Abstract
Background
Obesity is a disease that is caused by genetic and environmental factors. However, epigenetic mechanisms of obesity are less well known. DNA methylation provides a mechanism whereby environmental factors can influence gene transcription. The aim of our study was to investigate skeletal muscle DNA methylation of sorbin and SH3 domain containing 3 (SORBS3) with weight loss induced by Roux-en-Y gastric bypass (RYGB).
Results
Previously, we had shown increased methylation (5.0 to 24.4%) and decreased gene expression (fold change − 1.9) of SORBS3 with obesity (BMI > 30 kg/m2) compared to lean controls. In the present study, basal muscle biopsies were obtained from seven morbidly obese (BMI > 40 kg/m2) female subjects pre- and 3 months post-RYGB surgery, in combination with euglycemic-hyperinsulinemic clamps to assess insulin sensitivity. We identified 30 significantly altered promoter and untranslated region methylation sites in SORBS3 using reduced representation bisulfite sequencing (RRBS). Twenty-nine of these sites were decreased (− 5.6 to − 24.2%) post-RYGB compared to pre-RYGB. We confirmed the methylation in 2 (Chr.8:22,423,690 and Chr.8:22,423,702) of the 29 decreased SORBS3 sites using pyrosequencing. This decreased methylation was associated with an increase in SORBS3 gene expression (fold change + 1.7) post-surgery. In addition, we demonstrated that SORBS3 promoter methylation in vitro significantly alters reporter gene expression (P < 0.0001). Two of the SORBS3 methylation sites (Chr.8:22,423,111 and Chr.8:22,423,205) were strongly correlated with fasting plasma glucose levels (r = 0.9, P = 0.00009 and r = 0.8, P = 0.0010). Changes in SORBS3 gene expression post-surgery were correlated with obesity measures and fasting insulin levels (r = 0.5 to 0.8; P < 0.05).
Conclusions
These results demonstrate that SORBS3 methylation and gene expression are altered in obesity and restored to normal levels through weight loss induced by RYGB surgery.
Similar content being viewed by others
Background
One third of the US adult population is obese (body mass index [BMI] > 30 kg/m2), and the number of individuals entering into morbid obesity (BMI > 40 kg/m2) is on the rise [1, 2]. Roux-en-Y gastric bypass (RYGB) is one of the most common surgeries performed to treat obesity and combines restrictive and malabsorptive techniques [3]. Besides weight loss, other benefits of surgical intervention have included improved blood glucose levels and insulin sensitivity and secretion [3,4,5]. However, these studies do not completely explain the molecular basis of the metabolic improvements observed with weight loss induced by surgery.
One of the most studied epigenetic marks is DNA methylation, which is the addition of a methyl group to the fifth carbon of a cytosine, typically preceding a guanine, termed CpG dinucleotide [6]. The addition or removal of these marks has regulatory influence on gene expression [7]. Our previously published study identified a novel gene, sorbin and SH3 domain containing 3 (SORBS3), that was differentially methylated with obesity [8]. Specifically, we had shown an increase in skeletal muscle promoter methylation and a decrease in mRNA expression of SORBS3 with obesity [8]. The SORBS3 gene codes for the adapter protein vinexin and has been shown to play a role in growth-factor-induced signal transduction and cytoskeleton structure [9]. We have previously proposed a model where chronic inflammation from obesity may lead to insulin resistance by inducing changes to the extracellular matrix that are reminiscent of fibrosis and alter mechanosignal transduction mediated by cytoskeletal elements [10]. We believe that changes in SORBS3 expression may be connected to our proposed model since it is a cytoskeletal gene that was reduced with obesity [8]. However, it is unclear whether the reduction in SORBS3 observed in obesity could be rescued through surgical weight-loss interventions such as RYGB.
Here, we set out to determine if the changes in SORBS3 DNA methylation identified in obesity and its underlying insulin resistance can be altered in response to weight loss, using our previously described RYGB surgery cohort [11]. We hypothesized that 3 months post-surgery, SORBS3 methylation would be decreased and this would result in an increased gene expression, so it would be normalized to levels observed in lean controls.
Methods
Study design
Seven (one of which was diabetic, treated with metformin) morbidly obese (BMI > 40 kg/m2) females (ages 33–59 years) participated in this study pre- and 3 months post-RYGB surgery. The study was approved by the institutional review boards at the Mayo Clinic in Arizona and Arizona State University. The design of this study and surgical procedure have been previously described [11].
Muscle biopsy processing
Genomic DNA and total RNA were isolated from muscle biopsies as previously described [8].
Reduced representation bisulfite sequencing (RRBS)
RRBS sample preparation was performed on the pre- and 3-month post-surgery DNA by the Mayo Clinic Genotyping Shared Resource facility. The RRBS sample preparation has been described in detail elsewhere [8]. Sequence data was processed using the streamlined analysis and annotation pipeline for reduced representation bisulfite sequencing, SAAP-RRBS [8, 12].
Differentially methylated cytosine (DMC) analysis pre- and post-surgery
Differences in methylation sites were assessed in participants pre- and post-surgery. The aligned (Hg19) sequencing data was imported into the free open source R package, methylSig [13]. A minimum of five reads and the recovery of the site in all seven participants from pre- and post-surgery were required for the inclusion of a cytosine in subsequent analyses. The mean methylation differences (%) were determined and annotations were applied, as previously described [8]. DMCs within the promoter and untranslated regions for SORBS3 were extracted from the data set.
SORBS3 pyrosequencing
DNA methylation sites were confirmed using pyrosequencing, as previously described [8]. To assess SORBS3 DMCs at positions Chr.8:22,423,519 and Chr.8:22,423,529 on the sense strand, bisulfite-converted DNA was amplified by PCR using the following primers: forward 5′-AGTAGGGGGAGGAAGGAA-3′ and biotinylated reverse 5′-ACCCCCATCCTCTACTAAAAATTAAC-3′. For the DMCs at positions Chr.8:22,423,690 and Chr.8:22,423,702 on the antisense strand, bisulfite-converted DNA was amplified by PCR using the following primers: forward 5′-GGGTTTTGGGTTTTTTATAGGATG-3′ and biotinylated reverse 5′- CCACCCAAAACAACTAACTCCTAAC-3′. Pyrosequencing was performed using the PyroMark Q96 MD system and the Gold Q96 kit with sequencing primers for the sense 5′-GGGGGAGGAAGGAAT-3′ and antisense 5′-TGGGTTTTTTATAGGATGT-3′ strands according to the manufacturer’s instructions (Qiagen, Valencia, CA). Sequence analysis was performed using the PyroMark CpG SW 1.0 software (Qiagen, Valencia, CA).
SORBS3 quantitative real-time PCR (qRT-PCR)
Gene expression for SORBS3 pre- and post-surgery was detected using qRT-PCR on the ABI PRISM 7900HT sequence detection system (Life Technologies, Carlsbad, CA). The qRT-PCR analyses of the samples were performed using the TaqMan primer and probes as previously described [8].
Luciferase assay
An 811 bp fragment of the human SORBS3 promoter (Chr8:22,422,247–22,423,057) was cloned into a CpG-free luciferase reporter vector (pCpGL-basic). The SORBS3 construct was either mock methylated or methylated using 1600 μM S-adenosylmethionine (SAM) and two different DNA methyltransferases, SssI and HhaI (New England Biolabs, Frankfurt, Germany). Mouse muscle cell lines C2C12 were cultured in DMEM, supplemented with 10% serum and 1% of an antibiotic/antimycotic mixture. Cells were co-transfected with 100 ng of pCpGL-basic with the SORBS3 promoter insert or without (control) and 2 ng of pRL renilla luciferase control reporter vector using the Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA). Firefly luciferase activity was measured and normalized against the renilla luciferase activity using the Dual Luciferase Reporter Assay System (Promega, Madison, WI). The results presented are a mean of four independent experiments, containing the mean of five replicates in each experiment.
SORBS3 comparative DMC analysis
The RRBS data from our previous lean versus obese participant study [8] was used for comparative analysis. The data comprised of 11 lean (ages 21–43 years; 7 females/4 males; BMI 23.4 ± 2.4 kg/m2) and 9 obese (ages 32–52 years; 4 females/5 males; BMI 32.9 ± 2.3 kg/m2) participants.
Predictive transcription factor binding analysis
PROMO version 3.0.2 was used to perform transcription factor binding site analysis [14]. Sequences were analyzed with a 5% maximum matrix dissimilarity rate using TRANSFAC version 8.3 database. Analysis of the 30 SORBS3 DMCs was assessed as 10 separate sequences: Chr.8:22,411,723–22,411,734; Chr.8:22,422,932–22,422,973; Chr.8:22,423,009–22,423,025; Chr.8:22,423,086–22,423,116; Chr.8:22,423,181–22,423,215; Chr.8:22,423,219–22,423,256; Chr.8:22,423,514–22,423,573; Chr.8:22,423,684–22,423,695; Chr.8:22,423,697–22,423,741; and Chr.8:22,423,769–22,423,857.
Statistical analysis
Pre- and post-surgery comparisons were based on a paired Student t test. All phenotypic data was normally distributed and presented as a mean ± standard deviation (SD). Pearson correlation analysis was performed to determine the relationship between DNA methylation from RRBS or gene expression and the phenotypic data. A Bonferroni correction was applied to the Pearson correlation analysis performed on the RRBS 30 DMCs with the phenotype data. Therefore, for the correlation analysis of the RRBS DMCs with the phenotypic data, we considered a P ≤ 0.0167 to be significant. For all other correlations, we used the P ≤ 0.05 cutoff. See above for the statistical analysis of the methylation and qRT-PCR data.
Results
Participants
The metabolic data for these subjects have been described in a previous publication [11]. Briefly, 3 months post-surgery, significant improvements were observed (Table 1) in BMI, body fat percentage, cholesterol, low-density lipoprotein (LDL), fasting plasma glucose (FPG), fasting serum insulin (FSI), and homeostatic model assessment for insulin resistance (HOMA-IR). However, there were no significant improvements observed in blood pressure, triglycerides, high-density lipoprotein (HDL), hemoglobin A1c (HbA1c), endogenous glucose production (EGP), and insulin-stimulated glucose disposal (M-value).
SORBS3 differentially methylated cytosines (DMCs)
Methylation sites within the promoter (0 to − 1000 base pairs from transcription start site) and untranslated regions (5′ and 3′UTR) were used to detect sites that may lead to a change in SORBS3 mRNA expression. From the RRBS, 352 CpG sites associated with SORBS3 were captured. Of these sites, 8.5% were differentially methylated (30 DMCs). From the 30 DMCs, there were 20 DMCs in the sense strand and 10 DMCs in the antisense strand identified (Table 2). As shown in Table 2, 29 of the 30 sites were decreased post-RYGB. The loss of DNA methylation post-surgery was detected specifically for SORBS3 and was not the by-product of global losses in methylation (mean ± standard deviation, global pre 33.9 ± 39.9% versus global post 33.7 ± 39.7%).
SORBS3 validation
Pyrosequencing was used for confirmation of the 30 SORBS3 DMCs where primers could be designed. The SORBS3 region that we were targeting for pyrosequencing was difficult to validate. The sequence characteristics of that region resulted in amplicons that would be too long or would have too many CpG sites and as such resulted in the failure of the assay design. Regardless, we were able to design primers that captured the DMCs at positions Chr.8:22,423,519 and Chr.8:22,423,529 on the sense strand, as well as four additional CpG sites. All six sites were trending towards a decrease in methylation post-surgery; however, none were significant (Fig. 1a). The sequence that encompassed the DMCs at positions Chr.8:22,423,690 and Chr.8:22,423,702 on the antisense strand included two additional CpG sites. All four sites were decreased in methylation post-surgery, and changes in three of these sites were statistically significant (P < 0.05; Fig. 1b).
SORBS3 gene expression
The qRT-PCR results demonstrated an increase in gene expression of SORBS3 post-surgery compared to pre-surgery (+ 1.7 versus 1.0 fold change, respectively; P = 0.05).
SORBS3 correlation analysis
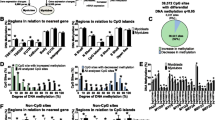
Pearson correlation analysis was performed to determine the relationship between the phenotypic data and SORBS3 changes observed with surgery. Of the DMCs detected using RRBS, we demonstrated two Bonferroni-corrected significant associations with fasting plasma glucose (FPG) at DMCs Chr.8:22,423,111 (r = 0.9, P = 0.00009) and Chr.8:22,423,205 (r = 0.8, P = 0.0010). Association of SORBS3 gene expression with phenotypic data was performed using the Ct values from qRT-PCR. Pearson’s correlation analysis identified a significant relationship between the gene expression data and BMI (r = 0.8, P = 0.00040), percent body fat (r = 0.6, P = 0.02), and FSI (r = 0.5, P = 0.04). Furthermore, an association between SORBS3 gene expression and methylation was identified at DMCs Chr.8:22,423,519 (r = 0.7, P = 0.004), Chr.8:22,423,689 (r = 0.5, P = 0.05), and Chr.8:22,423,702 (r = 0.6, P = 0.03).
SORBS3 promoter methylation in vitro alters reporter gene expression
The SORBS3 construct was created to test the effect of DNA methylation on transcriptional activity. Level of suppressed transcriptional activity, as measured by luciferase activity, was determined in comparison to the mock methylated control (Fig. 2). As shown in Fig. 2, when the SORBS3 construct was methylated in vitro using the HhaI enzyme (GCGC, n = 8 sites), transcriptional activity was not suppressed but was significantly suppressed with the SssI enzyme methylation (CG, n = 59 sites).
Predicted transcription factor binding analysis
To identify potential transcription factor binding that may be inhibited by SORBS3 methylation, we analyzed sequences containing DMCs using PROMO [14]. Transcription factor binding motifs were identified to overlap 13 of the 30 DMCs for SORBS3: Chr.8:22,422,937: AP-2alphaA; Chr.8:22,422,953: Sp1; Chr.8:22,423,014 and 22,423,020: GCF; Chr.8:22,423,100: CREB; Chr.8:22,423,204, 22,423,205, 22,423,206, and 22,423,210: GCF; Chr.8:22,423,235: Sp1, Pax-5, and p53; Chr.8:22,423,689 and 22,423,690: GCF; and Chr.8:22,423,736: RXR-alpha.
SORBS3 alterations with obesity and RYGB surgery
Increased DNA methylation in the promoter and 5′UTR of SORBS3 with obesity were originally identified in our previous study [8]. In the RYGB cohort, methylation levels of SORBS3 were found to decrease post-surgery. Upon comparing the 10 DMCs (9 increased and 1 decreased) from our previous study, and the 30 DMCs (29 decreased and 1 increased) identified with RYGB surgery, we found sites to cluster in the same region, but no sites were identical between studies (Fig. 3). We assessed the average methylation levels of all significant DMCs regardless of directional change (Fig. 4a). Moreover, we averaged methylation levels of the significant DMCs that were consistent in the direction of methylation change (9 increased sites were averaged from the lean and obese study and 29 decreased sites were averaged for the RYGB cohort) (Fig. 4b). Both analyses presented similar average methylation levels between the lean and post-surgery and the obese and pre-surgery (Fig. 4a, b).
Average methylation levels of SORBS3 DMCs from lean and obese participants in a previous study and the present study pre- and post-surgery levels. The average methylation was assessed with all DMCs, regardless of methylation direction (a) and of only the DMCs that were consistent in the direction of methylation (b). Data presented as mean ± SD
Discussion
Our previous study had identified SORBS3 as an obesity-associated gene, whose expression may be epigenetically regulated [8]. We set out to further establish the relationship between SORBS3 methylation and gene expression changes with obesity through a surgical weight-loss intervention. Three months following the RYGB surgery, there were significant reductions in weight and improvement of metabolic measures such as BMI, percent body fat, and fasting plasma insulin levels. However, we did not observe improvements in EGP or M-value as determined by the euglycemic-hyperinsulinemic clamp. Our observations are consistent with others [15, 16]. EGP has been shown to significantly decrease immediately after surgery and return to pre-surgery measures at 3 months [15]. The M-value has been shown to significantly improve at 12 months with major weight loss [16]. It could be hypothesized that we may have observed improvements in EGP and the M-value had we extended the study beyond the 3 months post-surgery.
Environmental factors can influence transcriptional regulation through DNA methylation. Following weight loss induced by surgery, we observed significant decreases in SORBS3 methylation and increased gene expression. When we applied a conservative Bonferroni correction, we demonstrated two highly significant associations between SORBS3 DMCs and fasting plasma glucose levels. Moreover, the gene expression data was significantly associated with BMI, percent body fat, and fasting insulin levels. In another study, Barres et al. identified significant positive correlations between PGC1α methylation levels and phenotypes, such as BMI and triglycerides 6 months post-surgery [17]. Both the Barres et al. study and our current study have observed that methylation changes are correlated with metabolic phenotypes [17]. This observation would suggest that the methylation changes are not contingent on the alteration of one phenotype, but changes in an individuals’ metabolic status. Collectively, our findings highlight the relationship between decreased SORBS3 DNA methylation in the presence of weight loss induced by surgery.
The changes observed in SORBS3 DNA methylation and gene expression complement our previous findings [8]. Interestingly, reduced methylation and increased expression post-surgery were relatively proportional to levels found in our lean individuals, even though the mean BMI for these groups were not comparable. This link between DNA methylation and gene expression changes was further established by in vitro measures. The luciferase assay has been used in this study and in others [17, 18] as a reliable means of providing evidence for the regulatory role of promoter methylation on gene expression. Typically, with the use of multiple methyltransferases, a stepwise decrease in luciferase expression is observed with an increasing number of methylated sites [18]. We observed decreased gene expression with the methylation set by SssI, but not with HhaI, suggesting the number and positioning of the sites in that promoter to be important. Coincidentally, the methylation changes identified in the current study were only represented within the SssI methylated sites and not the HhaI. However, the exact mechanism in which our DNA methylation sites regulate the transcription of SORBS3 has not been elucidated. We have identified potential transcription factor binding motifs that may be affected by the presence of methylation, but require further investigation. Specifically, the transcription factors that may be influenced in this assay that overlap with our sites of interest are GC-binding factor (GCF), specificity protein 1 (Sp1), and activating enhancer-binding protein 2-alpha (AP-2 Alpha).
The gene SORSB3 codes for the cytoskeletal adapter protein vinexin [19]. Previously, we have shown that cytoskeletal proteins are reduced in insulin resistance [20]. We have proposed a model in which the reduction in cytoskeletal elements can disrupt the sensing of contractile activity, leading to altered mechanosignaling for gene expression changes in mitochondrial biogenesis [10]. This can potentially lead to a reduction and abnormal function of mitochondria and ultimately result in cellular abnormalities (lipid accumulation, reduced fat oxidation, and insulin signaling) related to insulin resistance. It is tempting to speculate that a reduction in vinexin abundance may play a role in the altered cytoskeletal organization for mechanosignal transduction proposed with insulin resistance. Furthermore, our current findings with weight loss induced by surgery could ameliorate these changes associated with obesity and insulin resistance.
Conclusions
Collectively, the post-surgery findings present an exciting new addition to understanding the DNA methylation changes associated with SORBS3 expression. We have previously detected differences associated with SORBS3 in individuals with obesity and insulin resistance, and the present study provided further evidence of alterations in SORBS3 in response to weight loss by surgical intervention. However, we acknowledge the limitation of our sample size in the present study. Future studies will need to confirm our findings in a larger cohort. Moreover, we observed in vitro the suppression of SORBS3 promoter DNA methylation on transcriptional activity. The specific placement of these sites can play an important role on the binding ability of transcription factors. We identified potential transcriptional regulators overlapping our methylation sites; however, follow-up studies will be necessary to refine the specific interaction.
Abbreviations
- BMI:
-
Body mass index
- DMC:
-
Differentially methylated cytosine
- EGP:
-
Endogenous glucose production
- FPG:
-
Fasting plasma glucose
- FSI:
-
Fasting serum insulin
- HbA1c:
-
Hemoglobin A1c
- HDL:
-
High-density lipoprotein
- HOMA-IR:
-
Homeostatic model assessment for insulin resistance
- LDL:
-
Low-density lipoprotein
- M-value:
-
Insulin-stimulated glucose disposal
- RRBS:
-
Reduced representation bisulfite sequencing
- RYGB:
-
Roux-en-Y gastric bypass
- SORBS3 :
-
Sorbin and SH3 domain containing 3
- UTR:
-
Untranslated region
References
Sturm R, Hattori A. Morbid obesity rates continue to rise rapidly in the United States. Int J Obes. 2013;37:889–91.
Flegal KM, Carroll MD, Ogden CL, Curtin LR. Prevalence and trends in obesity among US adults, 1999-2008. JAMA. 2010;303:235–41.
Catoi AF, Parvu A, Muresan A, Busetto L. Metabolic mechanisms in obesity and type 2 diabetes: insights from bariatric/metabolic surgery. Obes Facts. 2015;8:350–63.
Knop FK, Taylor R. Mechanism of metabolic advantages after bariatric surgery: it’s all gastrointestinal factors versus it’s all food restriction. Diabetes Care. 2013;36(Suppl 2):S287–91.
Mazidi M, Gao HK, Li L, Hui H, Zhang Y. Effects of roux-en-Y gastric bypass on insulin secretion and sensitivity, glucose homeostasis, and diabetic control: a prospective cohort study in Chinese patients. Surgery. 2017;161:1423–9.
Ronn T, Ling C. DNA methylation as a diagnostic and therapeutic target in the battle against type 2 diabetes. Epigenomics. 2015;7:451–60.
Huidobro C, Fernandez AF, Fraga MF. The role of genetics in the establishment and maintenance of the epigenome. Cell Mol Life Sci. 2013;70:1543–73.
Day SE, Coletta RL, Kim JY, Campbell LE, Benjamin TR, Roust LR, De Filippis EA, Dinu V, Shaibi GQ, Mandarino LJ, Coletta DK. Next-generation sequencing methylation profiling of subjects with obesity identifies novel gene changes. Clin Epigenetics. 2016;8:77.
Kioka N, Ueda K, Amachi T. Vinexin, CAP/ponsin, ArgBP2: a novel adaptor protein family regulating cytoskeletal organization and signal transduction. Cell Struct Funct. 2002;27:1–7.
Coletta DK, Mandarino LJ. Mitochondrial dysfunction and insulin resistance from the outside in: extracellular matrix, the cytoskeleton, and mitochondria. Am J Physiol Endocrinol Metab. 2011;301:E749–55.
Campbell LE, Langlais PR, Day SE, Coletta RL, Benjamin TR, De Filippis EA, Madura JA 2nd, Mandarino LJ, Roust LR, Coletta DK. Identification of novel changes in human skeletal muscle proteome after Roux-en-Y gastric bypass surgery. Diabetes. 2016;65:2724–31.
Sun Z, Baheti S, Middha S, Kanwar R, Zhang Y, Li X, Beutler AS, Klee E, Asmann YW, Thompson EA, Kocher JP. SAAP-RRBS: streamlined analysis and annotation pipeline for reduced representation bisulfite sequencing. Bioinformatics. 2012;28:2180–1.
Park Y, Figueroa ME, Rozek LS, Sartor MA. MethylSig: a whole genome DNA methylation analysis pipeline. Bioinformatics. 2014;30:2414–22.
Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–4.
Bojsen-Moller KN, Dirksen C, Jorgensen NB, Jacobsen SH, Serup AK, Albers PH, Hansen DL, Worm D, Naver L, Kristiansen VB, et al. Early enhancements of hepatic and later of peripheral insulin sensitivity combined with increased postprandial insulin secretion contribute to improved glycemic control after Roux-en-Y gastric bypass. Diabetes. 2014;63:1725–37.
Albers PH, Bojsen-Moller KN, Dirksen C, Serup AK, Kristensen DE, Frystyk J, Clausen TR, Kiens B, Richter EA, Madsbad S, Wojtaszewski JF. Enhanced insulin signaling in human skeletal muscle and adipose tissue following gastric bypass surgery. Am J Physiol Regul Integr Comp Physiol. 2015;309:R510–24.
Barres R, Kirchner H, Rasmussen M, Yan J, Kantor FR, Krook A, Naslund E, Zierath JR. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3:1020–7.
Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, Nilsson E, Tornberg A, Dekker Nitert M, Eriksson KF, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572.
Kioka N, Sakata S, Kawauchi T, Amachi T, Akiyama SK, Okazaki K, Yaen C, Yamada KM, Aota S. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J Cell Biol. 1999;144:59–69.
Hwang H, Bowen BP, Lefort N, Flynn CR, De Filippis EA, Roberts C, Smoke CC, Meyer C, Hojlund K, Yi Z, Mandarino LJ. Proteomics analysis of human skeletal muscle reveals novel abnormalities in obesity and type 2 diabetes. Diabetes. 2010;59:33–42.
Acknowledgements
We thank the volunteers of the study and are grateful for their participation and cooperation. We thank the clinical studies infusion unit nurses and research staff for their excellent care of the participants. We thank the Mayo Clinic Genotyping Shared Resource facility for the RRBS next-generation methylation analysis. We thank Kara Peterson and Dr. Melanie Carless for their assistance with the pyrosequencing experiments. We thank Dr. Maja Klug and Dr. Michael Rheli for kindly providing the pCpG-basic vector.
Funding
This study was supported in part by a Mayo/Arizona State University seed grant (to LRR and DKC) and by the National Institutes of Health grant R01DK094013 (DKC).
Availability of data and materials
The RRBS data used for comparison to the data in the current study are available in the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/), accession GSE73304.
Author information
Authors and Affiliations
Contributions
DKC conceived the experiments. SED, LAG, RLC, LEC, and DKC carried out the experiments. TRB, LRR, and EADF performed the euglycemic-hyperinsulinemic clamps with muscle biopsies. JAM performed the Roux-en-Y gastric bypass surgery. SED, LAG, RLC, and DKC performed the analysis of the data with assistance from LJM. SED and DKC wrote the article. LAG, RLC, LEC, TRB, LRR, EADF, JAM, and LJM read the manuscript and provided comments. DKC is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All subjects gave informed written consent to participate in the study, which was approved by the Institutional Review Boards of the Mayo Clinic in Arizona and Arizona State University (IRB 12-009688 and IRB 11-007028).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Day, S.E., Garcia, L.A., Coletta, R.L. et al. Alterations of sorbin and SH3 domain containing 3 (SORBS3) in human skeletal muscle following Roux-en-Y gastric bypass surgery. Clin Epigenet 9, 96 (2017). https://doi.org/10.1186/s13148-017-0396-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13148-017-0396-5