed that the author’s name was spelt incorrectly. His name should be spelt as: Vladamir Chupakhin. The ‘h’ was missing from his surname.
An erratum to this article is available at http://dx.doi.org/10.1186/s13321-017-0222-2.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Sun, J., Jeliazkova, N., Chupakhin, V. et al. ExCAPE-DB: an integrated large scale dataset facilitating Big Data analysis in chemogenomics. J Cheminform 9, 17 (2017). https://doi.org/10.1186/s13321-017-0203-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13321-017-0203-5