Abstract
Background
To investigate the relationship between early-life stress and glucocorticoid receptor (GR) gene methylation, which may result in long-lasting neurodevelopmental impairment, we performed a longitudinal analysis of the methylation ratio within the GR gene promoter 1F region using next-generation sequencing in preterm infants.
Cell-free DNA was extracted from the frozen serum of 19 preterm birth infants at birth and at 1 and 2 months after birth. All were admitted to the neonatal intensive care unit of Juntendo University Shizuoka Hospital between August 2014 and May 2016 and suffered from chronic lung disease (CLD).
Through bisulfite amplicon sequencing using an Illumina Miseq system and Bismark-0.15.0 software, we identified the rate of cytosine methylation.
Results
Patients’ sex and body weight standard deviation were extracted as the associated independent variables at birth. Sex, glucocorticoid administration for treating CLD, and postnatal invasive procedures (surgical operation and blood sampling) were extracted as the associated independent variables at 1 month. Methylation rates increased significantly between postnatal 1 and 2 months at 9 of the 39 CpG sites. Postnatal glucocorticoid administration to treat circulatory collapse was the most-associated independent variable with a positive regression coefficient for a change in methylation rate at these nine CpG sites. It also influenced the methylation ratio at 22 of the 39 CpG sites at 2 months of age. The standard deviation (SD) score at birth was extracted as an independent variable, with a negative regression coefficient at 9 of the 22 CpG sites together with glucocorticoid administration.
Conclusions
The results of this study indicate that a prenatal environment that results in intrauterine growth restriction and postnatal relative adrenal insufficiency requiring glucocorticoid administration leads to GR gene methylation. That, in turn, may result in neurodevelopmental disabilities.
Similar content being viewed by others
Background
The improved survival of babies at early gestational ages (GAs) is considered a success of modern neonatology. In the current post-steroid and post-surfactant era of neonatology, extremely preterm babies who survive have non-negligible rates of neurological disabilities such as cerebral palsy, mental deficits, sensorineural impairment, and cognitive dysfunction ranging from mild to severe [1, 2]. Infants born with a birth weight < 10th percentile or small for gestational age (SGA) [3, 4] and infants with chronic lung disease (CLD) characterized by prolonged inflammation of lung tissue are at increased risk for neonatal mortality, and preterm infants suffer from both short- and long-term morbidities [5, 6].
The most commonly implicated mechanism of these long-term effects is the dysregulation of the hypothalamus–pituitary–adrenal (HPA) axis. Indeed, dysregulation of this axis has been noted in extremely low birth weight and very low birth weight (VLBW) survivors across their lifespan [7, 8]. An impaired HPA axis is an important risk factor for inflammatory disease, somatic fatigue, pain disorders, and psychiatric conditions such as depression and post-traumatic stress disorder [9, 10]. Activity of the HPA axis is regulated by the hypothalamic glucocorticoid receptor (GR) encoded by the nuclear receptor subfamily 3 group C member 1 (NR3C1) gene, which mediates a negative feedback loop [11].
Promoter DNA methylation is a well-established epigenetic regulator of gene expression. Prenatal stressors, such as maternal depression and anxiety and maternal exposure to stressors, are associated with GR gene methylation [12, 13]. Similarly, adults who retrospectively report a history of childhood maltreatment, early parental death, and childhood trauma show associations with GR gene methylation [14,15,16]. These links have also been demonstrated in postmortem brains from adult suicides [17] as well as from patients who suffer from depression or bipolar disorder [18]. In our previous study, we showed that a postnatal environment that includes the need for acute care and prolonged physical separation under neonatal intensive care affects epigenetic programming of GR expression through methylation of the NR3C1 promoter in premature infants, which might result in glucocorticoid resistance later in life [19]. These studies were performed by using several tissues, such as placenta [12], cord blood [13], peripheral blood [14, 19], saliva [15, 16, 18], and brain [17].
Several important regions are known to exist in the 39 CpG sites analyzed in the present study [20]. Among them, CpG 30–32 are the most important regions known to be binding sites for nerve growth factor inducible protein A (NGFI-A), which has emerged as a central regulator of early inflammatory and immune processes and potentiates GR 1-F promoter activity. The methylation rate in this region is influenced by an adverse environment during early life [17]. The high methylation status in this region is also known to result in low GR mRNA expression in an animal model in vitro [21]. CpG 35 has been reported to be associated with maternal stress, a low level of prenatal care, and childhood maltreatment. CpG 36 has been associated with maternal psychopathology during pregnancy. CpG 37 methylation has been associated with both early-life stress and maternal psychopathology. CpG 39 methylation has been associated with childhood maltreatment and prenatal stress. However, CpG site-specific methylation findings for CpG 1–29 are scarce.
Recently, Giarraputo et al. showed that the GR gene methylation in infants in neonatal intensive care unit (NICU) is correlated with high medical risk which is defined by many medical variables (Neonatal Therapeutic Intervention Scoring System (NTISS)) [22].
To investigate the relationship between the early-life environment and GR gene (NR3C1) methylation status, which may take part in long-lasting neurodevelopmental impairment, we performed a longitudinal analysis of methylation ratios within the GR gene promoter 1F region using next-generation sequencing in preterm infants with CLD. In this study, we used cell-free DNA which is thought to be derived from proliferating/apoptotic lymphocytes from the participant’s peripheral blood [23].
Methods
This study was approved by the Juntendo University Ethics Committee and conducted according to the principles of the Declaration of Helsinki. Nineteen infants admitted to the NICU of Juntendo University Shizuoka Hospital between August 2014 and May 2016 were enrolled in this study after written informed consent had been obtained. All serum samples were routinely collected at birth and at 1 and 2 months after birth and stored at − 80 °C until analysis. The criterion for CLD was the need for additional oxygen after the age of 28 days. Cell-free DNA was extracted from 100 μL frozen serum using a DNA Extractor SP Kit according to the manufacturer’s instructions (Wako Pure Chemical Industries, Ltd. Osaka, Japan), and bisulfite-treated DNA was obtained using an EZ DNA Methylation Direct Kit (Zymo Research Corp., Irvine, CA, USA). A 5-μL aliquot of the resulting 10-μL bisulfite-treated genomic DNA solution was subjected to polymerase chain reaction analysis to amplify the GR promoter 1F region, as previously described [19]. The amplicons were purified using a gel-based clean up, and ViewaBlue Stain KANTO (Kanto Chemical Co., Inc. Tokyo, Japan) was used for DNA staining. The purified amplicons were subjected to a NEBNext Ultra II Library Preparation Kit for Illumina (New England Biolabs Japan, Inc., Tokyo, Japan) including dual indexing. These libraries were multiplexed and sequenced on an Illumina MiSeq system (Illumina Inc., San Diego, CA, USA). The reads were aligned to an in silico converted reference using Bowtie2-2.2.1, and variant calling was used to identify the percentage of methylated cytosines using Bismark-0.15.0. [24]. We analyzed the epigenetic changes in the GR promoter 1F region (containing 39 CpG sites ranging from − 3466 to − 3189 bp upstream of the ATG start site).
Statistical analysis
We performed Wilcoxon’s signed-rank test to evaluate longitudinal differences between birth and at 1 and 2 months after birth. A stepwise multiple regression analysis was performed to investigate the relationships of methylation ratio at birth, 1 month, and 2 months with the amount of increase during months 1–2 at each CpG site as dependent variables and pre- and postnatal parameters in preterm infants as independent variables. To perform this analysis, several nominal variables were converted into categorical variables by grouping the values into two categories. The adjusted coefficient of determination (R)2 is the fraction of information of the dependent variable that is explained by the independent variables. A two-tailed p value < 0.05 was considered statistically significant. All of the statistical analyses were performed using SPSS v24.0 (IBM Corp., Armonk, NY, USA) software.
Results
The abbreviations in the present study are listed and described at the end of the main text and in Table 1.
Participant characteristics
Nineteen participants completed the analysis, and their characteristics are shown in Table 1. The GAs and birth weights of the infants ranged from 24 weeks + 3 days to 28 weeks + 2 days and from 316 to 1226 g, respectively. All infants received mechanical ventilation. Four were below − 2 SD of birth weight. Seven infants underwent an operation to close the patent ductus arteriosus (PDA). Twelve infants received antenatal steroid (AS) administration and 11 received postnatal steroid administration. Of these 11 infants, 9 were treated for circulatory collapse and 2 were treated for CLD. Fourteen infants received an opioid for sedation.
Relationship between prenatal parameters and GR gene methylation at birth
We examined the associations between the methylation ratios at the 39 CpG sites in the GR 1F promoter and GA at birth (weeks), body weight SD scores at birth calculated by the Japanese standard, AS administration (mg), and sex. Methylation rates in the GR 1F region were generally low at birth. At 3 of the 39 CpG sites analyzed, only sex (male = 1, female = 0) was correlated with the methylation ratio (CpG 12: p = 0.046, regression coefficient, − 0.463; CpG 19: p = 0.027, regression coefficient, 0.505; CpG 27: p = 0.030, regression coefficient, − 0.497). At 4 of the 39 CpG sites analyzed, only SD scores were correlated with the methylation ratios (CpG 7: p = 0.048, regression coefficient, − 0.460; CpG 25: p = 0.040, regression coefficient, − 0.475; CpG 36: p = 0.047, regression coefficient, − 0.461; CpG 38: p = 0.002, regression coefficient, − 0.675). The methylation ratio at CpG 24 was associated only with AS administration (p = 0.025, regression coefficient, − 0.512) (Fig. 3).
Relationship between prenatal and postnatal (0–1 month) parameters and GR gene methylation at 1 month
We analyzed the relationships of methylation rates at 1 month with prenatal and postnatal (0–1 month) parameters (sex, GA, SD, AS, Hc, Bf, Gc circ, Gc CLD, Opi, dSD, In, and PDA) at the 39 CpG sites. The results are shown in Fig. 3. Briefly, postnatal parameters such as glucocorticoid administration for CLD, surgical operation, or heel-cut blood sampling were extracted as independent variables with positive regression coefficients in CpG 5, 8, 14, 25, 32, 33, 34, 35, and 38, except that sex in CpG 19 had a negative regression coefficient.
Longitudinal changes in methylation status
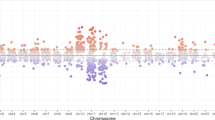
Methylation rates between birth and postnatal 1 month were stable at all of the 39 CpG sites examined. By contrast, methylation rates significantly increased from postnatal 1 to 2 months at 9 of the 39 CpG sites (CpG 3: p = 0.043; CpG 4: p = 0.047; CpG 5: p = 0.031; CpG 11: p = 0.014; CpG 15: p = 0.038; CpG 18: p = 0.047; CpG 19: p = 0.005; CpG 23: p = 0.0025; CpG 27: p = 0.03) (Fig. 1).
Longitudinal analysis of glucocorticoid receptor (GR) 1F promoter methylation. Methylation ratios at birth and at 1 and 2 months after birth in the 39 CpG sites analyzed are shown. The left, center, and right triplet bars indicate the methylation rates at birth, 1 month, and 2 months, respectively, with standard error bars. The asterisks under the CpG numbers indicate the methylation rates at CpG sites that significantly increased at between 1 and 2 months after birth
Relationships of the change in methylation ratio during months 1 and 2 with prenatal and postnatal parameters between birth and 2 months of age
We analyzed the relationship between changes in the methylation rates at CpG 3, 4, 5, 11, 15, 18, 19, 23, and 27, when significant increases in methylation rates were observed during months 1–2 after birth, and the prenatal and postnatal environments at 0–2 months of age. The independent variables analyzed at 0–2 months were sex, GA, SD, AS, days of mechanical ventilation (Mv), number of heel-cut (Hc) procedures, breast milk intake (mL/kg) (breast-fed [Bf] baby), amount of glucocorticoid administered (mg/kg prednisolone) for treatment of circulatory collapse (Gc circ), and CLD (Gc CLD), amount of opioid (Opi) administered (times 0.1 mL fentanyl), change in body weight SD scores (dSD), amount of indomethacin (In) administered (mg/kg), and experience of receiving surgery to close a patent ductus arteriosus (PDA) (yes = 1, no = 0).
At seven of the nine CpG sites, Gc circ was solely extracted as an efficient independent variable with a positive regression coefficient. CpG 3 (Gc circ [regression coefficient, 0.885, overall p < 0.001, adjusted R2 = 0.771]); CpG 4 (Gc circ [regression coefficient, 0.871, overall p < 0.001, adjusted R2 = 0.744]); CpG 5 (Gc circ [regression coefficient, 0.687, overall p = 0.001, adjusted R2 = 0.440]); CpG 15 (Gc circ [regression coefficient, 0.895, overall p < 0.001, adjusted R2 = 0.790]); CpG 18 (Gc circ [regression coefficient, 0.926, overall p < 0.001, adjusted R2 = 0.848]); CpG 19 (Gc circ [regression coefficient, 0.689, overall p = 0.001, adjusted R2 = 0.444]); and CpG 23 (Gc circ [regression coefficient, 0.800, overall p < 0.001, adjusted R2 = 0.619]). None of the eight parameters examined was extracted as an efficient independent variable at the remaining two CpG sites (CpG 11 and CpG 27).
Relationship between methylation rate at postnatal 2 months and prenatal and postnatal parameters
Next, we analyzed the relationship between methylation rates at 2 months after birth and prenatal and postnatal parameters, e.g., sex, GA, SD, AS, Hc, Bf, Gc circ, Gc CLD, Opi dSD, In, and PDA at all of the 39 CpG sites. The results are shown in Table 2. Briefly, some independent variables were extracted from 24 of the 39 CpG sites examined. Gc circ was extracted as an independent variable at 22 CpG sites with a positive regression coefficient. The volume of breast milk consumed was extracted as the sole independent variable at the remaining two CpG sites, with a negative regression coefficient (CpG 11 and CpG 36). SD scores at birth were simultaneously extracted as independent variables at 7 of 22 sites at which Gc circ was extracted, with a negative regression coefficient. AS administration was extracted as an independent variable at 3 of the 22 sites, with a negative regression coefficient. Glucocorticoid administration to treat CLD was extracted as an independent variable at CpG 2 with a negative regression coefficient and at CpG 8 with a positive regression coefficient. Figure 2 is a heat map of the methylation status focused on the effect of three major independent variables (Gc circ, AS, and SD).
Heat map of GR 1F promoter methylation at 2 months old. Patients who received a glucocorticoid for circulatory collapse (Gc circ), whose bodyweights were less than − 2 SD (BW < − 2 SD), and who were not administered an antenatal steroid (AS) are marked by the double circle in the right area of the heat map
Discussion
This is the first longitudinal report to demonstrate that prenatal and postnatal environments induce gene methylation. The results of this study show that postnatal Gc circ had positive and strong effects on GR gene methylation at 1–2 months of age, which may result in neurodevelopmental disability in later life due to dysfunction in the HPA axis. Simultaneously, prenatal factors such as SD scores at birth and antenatal glucocorticoid administration had moderate negative effects on GR gene methylation.
In the present study, glucocorticoid administration at 0–1 month influenced GR gene methylation at 1 month (Fig. 3). Our criteria for using glucocorticoid for CLD are as follows: mean airway pressure of mechanical ventilation > 10 mmHg and FiO2 > 0.4, which indicate a severe lung condition due to relative adrenal insufficiency.
Neonatal hypotension is commonly seen in premature neonates, and its incidence is inversely related to GA at delivery. Hypotension in preterm infants is predominantly due to either abnormal peripheral vasoregulation or myocardial dysfunction. Another etiology of hypotension in this population is physiological factors, including the presence of relative adrenal insufficiency secondary to an immature HPA axis [25]. Ng et al. showed an increase in plasma adrenocorticotrophic hormone with the use of inotropes and volume expanders in VLBW infants with hypotension [26]. In addition, low serum cortisol levels were also found in these patients [27]. These findings suggest that the adrenal cortex may be the primary site of dysfunction in this patient population, and not the hypothalamus or pituitary gland. Overall, these findings imply a normal response of the pituitary gland to hypotension, but the adrenal glands are transiently unable to maintain cortisol secretion during the immediate postnatal period. Hydrocortisone is the recommended treatment for hypotensive neonates if volume resuscitation with isotonic saline and dopamine treatment (10 μg/kg/min) are unsuccessful [28]. Our strategy to treat neonatal hypotension, which is defined as a mean blood pressure lower than the neonate’s GA, is described above. We also confirmed the effects of glucocorticoids by improvements in blood pressure and urine output soon after administration. We conclude that the hypotension observed in our study population resulted from relative adrenal insufficiency.
Postnatal steroids are widely used to manage preterm infants with CLD. Several reports have indicated that infants with CLD may develop adrenal insufficiency during the first 1–2 weeks of life [26, 29]. All of the infants in this study developed CLD, implying that they may have had some degree of adrenal insufficiency.
Taken together, these findings led us to hypothesize that severe adrenal insufficiency followed by both CLD and circulatory collapse results in GR gene methylation. Adrenal insufficiency during the early stage of life results in upregulation of the HPA axis later in life to respond adequately to stress.
Antenatal glucocorticoid administration had a moderate negative effect on GR gene methylation. In contrast, postnatal glucocorticoid administration for CLD and circulatory collapse had a positive effect on GR gene methylation. Thus, glucocorticoid demand due to relative adrenal insufficiency, not glucocorticoid administration itself, may result in GR gene methylation.
SD scores at birth were related to GR gene methylation at birth and at 2 months of age. Notably, the CpG sites affected at 2 months by SD (CpG 10, 15, 16, 17, 24, and 25) were different from those affected at birth by SD (CpG 7, 25, 36, and 38), except CpG 25. All of the SD effects at birth had disappeared by 1 month, even those in CpG 25. In addition, SD was always affected together with postnatal glucocorticoid administration only at 2 months. Thus, it is possible that the long-lasting influence of low SD (e.g., intrauterine growth restriction) on GR gene methylation is indirect.
Permanent changes in gene expression have been observed in multiple genes as a consequence of SGA. Dysregulation of the epigenome may explain changes that are propagated from parent to daughter cells in SGA offspring throughout life [30]. The results of this study suggest that dysregulation of several genes induced by intrauterine growth restriction may increase susceptibility to circumstances after birth, particularly relative adrenal insufficiency, leading to GR gene methylation, which in turn results in neurodevelopmental disabilities. Stressful postnatal procedures, such as the Hc procedure or Ope, were correlated with significant positive regression coefficients at three CpG sites at 1 month. Taken together, these findings suggest that relative adrenal insufficiency, which leads to severe CLD and circulatory collapse and the inability to respond to a stressful postnatal environment in the NICU, may result in GR gene methylation at 2 months after birth.
In this study, well-known CpG sites, which are methylated by early-life stress directly, such as CpG 30–32, CpG 35–37, and CpG 39, were affected at 2 months almost only by Gc circ. In contrast, CpG 1–29, which are not well-known as sites methylated by early-life stress, were affected by Gc circ and low SD. CpG 1–29 may be methylated by both the intrauterine and the postnatal environment. Figure 2 shows that CpG 30–39 and CpG 1–29 are widely methylated, which may result in a change in three-dimensional DNA structure.
It should be noted that the volume of mothers’ milk fed to infants during the first 2 months after birth had a weak but significant negative effect on GR methylation (CpG 11 and 36). Maternal breastfeeding has been emphasized as an influential factor in early childhood development [31]. However, empirical evidence for the effects of breastfeeding on children’s cognitive development has been conflicting. A comprehensive review by the Agency for Healthcare Research and Quality summarized 400 articles and found that breastfeeding had few or small effects on children’s cognitive ability [32]. This study may provide new insights into the effects of breastfeeding.
Because epigenetic modifications occur in a tissue-specific manner, it remains unclear whether DNA methylation measured in blood reflects DNA methylation patterns in other tissues including the brain. We used cell-free DNA, which is thought to be derived from proliferating/apoptotic lymphocytes in the participant’s peripheral blood [23].
Although research conducted on the effects of early-life stress on methylation of the alternate GR promoter 1F in blood, brain, saliva, and placenta produced similar results [12,13,14,15,16,17,18,19, 33], it should be noted that the correlation of methylation between blood and brain is still controversial. Watson et al. showed that only 7.9% of CpG sites were statistically significant, showing a large correlation between blood and brain tissue [34]. Tylee et al. reviewed seven studies comparing patterns of DNA methylation between the blood and brain and suggested that CpG island methylation levels are generally highly correlated (r = 0.90) between them [35].
This study has some limitations. First, cell type and tissue-specific diversity of methylation of the GR gene are noted problems. This study did not include a control for variations in cell type. Second, the participants were homogeneous in race, limiting generalizability of the findings. Another limitation is the lack of information about endogenous glucocorticoid secretion. Further investigations including a longitudinal follow-up study and cell type-specific investigation will enhance our understanding of the relationships between early-life experiences and long-lasting neurodevelopmental disability.
Conclusions
This study revealed that adverse pre- and postnatal environments, such as intrauterine growth restriction and postnatal relative adrenal insufficiency, increased glucocorticoid receptor gene methylation. This, in turn, may result in neurodevelopmental disabilities.
Abbreviations
- CLD:
-
Chronic lung disease
- GR:
-
Glucocorticoid receptor
- HPA:
-
Hypothalamus–pituitary–adrenal gland
- SGA:
-
Small for gestational age
- AS:
-
Antenatal steroid administration (mg)
- Bf:
-
Breast fed volume between 0 and 1 month after birth (mL/kg) (kg was calculated by birthweight + bodyweight at 2 months/2)
- BW:
-
Birth weight (g)
- dSD:
-
Change of SD scores between 0 and 2 months
- F:
-
Female
- GA:
-
Gestational age
- Gc circ:
-
Glucocorticoid administration between 0 and 2 months after birth for treatment of circulatory collapse (mg/kg prednisolone)
- Gc CLD:
-
Glucocorticoid administration between 0 and 2 months after birth for treatment or prevention of CLD (mg/kg prednisolone)
- Hc:
-
Heel cut procedure for blood examination between 0 and 2 months after birth (times)
- In:
-
Indomethacin administration for PDA closure (mg/kg)
- M:
-
Male
- Mv:
-
Duration of mechanical ventilation with intra-tracheal intubation between 0 and 2 months after birth (days)
- Opi:
-
Opioid administration between 0 and 2 months after birth (mg fentanyl)
- PDA:
-
Experience of receiving surgery to close a patent ductus arteriosus between 0 and 2 month after birth
- SD:
-
Standard deviation score of body weight
References
Marlow N, Wolke D, Bracewell MA, Samara M, EPICure Study Group. Neurologic and developmental disability at six years of age after extremely preterm birth. N Engl J Med. 2005;352:9–19.
Larroque B, Ancel PY, Marret S, Marchand L, André M, Arnaud C, EPIPAGE Study Group, et al. Neurodevelopmental disabilities and special care of 5-year-old children born before 33 weeks of gestation (the EPIPAGE study): a longitudinal cohort study. Lancet. 2008;371:813–20.
Guellec I, Lapillonne A, Renolleau S, Charlaluk ML, Roze JC, Marret S, et al. Neurologic outcomes at school age in very preterm infants born with severe or mild growth restriction. Pediatrics. 2011;127:e883–91.
von Ehrenstein OS, Mikolajczyk RT, Zhang J. Timing and trajectories of fetal growth related to cognitive development in childhood. Am J Epidemiol. 2009;170:1388–95.
Anderson PJ, Doyle LW. Neurodevelopmental outcome of bronchopulmonary dysplasia. Semin Perinatol. 2006;30:227–32.
Short EJ, Klein NK, Lewis BA, Fulton S, Eisengart S, Kercsmar C, et al. Cognitive and academic consequences of bronchopulmonary dysplasia and very low birth weight: 8-year-old outcomes. Pediatrics. 2003;112:e359.
Waxman JA, Lieshout RJ, Boyle MH, Saigal S, Schmidt LA. Linking extremely low birth weight and internalizing behaviors in adult survivors: influences of neuroendocrine dysregulation. Dev Psychobiol. 2015;57:486–96.
Kaseva N, Wehkalampi K, Pyhälä R, Moltch-anova E, Feldt K, Pesonen AK, et al. Blunted hypothalamic-pituitary-adrenal axis and insulin response to psycosocial stress in young adults born preterm at very low birth weight. Clin Endocrinol. 2014;80:101–6.
Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interaction, and epigenetics. Exp Neurol. 2012;233:102–11.
Buitelaar JK. The role of the HPA-axis in understanding psychopathology: cause, consequence, mediator, or moderator? Eur Child Adoles Psy. 2013;22:387–9.
Jacobson L. Hypothalamic-pituitary-adrenocortical axis regulation. Endocrin Metab Clin. 2005;34:271–92.
Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The roles of DNA methylation of NR3C1 and 11beta-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics. 2013;8:1321–9.
Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106.
Perroud N, Paoloni-Giacobino A, Prada P, Olié E, Salzmann A, Nicastro R, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry. 2011;1:e59.
Tyrka AR, Parade SH, Eslinger NM, Marsit CJ, Lesseur C, Seifer R. Methylation of exons 1D, 1F, and 1H of the glucocorticoid receptor gene promoter and exposure to adversity in preschool-aged children. Dev Psychopathol. 2015;27:577–85.
Parade SH, Ridout KK, Seifer R, Armstrong DA, Marsit CJ, McWilliams MA, et al. Methylation of the glucocorticoid receptor gene promoter in preschoolers: links with internalizing behavior problems. Child Dev. 2016;87:86–97.
McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8.
Melas PA, Wei Y, Wong CC, Sjöholm LK, Åberg E, Mill J, et al. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int J Neuropsychopharmacol. 2013;16:1513–28.
Kantake M, Yoshitake H, Ishikawa H, Araki Y, Shimizu T. Postnatal epigenetic modification of glucocorticoid receptor gene in preterm infants: a prospective cohort study. BMJ Open. 2014;4:e005318.
Palma-Gudiel H, Córdova-Palomera A, Leza JC, Fananás L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: a critical review. Neurosci Biobehav Rev. 2015;55:520–35.
Weaver ICG, D’Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S, Szyf M, Meaney MJ. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–68.
Giarraputo J, DeLoach J, Padbury J, Uzun A, Marsit C, Hawes K, Lester B. Medical morbidities and DNA methylation of NR3C1in preterm infants. Ped Res. 2017;81:68–74.
Tuaeva NO, Abramova ZI, Sofronov VV. The origin of elevated levels of circulating DNA in blood plasma of premature neonates. Ann N Y Acad Sci. 2008;1137:27–30.
Masser DR, Berg AS, Freeman WM. Focused, high accuracy 5-methylcytosine quantitation with base resolution by benchtop next-generation sequencing. Epigenetics Chromatin. 2013;6:33.
Seri I. Management of hypotension and low systemic blood flow in very low birth weight neonate during the first postnatal week. J Perinatol. 2006;26(Suppl 1):S8–S13.
Ng PC, Lee CH, Lam CW, Ma KC, Chan IH, Wong E, et al. Early pituitary-adrenal response and respiratory outcomes in preterm infants. Arch Dis Child Fetal Neonatal Ed. 2004;89:F127–30.
Helbock HJ, Insoft RM, Conte FA. Glucocorticoid-responsive hypotension in extremely low birth weight newborns. Pediatrics. 1993;92:715–6.
Johnson PJ. Hydrocortisone for treatment of hypotension in the newborn. Neonatal Netw. 2015;34:46–50.
Watterberg KL, Scott SM. Evidence of early adrenal insufficiency in babies who develop bronchopulmonary dysplasia. Pediatrics. 1995;95:120–5.
Einstein F, Thompson RF, Bhagat TD, Fazzari MJ, Verma A, Barzilai N, et al. Cytosine methylation dysregulation in neonates following intrauterine growth restriction. PLoS One. 2010;5:8887.
Kramer MS, Aboud F, Mironova E, Vanilovich I, Platt RW, Matush L, et al. Breastfeeding and child cognitive development: new evidence from a large randomized trial. Arch Gen Psychiatry. 2008;65:578–84.
Ip S, Chung M, Raman G, Chew P, Magula N, DeVine D, et al. Breastfeeding and maternal and infant health outcome in developed countries. Evid Rep Technol Assess (Full Rep). 2007;153:1–186.
Radtke KM, Schauer M, Gunter HM, Ruf-Leuschner M, Sill J, Meyer A, et al. Epigenetic modifications of the glucocorticoid receptor gene are associated with the vulnerability to psychopathology in childhood maltreatment. Transl Psychiatry. 2015;5:e571.
Walton E, Hass J, Liu J, Roffman L, Bernardoni F, Roessner V, et al. Correspondence of DNA Methylation Between blood and Brain Tissue and Its Application to Schizophrenia Research. Schizophrenia Bulletin. 2016;42:406–14.
Tylee DS, Kawaguchi DM, Glatt SJ. On the Outside, Looking In: A Review and Evaluation of the Comparability of Blood and Brain “-omes”. Am J Med Genet Part B. 2013;162B:595–603.
Acknowledgements
Thank you to Drs. Ryo Matoba and Noriko Ito for the technical assistance. Thank you also to Drs. Koichi Sato and Hajime Orita for their contribution in Shizuoka Medical Research Center for Disaster.
Funding
This study was supported by Ministry of Education, Culture, Sports, Science and Technology (MEXT)-Supported Program for the Strategic Research Foundation at Private Universities, 2015–2019, Japan.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on a reasonable request.
Author information
Authors and Affiliations
Contributions
MK conceptualized and designed the study, drafted the initial manuscript, and approved the final manuscript as submitted. NO, TI, AA, and NS designed the data collection instruments, coordinated and supervised the data collection, and approved the final manuscript as submitted. NI, HS, and TS carried out the statistical analysis, reviewed and revised the manuscript, and approved the final manuscript as submitted.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Juntendo University Ethics Committee and conducted according to the principles of the Declaration of Helsinki, and written informed consent from the parents of participants of this study had been obtained.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kantake, M., Ohkawa, N., Iwasaki, T. et al. Postnatal relative adrenal insufficiency results in methylation of the glucocorticoid receptor gene in preterm infants: a retrospective cohort study. Clin Epigenet 10, 66 (2018). https://doi.org/10.1186/s13148-018-0497-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-018-0497-9