Abstract
Background
The genus Chromobacterium, of which 12 species have been recognized, comprises bacteria that reside in tropical and subtropical environments. Of these species, Chromobacterium violaceum and Chromobacterium haemolyticum are known to cause infections in humans. There have been few reports of infections caused by Chromobacterium haemolyticum.
Case presentation
Chromobacterium haemolyticum was detected in spinal fluid and blood samples isolated from a 73-year-old Japanese male patient who fell into a canal in Kyoto City, Japan and developed bacteremia and meningitis. Although meropenem and vancomycin were administered, this patient died 9 days after admission. Although the infection was misidentified as being caused by Chromobacterium violaceum by conventional identification methods, average nucleotide identity analysis revealed that the causative pathogen was Chromobacterium haemolyticum. The same bacteria were also detected in the canal in which the accident occurred. Phylogenetic analysis of the strain isolated from the patient and the strain isolated from the canal suggested that the two strains were very closely related.
Conclusions
Chromobacterium haemolyticum can be misidentified as Chromobacterium violaceum by conventional identification methods and tends to be more resistant to β-lactams than Chromobacterium violaceum. Pigment production and β-hemolysis on blood sheep agar can provide clues for the early identification of Chromobacterium haemolyticum.
Similar content being viewed by others
Background
The genus Chromobacterium comprises bacteria that reside in tropical and subtropical environments, among which Chromobacterium violaceum and Chromobacterium haemolyticum are known to cause infections in humans [1]. C. haemolyticum is a facultative anaerobic Gram-negative rod that is known for producing hemolysin and showing a distinct β-hemolytic zone around the colony on sheep blood agar medium [2]. A few cases of infectious diseases, including pneumonia, bacteremia, necrotizing fasciitis, and pediatric proctitis, caused by C. haemolyticum have been reported [1,2,3,4,5,6]. While the worldwide geographic distribution of C. violaceum has been reported to be concentrated in tropical and subtropical regions [7], that of C. haemolyticum is not well known. In this study, C. haemolyticum was detected in spinal fluid specimens isolated from a patient who developed meningitis after falling into a canal in Kyoto City, Japan, which is a temperate region.
Case presentation
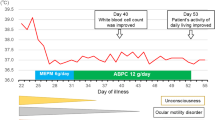
A 73-year-old Japanese male patient fell into a canal in Kyoto City in the summer of 2017 and was transported to an emergency department. He had a medical history of alcoholic hepatitis, hypertension, and hyperuricemia. He had no significant family medical history. He was an ex-smoker (stopped smoking 10 years prior) and had a history of alcohol consumption, drinking 50 g of alcohol daily. He had a 10 cm contusion wound on the back of the head and multiple fractures of the spinous process of the cervical vertebrae. On examination, his temperature was 36.1 °C, heart rate was 88 beats per minute, blood pressure was 170/96 mmHg, and respiratory rate was 18 breaths per minute. On admission, the physical and neurological examination revealed no abnormalities except for neck pain or the pre-existing mild ptosis in the left eye. He vomited on day 5 and had diminished consciousness on day 6. Computed tomography (CT) scan of the head revealed no obvious abnormalities, and a spinal tap was performed. Laboratory findings on day 6 of admission are presented in Table 1. The analysis revealed findings of bacterial meningitis with increased numbers of neutrophils, decreased glucose levels, and increased protein levels. The spinal fluid specimen exhibited a cloudy yellow appearance, and Gram staining revealed numerous Gram-negative rods along with inflammatory findings and increasing neutrophil levels (Fig. 1A, B). On the same day, blood culture samples were collected with two sets of aerobic bottles (BacT/ALERT SA; BioMerieux, Marcy l’Etoile, France) and anaerobic bottles (BacT/ALERT SN; BioMerieux) and tested by BacT/ALERT 3D (BioMerieux). Meropenem (2 g q8h) and vancomycin (1.25 g q8h) administration by infusion was initiated, and vancomycin administration was stopped after 3 days. Gram-negative rods were detected in all blood culture bottles, which all exhibited the same appearance as the spinal fluid. Cultures of spinal fluid and subcultures from the blood culture bottle were performed on sheep blood agar (Becton Dickinson: BD, Franklin Lakes, NJ, USA), Drigalski modified agar (Eiken Chemical Co., Ltd., Tokyo, Japan), and McConkey II agar medium (BD) at 37 °C for 48 hours (Fig. 1C, D). From each specimen, yellow colonies with β-hemolytic rings developed on blood agar, and green colonies developed on Drigalski modified agar. Small colonies developed on McConkey II agar medium (BD) (Fig. 1E). These colonies were nonproductive pigments. They were identified with the MicroScan WalkAway 96Plus (Beckman Coulter, Brea, CA, USA), resulting in a biotype of 40002007 with a 99.65% probability of being C. violaceum (the second candidate was Vibrio damsel with a probability of 0.35%). Identification testing by BBL CRYSTAL/NF (BD, version 5,4,2) resulted in the identification of biotype 3001310113, with only C. violaceum as a candidate for identification; MALDI Biotyper (Bruker Daltonics Inc., Billerica, MA, USA) yielded no peaks, and microbes were not identifiable. The main biochemical characteristics of this strain were oxidase (+), indole (−), mannose (−), and citric acid (+). Susceptibility testing was performed using the EN1J panel of the MicroScan WalkAway 96Plus. The results showed high minimum inhibitory concentration (MIC) values for penicillins, first- and second-generation cephalosporins, and ceftriaxone (Table 2). On day 8, the patient had frequent generalized convulsions, and he died on day 9. An autopsy was performed, and infiltration of inflammatory cells was detected in the subarachnoid space, which supported the diagnosis of meningitis.
Microbiological results. A The spinal fluid specimen appeared cloudy. B Numerous Gram-negative rods were present in the spinal fluid along with inflammatory findings with increasing numbers of neutrophils (Gram stain, ×1000 original magnification). C Results of assessments of Chromobacterium haemolyticum colonies on TSA II 5% sheep blood agar after a 48-h culture at 37 °C, showing yellow colonies with clear β-hemolysis, D Presence of lactose nondegradable colonies on BTB lactose-added agar, E Presence of transparent microcolonies on MacConkey II agar
A water culture survey was conducted at the site of the accidental fall, and the canal was approximately 1 m wide and 10 cm deep. Water samples were taken from eight locations over a 10 m stretch, and 50 µL of each sample was cultured on BTB lactose-supplemented agar medium. As a result, 20–30 colony-forming units (CFUs) of colony growth were suspected to comprise Chromobacterium spp. and were observed in samples from all locations, and these were identified as C. violaceum by MicroScan WalkAway 96Plus.
Whole-genome sequencing was performed with 151-cycle paired-end reads on an Illumina NextSeq 550 platform (Illumina, San Diego, CA, USA). All of the Illumina read data obtained from clinical and environmental isolates were deposited into the National Center for Biotechnology Information Sequence Read Archive database, SRA (accession numbers SRR20306466 and SRR20306467). Average nucleotide identity (ANI) analysis using JSpecies V1.2.1 [8] showed that the concordance rates between the clinical isolate strain and C. violaceum (ATCC12472), C. haemolyticum (DSM19808), and the strain from the canal were 85.42%, 96.49%, and 99.89%, respectively, and these clinical and environmental strains, each identified as C. violaceum by conventional methods, were C. haemolyticum. Then, phylogenetic analysis was performed using kSNP v3.1 with default parameters (k-mer = 21) [9]. There were only three single nucleotide polymorphisms (SNPs) between the strain from the patient and the strain from the canal. This finding indicates that both analyzed strains may have been derived from the same strain.
Discussion and conclusions
We report a fatal case of bacterial meningitis caused by C. haemolyticum acquired from a canal. C. haemolyticum was isolated from both this patient and the canal water, and the isolates were genomically closely related. This rare case revealed evidence supporting the development of severe meningitis accompanied by the invasion of environmental bacteria into the human body.
Twelve species of Chromobacterium spp. have been recognized [10]. Of these species, only C. violaceum and C. haemolyticum have been reported to infect humans [4]. C. violaceum is characterized by producing a purple pigment called violacein, and a high mortality rate of 53% resulting from infections caused by C. violaceum has been reported [7]. Alisjahbana et al. reviewed 132 cases of disease caused by C. violaceum in humans reported between 1953 and 2021, and the results showed that the organism is distributed worldwide in warm regions between 35° N and 35° S latitudes and causes human infections [11]. However, only six cases of infections caused by C. haemolyticum were reported, three of which were in Japan [1,2,3,4,5,6]. Although there have been six cases of infection caused by C. haemolyticum, this is the first case of meningitis caused by this bacterium that we know of (Table 3). In two of six cases observed in Japan, phylogenetic analysis was conducted using pulsed-field gel electrophoresis or whole-genome sequencing to compare the patient-derived strain and the strain derived from river water that was considered the source of infection. Nevertheless, in each study, there was some discordance between the patient-derived strains and the river-derived strains [3, 5]. In this study, we found that the strain isolated from the patient and the strain isolated from the environment were derived from a phylogenetically close strain. This finding indicates that the patient was infected by this organism living in the environment in Japan located in a temperate zone.
It should be noted that this is the only fatal case among C. haemolyticum infection reported. This patient’s comorbidities could explain his poor outcome. Both alcoholic hepatitis and hyperglycemic status are related to immunosuppression and poor outcome of infectious diseases [12, 13].
It has been noted that C. haemolyticum may be misidentified as pigment-nonproducing C. violaceum or that it may fail to be identified when conventional identification systems that use biochemical properties or other systems, such as matrix-assisted laser desorption ionization-time of flight–mass spectrometry (MALDI-TOF–MS), are used for identification [1,2,3,4,5]. We experienced a similar situation in this study. C. haemolyticum has characteristics that are easy to distinguish from those of C. violaceum, such as a β-hemolytic ring on sheep blood agar (+), poor growth on MacConkey agar, oxidase (+), indole (−), mannose (−), citric acid (+), and developmental ability at 32 °C [1, 2, 11]. Because the name of this organism was not present in the database used by identification instruments, kits, and TOF–MS, it was labeled C. violaceum despite having these characteristics. Other hemolytic strains in the genus Chromobacterium have been isolated in recent years. For example, C. paludis shows weak hemolysis on 5% sheep blood agar after 4–5 days of incubation at 25 °C [10]. Although human infections by C. paludis have not been reported, attention should be given to misidentification with C. haemolyticum.
Regarding susceptibility testing, C. haemolyticum has been reported to be more resistant to β-lactams than C. violaceum [1, 2]. The strains detected in this study also showed high MIC values against penicillins and cephalosporins. Therefore, it should be noted that C. haemolyticum could be misidentified as C. violaceum by conventional methods, and it is important to identify C. haemolyticum appropriately. The findings of hemolysis on sheep blood agar and pigment production could distinguish C. violaceum and C. haemolyticum.
In summary, C. haemolyticum could be misidentified as C. violaceum by conventional identification methods and tends to be more resistant to β-lactams than C. violaceum. Pigment production and β-hemolysis on blood sheep agar can provide clues for the early identification of C. haemolyticum.
Availability of data and materials
All data are contained in the manuscript.
Abbreviations
- ANI:
-
Average nucleotide identity
- CFU:
-
Colony-forming unit
- MALDI-TOF–MS:
-
Matrix-assisted laser desorption ionization-time of flight–mass spectrometry
- MIC:
-
Minimum inhibitory concentration
- SNP:
-
Single nucleotide polymorphism
References
Okada M, Inokuchi R, Shinohara K, Matsumoto A, Ono Y, Narita M, Ishida T, Kazuki C, Nakajima S, Yahagi N. Chromobacterium haemolyticum-induced bacteremia in a healthy young man. BMC Infect Dis. 2013;13:406.
Han XY, Han FS, Segal J. Chromobacterium haemolyticum sp. nov., a strongly haemolytic species. Int J Syst Evol Microbiol. 2008;58(Pt 6):1398–403.
Kanamori H, Aoyagi T, Kuroda M, Sekizuka T, Katsumi M, Ishikawa K, Hosaka T, Baba H, Oshima K, Tokuda K, et al. Chromobacterium haemolyticum pneumonia associated with near-drowning and River Water, Japan. Emerg Infect Dis. 2020;26(9):2186–9.
Harmon N, Mortensen JE, Robinette E, Powell EA. Pediatric bacteremia caused by Chromobacterium haemolyticum/Chromobacterium aquaticum. Diagn Microbiol Infect Dis. 2016;86(1):108–11.
Takenaka R, Nureki S, Ueno T, Shigemitsu O, Miyazaki E, Kadota J, Miki T, Okada N. Chromobacterium haemolyticum pneumonia possibly due to the aspiration of runoff water. Jpn J Infect Dis. 2015;68(6):526–9.
Tanpowpong P, Charoenmuang R, Apiwattanakul N. First pediatric case of Chromobacterium haemolyticum causing proctocolitis. Pediatr Int. 2014;56(4):615–7.
Yang CH, Li YH. Chromobacterium violaceum infection: a clinical review of an important but neglected infection. J Chin Med Assoc. 2011;74(10):435–41.
Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106(45):19126–31.
Gardner SN, Hall BG. When whole-genome alignments just won’t work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS ONE. 2013;8(12): e81760.
Blackburn MB, Farrar RR, Sparks ME, Kuhar D, Mowery JD, Mitchell A, Gundersen-Rindal DE. Chromobacterium paludis sp. nov., a novel bacterium isolated from a Chesapeake Bay marsh. Int J Syst Evol Microbiol. 2020;70(12):6142–6.
Alisjahbana B, Debora J, Susandi E, Darmawan G. Chromobacterium violaceum: a review of an unexpected scourge. Int J Gen Med. 2021;14:3259–70.
Kaur B, Rosenblatt R, Sundaram V. Infections in alcoholic hepatitis. J Clin Transl Hepatol. 2022;10(4):718–25.
Jay CA, Solbrig MV. Neurologic infections in diabetes mellitus. Handb Clin Neurol. 2014;126:175–94.
Acknowledgements
None.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
This patient was critically ill, and he needed treatment by a team made up of many physicians and other healthcare professionals, including neurologists (AY and TS), an infection control team (KI, TM, SeO, and YU), and a microbiological analysis team (KI, MY, TM, NM, HK, ShO, NI, YM, and MN). AY, TS, SeO, and YU contributed to patient management. KI, TM, NM, HK, ShO, and YU collected samples from the canal and performed microbiological procedures. YM, NI, YM, and MN performed microbiological procedures and molecular analysis. KI and MY drafted the initial manuscript. All authors contributed to writing the manuscript. All authors also read and approved all versions of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate:
The Ethics Committee of the Japanese Red Cross Kyoto Daiichi Hospital approved this study (no. 1470).
Consent for publication
Written informed consent was obtained from the patient’s next-of-kin for publication of this case report and any accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
Competing interests:
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Iwamoto, K., Yamamoto, M., Yamamoto, A. et al. Meningitis caused by Chromobacterium haemolyticum suspected to be derived from a canal in Japan: a case report. J Med Case Reports 17, 171 (2023). https://doi.org/10.1186/s13256-023-03913-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13256-023-03913-1