Abstract
Background
Satellite cells, a population of unipotent stem cells attached to muscle fibers, determine the excellent regenerative capability of injured skeletal muscles. Myogenic potential is also exhibited by other cell populations, which exist in the skeletal muscles or come from other niches. Mesenchymal stromal/stem cells inhabiting the bone marrow do not spontaneously differentiate into muscle cells, but there is some evidence that they are capable to follow the myogenic program and/or fuse with myoblasts.
Methods
In the present study we analyzed whether IGF-1, IL-4, IL-6, and SDF-1 could impact human and porcine bone marrow-derived mesenchymal stromal/stem cells (hBM-MSCs and pBM-MSCs) and induce expression of myogenic regulatory factors, skeletal muscle-specific structural, and adhesion proteins. Moreover, we investigated whether these factors could induce both types of BM-MSCs to fuse with myoblasts. IGF-1, IL-4, IL-6, and SDF-1 were selected on the basis of their role in embryonic myogenesis as well as skeletal muscle regeneration.
Results
We found that hBM-MSCs and pBM-MSCs cultured in vitro in the presence of IGF-1, IL-4, IL-6, or SDF-1 did not upregulate myogenic regulatory factors. Consequently, we confirmed the lack of their naïve myogenic potential. However, we noticed that IL-4 and IL-6 impacted proliferation and IL-4, IL-6, and SDF-1 improved migration of hBM-MSCs. IL-4 treatment resulted in the significant increase in the level of mRNA encoding CD9, NCAM, VCAM, and m-cadherin, i.e., proteins engaged in cell fusion during myotube formation. Additionally, the CD9 expression level was also driven by IGF-1 treatment. Furthermore, the pre-treatment of hBM-MSCs either with IGF-1, IL-4, or SDF-1 and treatment of pBM-MSCs either with IGF-1 or IL-4 increased the efficacy of hybrid myotube formation between these cells and C2C12 myoblasts.
Conclusions
To conclude, our study revealed that treatment with IGF-1, IL-4, IL-6, or SDF-1 affects BM-MSC interaction with myoblasts; however, it does not directly promote myogenic differentiation of these cells.
Similar content being viewed by others
Background
Satellite cells are skeletal muscle-specific unipotent stem cells, retained in a quiescent state, and characterized by the expression of PAX7 transcription factor. Their role in skeletal muscle regeneration is well characterized [1,2,3]. As a result of muscle injury, the numerous cytokines and growth factors are released by damaged muscle fibers, as well as endothelial cells, fibroadipogenic progenitors (FAP), fibroblasts, and inflammatory cells [4,5,6]. Some of these factors such as hepatocyte growth factor (HGF), insulin-like growth factor-1 (IGF-1), fibroblast growth factor-2 (FGF-2), and tumor necrosis factor-α (TNF-α) activate signaling pathways controlling cell cycle re-entry of satellite cells and enable their activation [2, 7]. As a result, satellite cells start proliferation and either generate myogenic progenitors/myoblasts, which undergo further differentiation, or return to the quiescent state and renew the satellite cell pool. The proliferation and differentiation of satellite cells and myoblasts are regulated inter alia by myogenic regulatory factors (MRFs) such as MYOD1, MYF5, myogenin (MYOG), and MFR4 (also known as MYF6) [8]. The activated satellite cells express both MYOD1, which induces the expression of genes encoding factors regulating cell cycle and MYOG, which mediates cell cycle exit and expression of genes necessary for differentiation into post-mitotic myocytes expressing MRF4. Then, in regenerating muscle, the fusion of myocytes leads to myotube and myofiber formation. Many intrinsic and extrinsic factors regulate the differentiation of myoblasts participating in skeletal muscle regeneration. In the present study we focused on the impact of IGF-1, interleukin-4 (IL-4), interleukin-6 (IL-6), or stromal derived factor-1 (SDF-1) on human and porcine bone marrow-derived mesenchymal stromal/stem cell (hBM-MSCs and pBM-MSCs) differentiation and fusion with myoblasts. These factors were selected on the basis of their role in embryonic myogenesis and skeletal muscle regeneration.
IGF-1, by interacting with its receptor—IGF-1R, activates at least two pathways involved in determination of cell fate, i.e., ERK1/ERK2 MAPK (extracellular signal-regulated protein kinases 1/extracellular signal-regulated protein kinases 2 mitogen-activated protein kinase) pathway which promotes cell proliferation [9] and p38 MAPK pathway which stimulates satellite cell differentiation [10,11,12]. IGF-1 can also induce phosphoinositide-3-kinase (PI3K)-dependent pathways, like PI3K/AKT (protein kinase B), PI3K/AKT/mTOR (mammalian target of rapamycin kinase), or PI3K/AKT/GSK3 (glycogen synthase kinase 3). Among the effects of PI3K/AKT action is the induction of the expression of MEF2 and MRFs, such as MYOD1 and MYOG [13]. Next, it was shown that stimulation of PI3K/AKT/GSK3 and PI3K/AKT/mTOR pathways by IGF-1 induces myotube hypertrophy by phosphorylation of downstream targets, such as p70S6 kinase, 4E-BP1, or eIF2, which are directly involved in the regulation of translation [14, 15]. The effect of IGF-1 was also tested in mdx mice. IGF-1 overexpression in mice skeletal muscles resulted in the reduction of myofiber atrophy, necrosis, and fibrosis [16, 17]. IGF-1 not only impacts myogenesis per se but also enhances the recruitment of stem cells from the bone marrow to the sites of muscle injury [18].
The next factor selected by us, i.e., IL-4, is a pleiotropic cytokine first described as a B cell stimulatory factor [19]. It also modulates the activity of other cell types, i.e., T cells and mast cells [20, 21]. The action of IL-4 can be transduced by two types of receptors: type I consisting of the IL-4Rα and γC subunits—expressed by hematopoietic cells, and type II consisting of the IL-4Rα and IL-13Rα1 subunits—expressed by non-hematopoietic cells, including myogenic cells, i.e., myoblasts, both in mouse and human [22]. In 2003, IL-4 was described as the myogenesis regulator engaged in recruiting mononuclear myoblasts to the newly formed myotubes and enabling their growth. Mice lacking IL-4 or IL-4Rα were characterized by a decreased number of nuclei present in myofibers as well as an increased proportion of smaller myofibers and a decreased proportion of larger ones [23]. Next, IL-4 was shown to promote migration of myogenic cells both in vitro and in vivo, i.e., during muscle regeneration, by increasing Itagb1 and Itagb3 expression [24]. IL-4 was also shown to play an important role in muscle growth during postnatal development. Mice lacking serum response factor (SRF), a transcription factor regulating expression of different muscle-specific genes such as muscle creatinine kinase and dystrophin, were characterized by strong downregulation of Il-4 expression and—as a consequence—impaired recruitment of myoblasts to myofibers, retarded postnatal muscle growth, and decreased muscle mass [25]. IL-4 possibly influences the expression of proteins localized on myogenic cell surface, similarly as it was described for smooth muscles [26], lymphocytes B [27], fibroblasts [28], and macrophages [29], but the precise mechanism of IL-4 action in myogenic cells is not known yet.
IL-6 is another pleiotropic cytokine classified both as pro- and anti-inflammatory protein. It is produced by many cell types, such as activated macrophages, vascular endothelial cells, and fibroblasts [30, 31] and locally in skeletal muscles where its level is related to the glycogen level, i.e., when the level of glycogen decreases the IL-6 production increases [32]. IL-6 is also secreted in vitro by human primary myoblasts and mouse C2C12 myoblasts [33, 34] and promotes their differentiation. Silencing of Il-6 gene expression in myoblasts leads to the downregulation of muscle-specific genes—α-actin and Myog [35]. Next, mice lacking IL-6 were characterized by defective muscle growth resulting from impaired proliferation and migration of satellite cells which indicates the role of this cytokine in satellite cell-mediated hypertrophy [36]. Thus, IL-6 is a significant mediator of both cell proliferation and myogenic differentiation and through these effects plays an important role in skeletal muscle growth and regeneration.
The last factor selected by us—stromal derived factor-1 (SDF-1, also known as CXCL12)—is a CXC chemokine, which binds to CXCR4 or CXCR7 receptors or CXCR4/CXCR7 heterodimers [37,38,39]. SDF-1 binding to CXCR4 leads to a number of cellular events, such as inhibition of adenylate cyclase, PI3K, and Rho pathways as well as ERK1/ERK2 MAPK and p38 MAPK independently of G-proteins [40]. The SDF-1 expression increases in injured tissues as it was shown for skeletal muscles [41,42,43] where SDF-1 is mainly produced by damaged myofibers [43]. We previously observed that SDF-1 treated muscles regenerated more efficiently due to CXCR4+ and CD34+ cell mobilization [42, 44]. Unquestionably, SDF-1 promotes the migration of myoblasts [42, 45,46,47,48] by inducing changes in actin organization via activation of FAK (focal adhesion kinase), CDC42 (cell division control protein 42), and RAC-1 (Ras-related C3 botulinum toxin substrate 1) [47]. Importantly, SDF-1 treatment of myoblasts also increased the expression of the tetraspanin CD9 that plays a crucial role in cell migration and fusion [49] as well as led to MYHC accumulation and increased myotube formation [50]. Interestingly, the inhibition of Myog and MYHC expression was also reported for SDF-1-treated myoblasts [43], which stays in contrast to our results showing no significant impact of SDF-1 on neither myoblast proliferation nor MRF expression nor fusion [42].
On the basis of the abovementioned information, we hypothesized that factors involved in the regulation of myogenesis and skeletal muscle regeneration could impact the myogenic identity, migration, and fusion of human and porcine BM-MSCs with mouse myoblasts. It was shown previously that hBM-MSCs characterized by CD146 expression are not able to undergo spontaneous myogenic differentiation [51]; thus, these cells do not present naïve myogenic potential. However, it was also documented that in the presence of exogenous myoblasts, BM-MSCs can fuse with them, although with low efficiency [52]. Moreover, modification of BM-MSCs by Pax3, β-catenin, or NICD (Notch intracellular domain) overexpression can reprogram them to start myogenic differentiation and fuse to form myotubes [53,54,55,56]. In our previous study, we showed that interactions of bone marrow isolated stem cells with satellite cell niche can enhance their myogenic identity [57]. Thus, in the current study, we analyzed the influence of IGF-1, IL-4, IL-6, or SDF-1 on the BM-MSC proliferation, migration, myogenic potential, and fusion in vitro.
Methods
BM-MSC culture
Human BM-MSC commercial cell line was obtained from Lonza (Lonza, PT-2501). Porcine BM-MSCs were isolated from transgenic porcine embryos which constitutively expressed green fluorescent protein (GFP). Both human and porcine BM-MSCs were cultured in high-glucose DMEM (Dulbecco’s modified Eagle’s medium, Invitrogen) supplemented with 15% FBS (fetal bovine serum, Invitrogen) and 10 μg/ml gentamycin (Sigma Aldrich) in 5% CO2 at 37 °C. Culture medium was replaced every 2 days, and cells were passaged after reaching confluency. For further analysis, cells from passages 4–9 were used. Depending on the type of analysis, cells were collected and frozen (for further RNA isolation) or cultured on cover slides covered with a 1% gelatin solution and fixed with a 3% PFA (for protein immunolocalization).
BM-MSC treatment or pre-treatment with IL-4, IL-6, IGF-1, or SDF-1
Human and porcine BM-MSCs were plated on 60-mm wells at a density of 3 × 103 cells per 1 cm2 and cultured in high-glucose DMEM supplemented with 15% FBS and 10 μg/ml gentamycin in 5% CO2 at 37 °C for 3 days. At the fourth day of culture, the so-called proliferating medium, i.e., high-glucose DMEM containing 15% FBS and 10 μg/ml gentamycin (PM), or differentiating medium, i.e., high-glucose DMEM containing 10% FBS, 10% horse serum, and 10 μg/ml gentamycin (DM) supplemented with 50 ng/ml IGF-1 or IL-4 or IL-6 or SDF-1 [recombinant human IGF-1 protein ab155614 (Abcam); recombinant human IL-4 protein ab83686 (Abcam); recombinant human IL-6 protein ab119444 (Abcam); recombinant human SDF-1 alpha protein ab73461 (Abcam)], was added to cells. From that moment, PM or DM containing selected factors was replaced every day, for the next 7 days. Such protocol allowed to keep the constant concentration of analyzed factors. To follow the number of cells in culture, they were counted every day from days 1 to 7. The cells were washed with PBS, trypsinized, suspended in a culture medium, and counted in a hemocytometer. Cells treated for 7 days were also collected and frozen in − 80 °C or fixed with 3% PFA for further analyses. In another set of experiments, BM-MSCs were transiently treated with selected factors. Thus, BM-MSCs were cultured for 3 days in control PM and then the medium was changed to PM supplemented with 50 ng/ml of either IGF-1 or IL-4 or IL-6 or SDF-1 for 3 days. Next, the medium was changed to control PM and pre-treated hBM-MSCs were analyzed (including cell counting) after 1, 3, and 7 days of culture.
Co-culture of human and porcine BM-MSCs and C2C12 myoblasts
C2C12 myoblasts, human BM-MSCs, and porcine BM-MSCs were separately cultured in high-glucose DMEM supplemented with 15% FBS and 10 μg/ml gentamycin in 5% CO2 at 37 °C. Then, each type of cells was trypsinized, washed, and suspended in the medium. Next, 25 × 103 of BM-MSCs and 25 × 103 of C2C12 myoblasts were plated together and co-cultured in 2 ml of high-glucose DMEM supplemented with 10 μg/ml gentamycin and 15% FBS (PM) or 10% FBS and 10% HS (DM). For the cell treatments, PM and DM were supplemented with 50 ng/ml IGF-1, IL-4, IL-6, or SDF-1. From that moment, culture medium (PM or DM) containing one of the factors was replaced every day for the next 7 days which allowed to keep the required concentration of analyzed factors. After 7 days of treatment, cells were collected and frozen or fixed with 3% PFA for further RNA analyses or immunostaining. In pre-treated co-cultures, the BM-MSCs were cultured in the presence of 50 ng/ml IGF-1, IL-4, IL-6, or SDF-1 in PM for 3 days. Then, the 25 × 103 of pre-treated BM-MSCs and 25 × 103 of C2C12 myoblasts were plated together and co-cultured in 2 ml of high-glucose DMEM supplemented with 15% FBS and 10 μg/ml gentamycin (PM) for 1, 3, and 7 days when they were collected and frozen or fixed with 3% PFA for further RNA analyses or immunostaining.
The cell migration assay
The 1 × 104 hBM-MSCs were plated in the culture dish (35 mm) and cultured until they reached 90% of confluency. Linear scratches were made in cell sheets using a 200μl pipette tip. The culture medium was replaced with a fresh control culture medium or the one containing either SDF-1, IGF-1, IL-4, or IL-6 in concentrations of 10, 50, and 100 ng/ml. The scratch area was photographed directly after the scratch and after 6 h and 12 h. Next, the area of the scratch was measured for control and each experimental group. Results were presented as relative mobility of treated cells compared to control, i.e., untreated cells and represent four independent experiments with three technical repeats each.
Quantitative reverse transcriptase real-time PCR
RNA was isolated from human and porcine BM-MSCs, mouse C2C12 myoblasts, and co-cultures of C2C12 myoblasts with either hBM-MSCs or pBM-MSCs using High Pure Isolation Kit (Roche Applied Science). Three independent cell cultures were used per each experiment). The cDNA was obtained using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) in accordance with the manufacturers’ protocols. Then, using TaqMan® Gene Expression Master Mix and TaqMan assays (Thermo Fisher Scientific), the quantitative real-time PCR was performed. Hypoxanthine phosphoribosyltransferase 1 (HPRT1) was used as the reference gene. All reactions were performed in technical duplicates. The conditions of RT-qPCR were as follows: reverse transcription: 25 °C for 10 min, 42 °C for 60 min, 85 °C for 5 min; qPCR: 50 °C for 2 min, template denaturation 95 °C for 10 min, 45 cycles of 95 °C for 15 s and 60 °C for 60 s. Threshold-cycle (Ct) values of the analyzed amplicons were determined with LightCycler®480 Software (Roche Applied Science). Expression levels were calculated with the 2-(ΔΔCt) formula using a relative quantification tool in LightCycler® 480 Software [58]. The samples were compared to non-treated cells (NT). The Ct over 32 cycles were not analyzed. TaqMan assays: hADAM9 (Hs00177638_m1); mADAM9 (Mm00475770_m1), hCD9 (Hs00233521_m1); hCDH15 (m-cadherin) (Hs00170504_m1); hCXCR4 (Hs00607978_s1), hCXCR7 (Hs00664172_s1), hDES (desmin) (Hs00157258_m1); hHPRT1 (Hs02800695_m1); mHPRT1 (Mm03024075_m1); hIGF-1 (Hs01651089_g1); hIGFR (Hs00609566_m1); hIL-4 (Hs00598625_m1); hIL-4R (Hs00965056_m1); hIL-6 (Hs00174131_m1); hIL-6R (Hs01075664_m1); hMYF5 (Hs00929416_g1); hMYOD1 (Hs00159528_m1); hMYOG (myogenin) (Hs01072232_m1); mMYOG (myogenin) (Mm00446194_m1); hMYH3 (MyHC3) (Hs01074230_m1); mMYH3 (MyHC3) (Mm01332463_m1); hNCAM1 (Hs00941830_m1); hSDF-1 (Hs00664172_s1); hVCAM (Hs01003372_m1).
Microarray analysis
Human BM-MSCs were cultured in high-glucose DMEM supplemented with 15% FBS and 10 μg/ml gentamycin and non-treated or treated with IL-4 (50 ng/ml) for 7 days. Total RNA was isolated using the mirVana Isolation Kit (Thermo Fisher Scientific). Next, its integrity was checked with 2100 Bioanalyzer (Agilent Technologies) using RNA 6000 NAno LAb Chip kit (Agilent Technologies). All RNA samples had integrity number above 8.5. One hundred nanograms of total RNA for each sample was biotin labeled with the TargetAmpTM-Nano Labeling Kit for Illumina Expression BeadChip (Epicentre Biotechnologies). Labeled RNA was purified with RNeasy MinElute Cleanup Kit (Qiagen) and hybridized onto MouseRef-8 v2.0 Expression BeadChip (illumina) according to the manufacturer’s instructions. Arrays were scanned with a HiScanSQ System (illumina). Raw data were imported to GenomeStudio (illumina), and the average signal intensities were analyzed in Partek Genomic Suite (Partek, Inc.) v. 6.6 after quantile normalization and Log2 transformation. Qualitative analysis was performed, e.g., principal component analysis, in order to identify outliers and artifacts on the microarray. After the quality check, the two-way ANOVA (analysis of variance) model by using Method of Moments was performed on the data and lists of significantly and differentially expressed genes between biological variants [with the cutoff values: p value < 0.05, (1.3 ≥ fold change ≥ 1.33)] were created. Fisher’s least significant difference (LSD) was used as the contrast method to compare non-treated hBM-MSCs vs treated with IL-4 hBM-MSCs. Unsupervised hierarchical clustering was performed on the selected lists in order to find genes and samples with similar profiles. Gene networks were created by interposing the results onto the database of Ingenuity containing information about gene functions with the use of Ingenuity Pathway Analysis tool.
Protein immunolocalization
Human and porcine BM-MSCs, mouse C2C12 myoblasts, and co-cultures of BM-MSCs and C2C12 myoblasts were fixed with 3% PFA in PBS, washed in PBS, permeabilized by incubation in 0.05% Triton X100 in PBS, washed in PBS, then incubated in 0.15% glycine in PBS, and blocked in 3% BSA in PBS. Then, cells were incubated overnight with primary antibodies diluted 1:100 in 3% BSA at 4 °C. Next, cells were washed with PBS and incubated with fluorochrome-conjugated secondary antibodies diluted 1:200 in 3% BSA in PBS for 2 h at room temperature. After washing in PBS, cells were incubated with DRAQ5 (Biostatus) diluted 1:1000 in PBS for 5 min, washed in PBS, and mounted with Dako Cytomation Fluorescent Mounting Medium. Fluorescence was analyzed using confocal microscopy (LSM 500, Zeiss) and ZEN application (Zeiss). Antibodies used were: anti-human nuclear antigen antibody [235-1] (Abcam); anti-myosin (Skeletal), antibody produced in rabbit (Sigma-Aldrich); donkey anti-mouse IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 488 (Thermo Fisher Scientific Scientific); and donkey anti-rabbit IgG (H+L) highly cross-adsorbed secondary antibody, Alexa Fluor 594 (Thermo Fisher Scientific Scientific).
Giemsa–May–Grünwald staining
Co-cultures of hBM-MSCs or pBM-MSCs and C2C12 myoblasts were fixed with cold methanol (− 20 °C). Then, cells were incubated with May–Grünwald and Giemsa dyes according to the manufacturer’s protocol (Merck). The fusion index of cells was calculated as the percentage of nuclei in myotubes to all nuclei visible in the field. Three independent experiments were performed.
Statistical analysis
Results were analyzed using GraphPad Software. The mean values and standard deviations were presented, and the non-paired t test was performed to compare treated cells with control cells. Differences were considered statistically significant when p < 0.05 (marked on charts with asterisks).
Results
Expression of IGF-1, IL-4, IL-6, and SDF-1 and their receptors in human BM-MSCs
In the present study we analyzed human BM-MSC cell line (further described as hBM-MSCs) according to the experimental scheme involving treatment with various factors (Fig. 1a). The cells were cultured in the presence of 50 ng/ml of either IGF-1, IL-4, IL-6, or SDF-1 for 7 days in either the so-called proliferating medium (PM) or differentiating medium (DM). Control cells consisted of non-treated hBM-MSCs. Cells analyzed at day 7 of culture were characterized by the expression of standard mesenchymal stromal cell markers, such as CD73, CD90, and CD105 (Fig. 1b) [59]. Moreover, we confirmed that these cells expressed CD146 that was recently described as a marker of human bone marrow-derived mesenchymal stem cells (Fig. 1b) [51]. hBM-MSCs analyzed by us also expressed both SDF-1 receptors, i.e., CXCR4 and CXCR7 (Fig. 1b), but the relative expression level of CXCR4 mRNA was extremely low (data not shown).
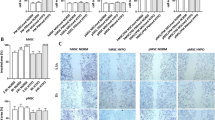
Expression of IGF-1, IL-4, IL-6, and SDF-1 and their receptors in human BM-MSCs (hBM-MSCs) non-treated (NT) and treated with either IGF-1, IL-4, IL-6, or SDF-1 cultured in the proliferating medium (PM) or differentiating medium (DM) - qRT-PCR analysis. a The experimental design. b Expression of mesenchymal stromal/stem cell markers in non-treated (NT) hBM-MSCs cultured in PM or DM for 7 days (n = 3). c Expression of cytokines and their receptors in hBM-MSCs non-treated (NT) or 7 days treated with either IGF-1 or IL-4 or IL-6 or SDF-1 cultured in PM (n = 3). d Expression of cytokines and their receptors in hBM-MSCs non-treated (NT) or 7 days treated with either IGF-1 or IL-4 or IL-6 or SDF-1 cultured in DM (n = 3); *p < 0.05, **p < 0.01, ***p < 0.005
Next, we analyzed the expression of mRNAs encoding endogenous growth factors and cytokines (IGF-1, IL-4, IL-6, and SDF-1) as well as their receptors in non-treated, i.e., control, and continuously treated cells (Fig. 1c, d). hBM-MSCs cultured in PM or DM expressed IGF-1, IL-4, IL-6, SDF-1, and IGF-1 receptor (IGF-1R), IL-4 receptor (IL-4Rα), IL-6 receptor (IL-6R), and SDF-1 receptor (CXCR4; Fig. 1c, d). Next, we compared the level of all analyzed growth factors and cytokines between non-treated cells and those ones treated with IGF-1, IL-4, IL-6, or SDF-1 and cultured in PM or DM (Fig. 1c, d). IL-4 treatment increased the expression of IL-4 and IL-6 in hBM-MSCs cultured in DM while in cells cultured in PM it increased the expression of IL-4Rα but decreased the expression of IGF-1. IL-6 increased IL-4, IL-6, and CXCR4 expression in hBM-MSCs cultured in DM. Thus, IL-4 and IL-6 changed the expression of endogenous IGF-1, IL-4, IL-6, IL-4Rα, and CXCR4 in treated hBM-MSCs but these changes were dependent of culture conditions, i.e., the presence of PM or DM.
The proliferation and migration of human BM-MSCs in the presence of IGF-1, IL-4, IL-6, and SDF-1
To follow the influence of IGF-1, IL-4, IL-6, and SDF-1 on hBM-MSC proliferation, we analyzed the cell number each day during a 7-day long culture. We examined hBM-MSCs cultured according to two experimental schemes (Fig. 2a). The first set of experiments focused on hBM-MSCs treated with selected factors continuously for 7 days and then analyzed after 7 days of such culture in PM or DM. The second set of experiments focused on hBM-MSCs which were for 3 days pre-treated with the studied factors and then cultured in the proliferating medium (PM) and analyzed after 1, 3, and 7 days (Fig. 2a). In the case of continuously treated cells, their number increased regardless of the medium type used (Fig. 2b, c). After 7 days of the culture, the number of hBM-MSCs treated with IL-4 and IL-6 was significantly lower while cells treated with SDF-1 higher when compared to non-treated cells, cultured in either PM or DM (Fig. 2b, c). In the case of pre-treated cells, no significant differences were found after 7 days of the culture (Fig. 2d).
Proliferation and migration of human BM-MSCs (hBM-MSCs) treated or pre-treated with IGF-1, IL-4, IL-6, or SDF-1. a The experimental design. b The number of hBM-MSCs non-treated (NT) or treated with either IGF-1, IL-4, IL-6, or SDF-1 and cultured in the proliferating medium (PM) for 7 days (n = 3). c The number of hBM-MSCs non-treated (NT) or treated with either IGF-1, IL-4, IL-6, or SDF-1 and cultured in the differentiating medium (DM) for 7 days (n = 3). d The number of hBM-MSCs pre-treated with either IGF-1, IL-4, IL-6, or SDF-1 for 3 days and cultured in the proliferating medium (PM), analyzed after 1, 3, and 7 days (n = 3). e The migration of hBM-MSCs non-treated (NT) and treated either with IGF-1, IL-4, IL-6, or SDF-1 cultured in the proliferating medium (PM) and analyzed after 6 h and 12 h (n = 3). *p < 0.05, **p < 0.01, ***p < 0.005
Next, we performed a scratch test to assess the hBM-MSC migration. To this point, we exposed hBM-MSCs to IGF-1, IL-4, IL-6, and SDF-1 for 6 h and 12 h. SDF-1, IL-4, and IL-6 used in the concentration of 10 ng/μl significantly increased the cell migration after 6 h as well as after 12 h compared to other tested concentrations, i.e., 50 ng/μl and 100 ng/μl (Fig. 2e). IGF-1 significantly impacted hBM-MSC migration only after 6 h (Fig. 2e). Thus, IL-4 and IL-6 significantly reduced hBM-MSC proliferation but induced their migration. Moreover, SDF-1 induced migration of the analyzed cells.
The impact of IGF-1, IL-4, IL-6, or SDF-1 treatment on MRFs, adhesion, and structural protein expression in human BM-MSCs
In the next step, we analyzed if factors selected by us influence the expression of MRFs, as well as adhesion and structural protein characteristic for cells undergoing myogenic differentiation in treated hBM-MSCs (Fig. 3). First, we analyzed hBM-MSCs cultured in either PM or DM in the constant presence of IGF-1, IL-4, IL-6, or SDF-1 for 7 days (experimental scheme on Fig. 2a) but did not detect expression of MYF5, MYOD, and MYOG (myogenin), regardless of culture conditions (data not shown). Since MYOD, MYF5, and myogenin are involved in the specification of the muscle cell lineage [8], we concluded that hBM-MSCs were not able to follow myogenic program in response to the treatment applied by us. Then, we followed the expression of mRNAs encoding adhesion proteins that are engaged in cell adhesion, migration, and myoblast fusion, i.e., ADAM9 (disintegrin and metalloproteinase), tetraspanin CD9, NCAM (neural cell adhesion molecule), VCAM (vascular cell adhesion molecule), m-cadherin (CDH15), and structural muscle proteins, i.e., desmin and muscle embryonic myosin heavy chains 3 (MYH3). IGF-1 treatment led to a CD9 mRNA level increase in hBM-MSCs cultured in PM (Fig. 3b) while IL-4 significantly increased expression of CD9, NCAM, VCAM, and CDH15 in hBM-MSCs cultured in DM (Fig. 3b).
The IGF-1, IL-4, IL-6, or SDF-1 impact on adhesion and structural protein expression in human BM-MSCs (hBM-MSCs). a The expression of ADAM9, CD9, NCAM, VCAM, CDH15 (m-cahderin), desmin, and MYH3 (muscle embryonic myosin heavy chain 3) in hBM-MSCs non-treated (NT) or treated with either IGF-1, IL-4, IL-6, or SDF-1, cultured for 7 days in the proliferating medium (PM) (n = 3). b The expression of mRNAs encoding ADAM9, CD9, NCAM, VCAM, m-cadherin (CDH15), desmin, and MYH3 in hBM-MSCs non-treated (NT) or treated with either IGF-1, IL-4, IL-6, or SDF-1, cultured for 7 days in the differentiating medium (DM) (n = 3). c The expression of ADAM9, CD9, and CDH15 in hBM-MSCs non-treated (NT) or pre-treated with either IGF-1, IL-4, IL-6, or SDF-1 for 3 days, then cultured in the proliferating medium (PM) and analyzed after 1, 3, and 7 days (n = 3); *p < 0.05, **p < 0.01, ***p < 0.005
Next, we studied the expression of MRFs, ADAM9, CD9, and CDH15 in pre-treated cells cultured in PM (experimental scheme on Fig. 2a). Similarly to continuously treated hBM-MSCs, pre-treated ones did not express mRNAs encoding MRFs; however, IGF-1 pre-treatment led to CD9 and CDH15 mRNA increase which was noticed already 1 day after the end of the pre-treatment and was observed also at days 3 and 7 (Fig. 3c). The other pre-treated hBM-MSCs did not present any significant changes in adhesion protein expression (Fig. 3c).
Then, we analyzed treated and pre-treated hBM-MSCs that were co-cultured with mouse C2C12 myoblasts in PM or DM. In the first set of the experiments, hBM-MSCs and C2C12 myoblasts were co-cultured in the constant presence of IGF-1, IL-4, IL-6, or SDF-1 (Fig. 4a). In the second set of the experiments, hBM-MSCs were first pre-treated with the abovementioned factors and then the co-culture with C2C12 myoblasts was established and conducted in PM (Fig. 4a). The use of mouse myoblasts allowed us to distinguish human and mouse transcripts. Again, we did not notice any significant changes in the MRF mRNA level, regardless of the treatment and co-culture conditions (data not shown). However, we observed the increase of human ADAM9 expression level in the co-cultures treated with IL-4 and cultured in PM and ADAM9, CDH15, and VCAM upregulation in the co-cultures treated with IL-4 in DM (Fig. 4b, c). The level of mouse transcripts was also analyzed to verify if human IGF-1, IL-4, IL-6, or SDF-1 impacted the expression of mRNAs encoding MRFs and structural proteins in C2C12 myoblasts. Except changes in the Adam9 level which increased in the co-cultures in PM continuously treated with IL-4, we did not detect any significant differences (Fig. 4d, e). In the co-cultures of pre-treated hBM-MSCs and C2C12 myoblasts, any significant changes in adhesion protein mRNA level between non-treated and pre-treated co-cultures were found (Fig. 4f).
The IGF-1, IL-4, IL-6, or SDF-1 impact on MRF and structural protein expression in co-cultures of human BM-MSCs (hBM-MSCs) and mouse C2C12 myoblasts. a The experiment design. b The expression of human ADAM9, CD9, CDH15 (m-cadherin), VCAM, and MYH3 (muscle embryonic myosin heavy chain 3) in hBM-MSCs non-treated (NT) and treated either with IGF-1, IL-4, IL-6, or SDF-1 co-cultured with C2C12 for 7 days in the proliferating medium (PM) (n = 3). c The expression of human ADAM9, CD9, CDH15 (m-cadherin), VCAM, and MYH3 (muscle embryonic myosin heavy chain 3) in hBM-MSCs non-treated (NT) or treated with either IGF-1, IL-4, IL-6, or SDF-1 co-cultured with C2C12 myoblasts for 7 days in the differentiating medium (DM) (n = 3). d The expression of mouse Adam9, Myog, and Myh3 mRNA in C2C12 myoblasts non-treated (NT) and treated with either IGF-1, IL-4, IL-6, or SDF-1 co-cultured with hBM-MSCs for 7 days in the proliferating medium (PM) (n = 3). e The expression of mouse Adam9, Myog, and Myh3 mRNA in C2C12 myoblasts non-treated (NT) and treated with either IGF-1, IL-4, IL-6, or SDF-1 co-cultured with hBM-MSCs for 7 days in the differentiating medium (DM) (n = 3). f The expression of human ADAM9, CD9, and CDH15 (m-cadherin) in hBM-MSCs pre-treated for 3 days with either IGF-1, IL-4, IL-6, or SDF-1 and then co-cultured with C2C12 myoblasts in the proliferating medium (PM) for 7 days (n = 3); *p < 0.05, **p < 0.01, ***p < 0.005
Formation of hybrid myotubes between human or porcine BM-MSCs with C2C12 myoblasts
Our experiments showed that IL-4 or IGF-1 treatment increased expression of mRNAs encoding adhesion proteins in hBM-MSCs. Moreover, IL-4 and IL-6 influenced hBM-MSC proliferation, while SDF-1 enhanced their migration. In the next step, we tested if observed changes translated to the ability of human and porcine BM-MSCs to fuse with myoblasts. We decided to analyze porcine BM-MSCs (pBM-MSCs), in addition to hBM-MSCs, to verify if these mechanisms were species-specific. Moreover, pig already serves as an animal model in numerous studies but the understanding of porcine cell biology is limited. To this point, we analyzed co-cultures of hBM-MSCs or pBM-MSCs with mouse C2C12 myoblasts and presented the fusion index, as well as proportion of hybrid myotubes formed by human or porcine BM-MSCs and mouse C2C12 myoblasts (Fig. 5, Additional file 1: Figure S1). The use of human or porcine and mouse cells allowed to distinguish the hybrid myotubes in co-cultures. The human cells were detected on the basis of human nuclear antigen expression; porcine cells were GFP positive. Finally, myotubes were identified by myosin expression. The C2C12 cells are the ones typically used in studies involving hybrid myotube formation [51, 60]. The co-cultures were treated, i.e., cultured in constant presence with IGF-1, IL-4, IL-6, or SDF-1, or the hBM-MSCs were first pre-treated for 3 days with selected factors and then co-cultured with myoblasts in the absence of these factors (experimental scheme on Fig. 4a). Hybrid myotubes were present in both control and co-cultures treated with either IGF-1, IL-4, IL-6, or SDF-1 in PM or DM, but no significant effect of treatment was found (Fig. 5a, b, e). Similarly, the fusion index did not differ between control and continuously treated co-cultures (Fig. 5b). However, the proportion of hybrid myotubes as well fusion index was higher in the co-cultures of hBM-MSCs pre-treated with selected factors and C2C12 myoblasts, as compared to control ones (Fig. 5c). The pBM-MSCs were also able to form hybrid myotubes with C2C12 myoblasts; however, any of tested factors did not significantly impact the frequency of this process but IGF-1 and IL-4 slightly increased index of fusion in these co-cultures (Fig. 5d). Thus, the pre-treatment of hBM-MSCs with IGF-1, IL-4, or SDF-1 increased the effectivity of hybrid myotube formation by their fusion with C2C12 myoblasts.
The fusion of non-treated (NT) and IGF-1-, IL-4-, IL-6-, or SDF-1-treated or pre-treated human and pig BM-MSCs (hBM-MSCs and pBM-MSCs) with C2C12 myoblasts. a The percentage of hybrid myotubes in co-culture of treated hBM-MSCs and C12C12 myoblasts in the proliferating medium (PM) for 7 days (n = 3). b The percentage of hybrid myotubes and the fusion index in co-culture of treated hBM-MSCs and C12C12 myoblasts in the differentiating medium (DM) for 7 days (n = 3). c The percentage of hybrid myotubes and fusion index in co-culture of hBM-MSCs and C2C12 myoblasts pre-treated for 3 days in control (without selected factor supplementation) proliferating medium (PM) for 7 days (n = 3). d The percentage of hybrid myotubes and fusion index in co-culture of non-treated (NT) or treated pBM-MSCs and C2C12 myoblasts in the differentiating medium (DM) after 7 days (n = 3). e hBM-MSC localization in myotubes in co-cultures with C2C12 myoblasts after 7 days of IGF-1, IL-4, IL-6, or SDF-1 treatment in PM and DM. Scale bar 50 μm. Blue—cell nuclei, red—skeletal myosin, green—human cell nuclei. *p < 0.05, **p < 0.01, ***p < 0.005
IL-4 treatment modifies the transcriptome of human BM-MSCs
Important changes in gene expression in hBM-MSC treated with IL-4 were observed. Thus, we decided to compare the global transcriptome of control and IL-4 treated hBM-MSCs. Cells were treated for 7 days with IL-4 in PM, and then RNA was isolated and subjected to microarray analysis. Analysis of variance (ANOVA) allowed to create the lists of genes significantly upregulated or downregulated in IL-4-treated hBM-MSCs (with the cutoff values: p value < 0.05, − 1.3 ≥ fold change ≥ 1.3). This analysis showed that IL-4 treatment regulates the expression of 61 transcripts (Fig. 6). Using Ingenuity Pathway Analysis, we showed that IL-4 impacts the expression of many genes encoding proteins engaged in cell adhesion and migration such as VCAM (as showed also by qRT-PCR analysis), k-cadherin (cadherin-6, CDH6), and extracellular proteins such as collagen XIV (COL14A1; Fig. 6). The increase was also noticed in the transcript level for chemokine (C-C motif) ligand 26 and 11 (CL26, CCL11) and cytokine IL-6. Also, transcripts encoding signaling involved proteins were upregulated, among them were JAG1, i.e., membrane protein that interacts with Notch receptors, and IGFBP, i.e., insulin-like growth factor-binding protein. Summarizing, global transcriptome analysis confirmed the important role of IL-4 in the activation of adhesion and migration as well as regulation of inflammatory process and signaling in hBM-MSCs.
The changes in human BM-MSC (hBM-MSCs) transcriptome after IL-4 treatment. Human BM-MSCs were cultured for 48 h in the presence of IL-4 and then analyzed. a The changes in the level of transcripts between IL-4-treated and non-treated hBM-MSCs. b Gene networks created by interposing the results onto database of Ingenuity containing information about the gene function with the use of Ingenuity Pathway Analysis tool (red—upregulation, green—downregulation)
Discussion
Bone marrow mesenchymal stromal cells (BM-MSCs) were first identified in mouse by Friedenstein [61, 62] and described as multipotent progenitors on the basis of their ability to proliferate, form cell colonies, and generate bone, cartilage, and adipocytes after heterotopic transplantation in vivo [63,64,65,66,67,68,69]. Then, the presence of multipotent stem cells in the population of human BM-MSCs was also proved. These cells expressed CD146, were perivascular located, and able to differentiate into the bone, cartilage, and bone marrow stroma [51, 70, 71]. Moreover, human CD146+ stem cells, which were isolated from heterotopic ossicles formed after subcutaneous transplantation of BM-MSCs and then cultured at clonal density, were still able to generate colonies and formed next ossicles after serial transplantation [70, 71]. Thus, human CD146+ cells isolated from BM-MSCs fulfill the criteria of multipotent stem cells, i.e., they are able to self-renew and differentiate into several cell types. Cells with similar features present in mouse bone marrow were also described on the basis of nestin expression [72].
In our experiments, we used BM-MSC cell line derived from adult human bone marrow isolated as a result of bilateral punctures of the posterior iliac crests of healthy volunteers. These cells were characterized by expression of mRNAs encoding CD73, CD90, CD105, CD146, CXCR7, and at a very low level also CXCR4 [73, 74]. Furthermore, we showed that hBM-MSCs expressed IGF-1R, IL-4Rα, and IL-6R as well as the receptors for SDF-1, i.e., CXCR4 and CXCR7. The expression of CXCR4 by BM-MSCs is discussable as the proportion of BM-MSCs found to express CXCR4 oscillates between 0 and 27% for mouse cells and 0 and 96% for human cells [75]. It was also suggested that CXCR4 expression decreased during in vitro culture and passages of BM-MSCs [76, 77]. We showed that BM-MSCs produced also mRNAs encoding endogenous cytokines such as IL-4, IL-6, SDF-1, and IGF-1. Interestingly, the expression of transcripts for IL-4Rα as well as IL-4 and IL-6 increased in IL-4-treated BM-MSCs. This corresponds to previously published data on mouse T and B cells [78].
Human BM-MSCs characterized by CD146 expression are not able to undergo spontaneous myogenic differentiation [51]; thus, these cells do not present naïve myogenic potential. Nevertheless, it was revealed that human, rat, and mouse BM-MSCs can fuse, with low efficiency, with myoblasts [52, 79]. Additionally, modification of BM-MSCs by Pax3, β-catenin, or NICD (Notch intracellular domain) overexpression can reprogram them to start myogenic differentiation and fuse with myoblasts [53,54,55,56, 79]. Recently, we showed that satellite cell niche could induce myogenic identity of BM-MSCs. Since cytokines and growth factors are an important component of the niche, we hypothesized that factors such as IGF-1, IL-4, IL-6, and SDF-1, which are involved in the regulation of myogenesis and muscle regeneration, could impact the myogenic identity, proliferation, migration, and fusion of hBM-MSCs with myoblasts.
We showed that IL-4 and IL-6 limited proliferation but promoted migration of BM-MSCs. The SDF-1 also increased migration of BM-MSCs while IGF-1 was not a strong chemoattractant for these cells. This was in agreement with the study showing that SDF-1 stimulates migration of rat BM-MSCs but IGF-1 does not [80]. To follow the impact of selected factors on the ability of BM-MSCs to form myotubes, we co-cultured them with C2C12 myoblasts. Both human and porcine BM-MSCs were able to fuse with myoblasts, however, with low efficiency. We found that pre-treatment of human BM-MSCs with selected factors, such as IGF-1, IL-4, or SDF-1, boosts their fusion with myoblasts, while adding these factors during co-culture of both cell types has no significant effect on this process. It is a very important finding as such pre-treatment is a feasible procedure, which could be used to prime BM-MSCs prior to their transplantation.
Thus, the pre-treatment of human BM-MSCs and their further co-culture with myoblasts provided better signals increasing the hybrid myotube formation than continuous treatment of analyzed co-cultures. The analysis of changes in the expression of adhesion proteins involved in cell fusion showed that numerous factors, i.e., timing of treatment with IGF-1, IL-4, IL-6, or SDF-1, and also the type of culture medium, as well as the presence or absence of myoblasts, impact the level of mRNAs encoding adhesion proteins. IL-4 treatment and culture of human BM-MSCs in the differentiating medium increased CD9, NCAM, VCAM, and CDH15 mRNA expression. For cells cultured in the proliferating medium, this effect was observed only for VCAM mRNA. Thus, the IL-4 treatment led to VCAM mRNA upregulation independently of the type of the culture medium while CD9, NCAM, and CDH15 mRNA upregulation required additional signals provided by the differentiating medium, i.e., the presence of horse serum. The increase of VCAM and CDH15 expression was also noticed in the co-cultures of hBM-MSCs with myoblasts treated with IL-4 and cultured in the differentiating medium. However, these changes were not translated to the increase of hybrid myotube formation. Moreover, the presence of myoblasts and IL-4 treatment led to ADAM9 mRNA upregulation in case of hBM-MSCs in both proliferating and differentiating media. Interestingly, the increase of Adam9 mRNA level was also detected in C2C12 myoblasts treated with IL-4. In addition, IGF-1 treatment or pre-treatment was sufficient to increase the level of CD9 mRNA in hBM-MSCs (cultured separately, without myoblasts) but only in cells cultured in the proliferating medium. As mentioned above IGF-1 could induce the pathways leading to myogenic differentiation [81] and enhances regeneration of injured muscles [82]. The analysis of human placental mesenchymal stromal cells showed that IGF-1 treatment and culture of these cells in differentiation medium decreased MYOD1 expression level and did not change MYOG and MYHC level. The authors suggested that IGF-1 treatment maintains progenitor cell phenotype of mesenchymal stromal cells cultured under differentiating conditions [83]. It was also revealed that IGF-1 treatment of rat BM-MSCs did not impact at the expression level of MEF2, myogenin, alpha-sarcomeric actinin 2 (ACTN2), and desmin; however, the three-dimensional culture of rat BM-MSCs and primary myoblasts enhances their myogenic capacity [84].
Differences observed by us in the expression of mRNAs encoding adhesion proteins were translated to the ability of pre-treated hBM-MSCs to fuse with myoblasts. The cells pre-treated with IGF-1, IL-4, and SDF-1 fused more efficiently with myoblasts. This phenomenon could be connected inter alia with CD9 and CDH15 mRNA upregulation in IGF-1 pre-treated hBM-MSCs or increase in IL4Rα, VCAM, or NCAM expression in hBM-MSCs after incubation with IL-4. The improvement of hybrid myotube formation in the co-cultures of SDF-1 pre-treated hBM-MSCs with C2C12 myoblasts could be associated with their higher migration abilities, which we observed also in mouse BM-MSC and C2C12 myoblast co-cultures [49]. An important role of SDF-1 and its receptors in BM-MSC migration is well documented [47, 85,86,87]. Previously, we showed that SDF-1 altered actin organization via FAK, CDC42, and RAC-1 activation [47] and promoted mesenchymal stem cell, myoblast, and embryonic stem cell migration. However, treatment of hBM-MSCs with analyzed factors during co-culture with myoblasts did not result in the increase of hybrid myotube formation. This indicates that the upregulation of mRNAs encoding adhesion proteins should be induced in hBM-MSCs at early stages of cell differentiation and fusion since further treatment of cells does not increase their ability to fuse with myoblasts.
Conclusions
Selected factors impacted BM-MSC proliferation (IL-4 and IL-6) and improved migration (IL-4, IL-6, and SDF-1). IL-4 and IL-6 changed the expression of endogenous IGF-1, IL-4, IL-6, IL-4Rα, and CXCR4 in hBM-MSCs. IL-4 treatment resulted in the significant expression increase of CD9, NCAM, VCAM, and CDH15 (m-cadherin), i.e., factors engaged in cell fusion during myotube formation. On the other hand, IGF-1 treatment led to CD9 mRNA upregulation. However, hBM-MSCs were not able to follow myogenic program in response to any treatment applied by us. Nevertheless, the pre-treatment of hBM-MSCs with IGF-1, IL-4, or SDF-1 increased their effectivity to participate in hybrid myotube formation. Moreover, IL-4 treatment influenced the expression of cytokines that could result in inflammatory modification. Our study revealed that IGF-1, IL-4, IL-6, and SDF-1 could be important factors affecting BM-MSC adhesion, migration, and fusion; however, they could not initiate myogenic program in these cells.
Availability of data and materials
Department of Cytology, Faculty of Biology, University of Warsaw, Miecznikowa 1 St, 02-096 Warsaw, Poland
Abbreviations
- ADAM9:
-
A disintegrin and metalloproteinase domain-containing protein 9
- ALS:
-
Amytropic lateral sclerosis
- BAD:
-
BCL2-associated death promoter
- BM-MSC:
-
Bone marrow-derived mesenchymal stromal/stem cell
- CD9:
-
Tetraspanin CD9
- CDC42:
-
Cell division control protein 42
- CDH15:
-
M-cadherin
- CXCR4:
-
C-X-C chemokine receptor type 4 (SDF-1 receptor)
- CXCR7:
-
C-X-C chemokine receptor type 7 (SDF-1 receptor)
- DM:
-
Differentiating medium
- ERK1:
-
Extracellular signal-regulated protein kinases 1
- ERK2:
-
Extracellular signal-regulated protein kinases 2
- FAK:
-
Focal adhesion kinase
- FAP:
-
Fibroadipogenic progenitors
- FBS:
-
Fetal bovine serum
- FGF-2:
-
Fibroblast growth factor-2
- GSK3:
-
Glycogen synthase kinase 3
- HGF:
-
Hepatocyte growth factor
- HPRT1:
-
Hypoxanthine phosphoribosyltransferase 1
- HS:
-
Horse serum
- IGF-1:
-
Insulin-like growth factor-1
- IGF-1R:
-
Insulin-like growth factor-1 receptor
- IL-4:
-
Interleukin-4
- IL-4Rα:
-
Interleukin-4 receptor alpha
- IL-6:
-
Interleukin-6
- IL-6R:
-
Interleukin-6 receptor
- MAPK:
-
Mitogen-activated protein kinase
- MEF2:
-
Myocyte enhancer factor 2
- MRF:
-
Myogenic regulatory factor
- MuRF1:
-
Muscle RING-finger protein-1
- MYH3:
-
Muscle embryonic myosin heavy chain 3
- MYHC:
-
Myosin heavy chain
- MYOG:
-
Myogenin
- NCAM:
-
Neural cell adhesion molecule
- NICD:
-
Notch intracellular domain
- PI3K:
-
Phosphoinositide-3-kinase
- PM:
-
Proliferating medium
- RAC-1:
-
Ras-related C3 botulinum toxin substrate 1
- SDF-1:
-
Stromal derived factor-1
- TNF-α:
-
Tumor necrosis factor-α
- VCAM:
-
Vascular cell adhesion protein
References
Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139(16):2845–56 Epub 2012/07/27.
Dumont NA, Wang YX, Rudnicki MA. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development. 2015;142(9):1572–81 Epub 2015/04/30.
Costamagna D, Berardi E, Ceccarelli G, Sampaolesi M. Adult stem cells and skeletal muscle regeneration. Curr Gene Ther. 2015;15(4):348–63 Epub 2015/07/01.
Saclier M, Cuvellier S, Magnan M, Mounier R, Chazaud B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013;280(17):4118–30 Epub 2013/02/07.
Christov C, Chretien F, Abou-Khalil R, Bassez G, Vallet G, Authier FJ, et al. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol Biol Cell. 2007;18(4):1397–409 Epub 2007/02/09.
Brzoska E, Ciemerych MA, Przewozniak M, Zimowska M. Regulation of muscle stem cells activation: the role of growth factors and extracellular matrix. Vitam Horm. 2011;87:239–76 Epub 2011/12/01.
Ciemerych MA, Archacka K, Grabowska I, Przewozniak M. Cell cycle regulation during proliferation and differentiation of mammalian muscle precursor cells. In: Kubiak JZ, editor. Cell cycle in development, results and problems in cell differentiation. Berlin Heidelberg: Springer-Verlag; 2011. p. 498–507.
Zammit PS. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. 2017;72:19–32 Epub 2017/11/12.
Duan C, Ren H, Gao S. Insulin-like growth factors (IGFs), IGF receptors, and IGF-binding proteins: roles in skeletal muscle growth and differentiation. Gen Comp Endocrinol. 2010;167(3):344–51 Epub 2010/04/21.
Girardi F, Le Grand F. Wnt signaling in skeletal muscle development and regeneration. Prog Mol Biol Transl Sci. 2018;153:157–79 Epub 2018/02/02.
Palacios D, Mozzetta C, Consalvi S, Caretti G, Saccone V, Proserpio V, et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7(4):455–69 Epub 2010/10/05.
Lluis F, Ballestar E, Suelves M, Esteller M, Munoz-Canoves P. E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J. 2005;24(5):974–84 Epub 2005/02/19.
Mourkioti F, Rosenthal N. IGF-1, inflammation and stem cells: interactions during muscle regeneration. Trends Immunol. 2005;26(10):535–42 Epub 2005/08/20.
Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3(11):1014–9 Epub 2001/11/21.
Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI (3) K/Akt/mTOR and PI (3) K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3(11):1009–13 Epub 2001/11/21.
Barton ER, Morris L, Musaro A, Rosenthal N, Sweeney HL. Muscle-specific expression of insulin-like growth factor I counters muscle decline in mdx mice. J Cell Biol. 2002;157(1):137–48 Epub 2002/04/03.
Shavlakadze T, White J, Hoh JF, Rosenthal N, Grounds MD. Targeted expression of insulin-like growth factor-I reduces early myofiber necrosis in dystrophic mdx mice. Mol Ther. 2004;10(5):829–43 Epub 2004/10/29.
Musaro A, Giacinti C, Borsellino G, Dobrowolny G, Pelosi L, Cairns L, et al. Stem cell-mediated muscle regeneration is enhanced by local isoform of insulin-like growth factor 1. Proc Natl Acad Sci U S A. 2004;101(5):1206–10 Epub 2004/01/28.
Howard M, Farrar J, Hilfiker M, Johnson B, Takatsu K, Hamaoka T, et al. Identification of a T cell-derived b cell growth factor distinct from interleukin 2. J Exp Med. 1982;155(3):914–23 Epub 1982/03/01.
Mosmann TR, Bond MW, Coffman RL, Ohara J, Paul WE. T-cell and mast cell lines respond to B-cell stimulatory factor 1. Proc Natl Acad Sci U S A. 1986;83(15):5654–8 Epub 1986/08/01.
Fernandez-Botran R, Krammer PH, Diamantstein T, Uhr JW, Vitetta ES. B cell-stimulatory factor 1 (BSF-1) promotes growth of helper T cell lines. J Exp Med. 1986;164(2):580–93 Epub 1986/08/01.
Paul WE. History of interleukin-4. Cytokine. 2015;75(1):3–7 Epub 2015/03/31.
Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113(4):483–94.
Lafreniere JF, Mills P, Bouchentouf M, Tremblay JP. Interleukin-4 improves the migration of human myogenic precursor cells in vitro and in vivo. Exp Cell Res. 2006;312(7):1127–41 Epub 2006/02/10.
Charvet C, Houbron C, Parlakian A, Giordani J, Lahoute C, Bertrand A, et al. New role for serum response factor in postnatal skeletal muscle growth and regeneration via the interleukin 4 and insulin-like growth factor 1 pathways. Mol Cell Biol. 2006;26(17):6664–74 Epub 2006/08/18.
Blindt R, Krott N, Hanrath P, vom Dahl J, van Eys G, Bosserhoff AK. Expression patterns of integrins on quiescent and invasive smooth muscle cells and impact on cell locomotion. J Mol Cell Cardiol. 2002;34(12):1633–44 Epub 2002/12/31.
Elenstrom-Magnusson C, Chen W, Clinchy B, Obrink B, Severison E. IL-4-induced B cell migration involves transient interactions between beta 1 integrins and extracellular matrix components. Int Immunol. 1995;7(4):567–73 Epub 1995/04/01.
Doucet C, Brouty-Boye D, Pottin-Clemenceau C, Jasmin C, Canonica GW, Azzarone B. IL-4 and IL-13 specifically increase adhesion molecule and inflammatory cytokine expression in human lung fibroblasts. Int Immunol. 1998;10(10):1421–33 Epub 1998/10/31.
McNally AK, Anderson JM. Beta1 and beta2 integrins mediate adhesion during macrophage fusion and multinucleated foreign body giant cell formation. Am J Pathol. 2002;160(2):621–30 Epub 2002/02/13.
Akira S, Taga T, Kishimoto T. Interleukin-6 in biology and medicine. Adv Immunol. 1993;54:1–78 Epub 1993/01/01.
Hiscock N, Chan MH, Bisucci T, Darby IA, Febbraio MA. Skeletal myocytes are a source of interleukin-6 mRNA expression and protein release during contraction: evidence of fiber type specificity. FASEB J. 2004;18(9):992–4 Epub 2004/04/03.
Pedersen BK, Steensberg A, Fischer C, Keller C, Ostrowski K, Schjerling P. Exercise and cytokines with particular focus on muscle-derived IL-6. Exerc Immunol Rev. 2001;7:18–31 Epub 2001/10/03.
Bartoccioni E, Michaelis D, Hohlfeld R. Constitutive and cytokine-induced production of interleukin-6 by human myoblasts. Immunol Lett. 1994;42(3):135–8 Epub 1994/10/01.
Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol. 2000;18:217–42 Epub 2000/06/03.
Baeza-Raja B, Munoz-Canoves P. p38 MAPK-induced nuclear factor-kappaB activity is required for skeletal muscle differentiation: role of interleukin-6. Mol Biol Cell. 2004;15(4):2013–26 Epub 2004/02/10.
Serrano AL, Baeza-Raja B, Perdiguero E, Jardi M, Munoz-Canoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008;7(1):33–44 Epub 2008/01/08.
Pozzobon T, Goldoni G, Viola A, Molon B. CXCR4 signaling in health and disease. Immunol Lett. 2016;177:6–15 Epub 2016/07/02.
Murphy PM, Heusinkveld L. Multisystem multitasking by CXCL12 and its receptors CXCR4 and ACKR3. Cytokine. 2018;109:2–10 Epub 2018/02/06.
Janowski M. Functional diversity of SDF-1 splicing variants. Cell Adhes Migr. 2009;3(3):243–9 Epub 2009/03/17.
Masyuk M, Brand-Saberi B. Recruitment of skeletal muscle progenitors to secondary sites: a role for CXCR4/SDF-1 signalling in skeletal muscle development. In: Brand-Saberi B, editor. Vertebrate myogenesis: stem cells and precursors. Berlin, Heidelberg: Springer Berlin Heidelberg; 2015. p. 1–23.
Brzoska E, Grabowska I, Hoser G, Streminska W, Wasilewska D, Machaj EK, et al. Participation of stem cells from human cord blood in skeletal muscle regeneration of SCID mice. Exp Hematol. 2006;34(9):1262–70 Epub 2006/08/31.
Brzoska E, Kowalewska M, Markowska-Zagrajek A, Kowalski K, Archacka K, Zimowska M, et al. Sdf-1 (CXCL12) improves skeletal muscle regeneration via the mobilisation of Cxcr4 and CD34 expressing cells. Biol Cell. 2012;104(12):722–37 Epub 2012/09/18.
Hunger C, Odemis V, Engele J. Expression and function of the SDF-1 chemokine receptors CXCR4 and CXCR7 during mouse limb muscle development and regeneration. Exp Cell Res. 2012;318(17):2178–90 Epub 2012/07/07.
Kowalski K, Archacki R, Archacka K, Stremińska W, Paciorek A, Gołąbek M, et al. Stromal derived factor-1 and granulocyte-colony stimulating factor treatment improves regeneration of Pax7−/− mice skeletal muscles. J Cachexia Sarcopenia Muscle. 2015.
Griffin CA, Apponi LH, Long KK, Pavlath GK. Chemokine expression and control of muscle cell migration during myogenesis. J Cell Sci. 2010;123(Pt 18):3052–60.
Odemis V, Boosmann K, Dieterlen MT, Engele J. The chemokine SDF1 controls multiple steps of myogenesis through atypical PKCzeta. J Cell Sci. 2007;120(Pt 22):4050–9 Epub 2007/11/01.
Kowalski K, Kolodziejczyk A, Sikorska M, Placzkiewicz J, Cichosz P, Kowalewska M, et al. Stem cells migration during skeletal muscle regeneration - the role of Sdf-1/Cxcr4 and Sdf-1/Cxcr7 axis. Cell Adhes Migr. 2017;11(4):384–98 Epub 2016/10/14.
Dalonneau F, Liu XQ, Sadir R, Almodovar J, Mertani HC, Bruckert F, et al. The effect of delivering the chemokine SDF-1alpha in a matrix-bound manner on myogenesis. Biomaterials. 2014;35(15):4525–35 Epub 2014/03/13.
Brzoska E, Kowalski K, Markowska-Zagrajek A, Kowalewska M, Archacki R, Plaskota I, et al. Sdf-1 (CXCL12) induces CD9 expression in stem cells engaged in muscle regeneration. Stem Cell Res Ther. 2015;6(1):46 Epub 2015/04/19.
Melchionna R, Di Carlo A, De Mori R, Cappuzzello C, Barberi L, Musaro A, et al. Induction of myogenic differentiation by SDF-1 via CXCR4 and CXCR7 receptors. Muscle Nerve. 2010;41(6):828–35 Epub 2010/02/13.
Sacchetti B, Funari A, Remoli C, Giannicola G, Kogler G, Liedtke S, et al. No identical “mesenchymal stem cells” at different times and sites: human committed progenitors of distinct origin and differentiation potential are incorporated as adventitial cells in microvessels. Stem Cell Reports. 2016;6(6):897–913 Epub 2016/06/16.
Shi D, Reinecke H, Murry CE, Torok-Storb B. Myogenic fusion of human bone marrow stromal cells, but not hematopoietic cells. Blood. 2004;104(1):290–4 Epub 2004/03/11.
Dezawa M, Ishikawa H, Itokazu Y, Yoshihara T, Hoshino M, Takeda S, et al. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science. 2005;309(5732):314–7 Epub 2005/07/09.
Gang EJ, Bosnakovski D, Simsek T, To K, Perlingeiro RC. Pax3 activation promotes the differentiation of mesenchymal stem cells toward the myogenic lineage. Exp Cell Res. 2008;314(8):1721–33 Epub 2008/04/09.
Gang EJ, Jeong JA, Hong SH, Hwang SH, Kim SW, Yang IH, et al. Skeletal myogenic differentiation of mesenchymal stem cells isolated from human umbilical cord blood. Stem Cells. 2004;22(4):617–24.
Shang Y, Zhang C, Wang S, Xiong F, Zhao C, Peng F, et al. Activated beta-catenin induces myogenesis and inhibits adipogenesis in BM-derived mesenchymal stromal cells. Cytotherapy. 2007;9(7):667–81 Epub 2007/10/06.
Kowalski K, Dos Santos M, Maire P, Ciemerych MA, Brzoska E. Induction of bone marrow-derived cells myogenic identity by their interactions with the satellite cell niche. Stem Cell Res Ther. 2018;9(1):258 Epub 2018/09/29.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–8 Epub 2002/02/16.
Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7 Epub 2006/08/23.
Persichini T, Funari A, Colasanti M, Sacchetti B. Clonogenic, myogenic progenitors expressing MCAM/CD146 are incorporated as adventitial reticular cells in the microvascular compartment of human post-natal skeletal muscle. PLoS One. 2017;12(11):e0188844 Epub 2017/12/01.
Friedenstein AJ, Petrakova KV, Kurolesova AI, Frolova GP. Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation. 1968;6(2):230–47 Epub 1968/03/01.
Friedenstein AJ, Piatetzky S II, Petrakova KV. Osteogenesis in transplants of bone marrow cells. J Embryol Exp Morphol. 1966;16(3):381–90 Epub 1966/12/01.
Friedenstein A, Kuralesova AI. Osteogenic precursor cells of bone marrow in radiation chimeras. Transplantation. 1971;12(2):99–108 Epub 1971/08/01.
Friedenstein AJ. Stromal mechanisms of bone marrow: cloning in vitro and retransplantation in vivo. Haematology and blood transfusion. 1980;25:19–29 Epub 1980/01/01.
Friedenstein AJ, Chailakhyan RK, Gerasimov UV. Bone marrow osteogenic stem cells: in vitro cultivation and transplantation in diffusion chambers. Cell and tissue kinetics. 1987;20(3):263–72 Epub 1987/05/01.
Luria EA, Owen ME, Friedenstein AJ, Morris JF, Kuznetsow SA. Bone formation in organ cultures of bone marrow. Cell Tissue Res. 1987;248(2):449–54 Epub 1987/05/01.
Owen M, Friedenstein AJ. Stromal stem cells: marrow-derived osteogenic precursors. CIBA Found Symp. 1988;136:42–60 Epub 1988/01/01.
Friedenstein AJ, Latzinik NV, Gorskaya Yu F, Luria EA, Moskvina IL. Bone marrow stromal colony formation requires stimulation by haemopoietic cells. Bone and mineral. 1992;18(3):199–213 Epub 1992/09/01.
Kuznetsov SA, Friedenstein AJ, Robey PG. Factors required for bone marrow stromal fibroblast colony formation in vitro. Br J Haematol. 1997;97(3):561–70 Epub 1997/06/01.
Sacchetti B, Funari A, Michienzi S, Di Cesare S, Piersanti S, Saggio I, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131(2):324–36 Epub 2007/10/25.
Serafini M, Sacchetti B, Pievani A, Redaelli D, Remoli C, Biondi A, et al. Establishment of bone marrow and hematopoietic niches in vivo by reversion of chondrocyte differentiation of human bone marrow stromal cells. Stem Cell Res. 2014;12(3):659–72 Epub 2014/03/29.
Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466(7308):829–34 Epub 2010/08/13.
Samsonraj RM, Rai B, Sathiyanathan P, Puan KJ, Rotzschke O, Hui JH, et al. Establishing criteria for human mesenchymal stem cell potency. Stem Cells. 2015;33(6):1878–91 Epub 2015/03/11.
Lv FJ, Tuan RS, Cheung KM, Leung VY. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32(6):1408–19 Epub 2014/03/01.
Wu Y, Zhao RC. The role of chemokines in mesenchymal stem cell homing to myocardium. Stem Cell Rev. 2012;8(1):243–50 Epub 2011/06/28.
Wynn RF, Hart CA, Corradi-Perini C, O'Neill L, Evans CA, Wraith JE, et al. A small proportion of mesenchymal stem cells strongly expresses functionally active CXCR4 receptor capable of promoting migration to bone marrow. Blood. 2004;104(9):2643–5 Epub 2004/07/15.
Sordi V, Malosio ML, Marchesi F, Mercalli A, Melzi R, Giordano T, et al. Bone marrow mesenchymal stem cells express a restricted set of functionally active chemokine receptors capable of promoting migration to pancreatic islets. Blood. 2005;106(2):419–27 Epub 2005/03/24.
Renz H, Domenico J, Gelfand EW. IL-4-dependent up-regulation of IL-4 receptor expression in murine T and B cells. J Immunol. 1991;146(9):3049–55 Epub 1991/05/01.
Archacka K, Brzoska E, Ciemerych MA, Czerwinska AM, Grabowska I, Kowalski K, et al. Pluripotent and mesenchymal stem cells—challenging sources for derivation of myoblast. In: Brzozka Z, Jastrzebska E, editors. Cardiac cell culture technologies,2018.
Naaldijk Y, Johnson AA, Ishak S, Meisel HJ, Hohaus C, Stolzing A. Migrational changes of mesenchymal stem cells in response to cytokines, growth factors, hypoxia, and aging. Exp Cell Res. 2015;338(1):97–104 Epub 2015/09/04.
Lawlor MA, Rotwein P. Coordinate control of muscle cell survival by distinct insulin-like growth factor activated signaling pathways. J Cell Biol. 2000;151(6):1131–40 Epub 2000/12/21.
Lefaucheur JP, Sebille A. Muscle regeneration following injury can be modified in vivo by immune neutralization of basic fibroblast growth factor, transforming growth factor beta 1 or insulin-like growth factor I. J Neuroimmunol. 1995;57(1–2):85–91 Epub 1995/03/01.
Aboalola D, Han VKM. Different effects of insulin-like growth factor-1 and insulin-like growth factor-2 on myogenic differentiation of human mesenchymal stem cells. Stem Cells Int. 2017;2017:8286248 Epub 2018/02/02.
Witt R, Weigand A, Boos AM, Cai A, Dippold D, Boccaccini AR, et al. Mesenchymal stem cells and myoblast differentiation under HGF and IGF-1 stimulation for 3D skeletal muscle tissue engineering. BMC Cell Biol. 2017;18(1):15 Epub 2017/03/02.
Kucia M, Wojakowski W, Reca R, Machalinski B, Gozdzik J, Majka M, et al. The migration of bone marrow-derived non-hematopoietic tissue-committed stem cells is regulated in an SDF-1-, HGF-, and LIF-dependent manner. Arch Immunol Ther Exp. 2006;54(2):121–35.
Liu X, Duan B, Cheng Z, Jia X, Mao L, Fu H, et al. SDF-1/CXCR4 axis modulates bone marrow mesenchymal stem cell apoptosis, migration and cytokine secretion. Protein Cell. 2011;2(10):845–54 Epub 2011/11/08.
Nowakowski A, Walczak P, Lukomska B, Janowski M. Genetic engineering of mesenchymal stem cells to induce their migration and survival. Stem Cells Int. 2016;2016:4956063 Epub 2016/06/01.
Acknowledgements
The study was funded by the National Centre for Research and Development, grant number: STRATEGMED1/235773/19/NCBR/2016. Porcine BM-MSC cell line was kindly supplied by professor Joanna Wojtkiewicz from the Department of Pathophysiology, Faculty of Medical Sciences, University of Warmia and Mazury, Olsztyn, Poland.
Funding
The study was funded by National Centre for Research and Development, grant number: STRATEGMED1/235773/19/NCBR/2016.
Author information
Authors and Affiliations
Contributions
Conceptualization was done by EB, MAC, and MJ. Investigation was performed by PK, KK, BM, MP, AI, MR, MG, AR, WS, KJI, MK, KA, MZ, RIN, AF, PW, and IG. Data curation was performed by EB, PK, KK, KA, MZ, and IG. Writing of the manuscript and original draft preparation were performed by EB, KA, and BM. Writing, review, and editing were performed by MAC, EB, BM, PK, KA, MZ, IG, KK, PW, and MJ. Visualization was done by IG. Supervision was performed by BE, CMA, and MJ. Project administration was performed by WS, MAC, EB and MJ. Founding acquisition was done by MJ. All authors read and approved the final manuscript..
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
The hybrid myotubes in hBM-MSC and C2C12 myoblast co-cultures. Blue – cell nuclei, red – skeletal myosin, green – human cell nuclei. Scale bar 50 μm.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kasprzycka, P., Archacka, K., Kowalski, K. et al. The factors present in regenerating muscles impact bone marrow-derived mesenchymal stromal/stem cell fusion with myoblasts. Stem Cell Res Ther 10, 343 (2019). https://doi.org/10.1186/s13287-019-1444-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13287-019-1444-1