Abstract
Background
Salmonella Enteritidis causes intestinal and extra-intestinal infections, but rarely cutaneous infections. It has never been reported to cause carbuncle (a collection of interconnected furuncles with multiple pustular openings). We report a case of carbuncle due to S. Enteritidis.
Case presentation
An adult Bangladeshi patient with type 2 diabetes presented with a carbuncle on the left-side of his neck. A pure culture of S. Enteritidis was grown from the pus of the carbuncle. The patient was successfully treated with ciprofloxacin to which the isolate was susceptible. Whole genome sequencing of the strain showed that it possessed three additional virulence genes—pef (for plasmid-encoded fimbriae), spv (for salmonella plasmid virulence), rck (for resistance to complement killing) -responsible for systemic infections that were absent in the genome of a reference S. Enteritidis strain. In phylogenetic analysis, the strain clustered with other S. Enteritidis strains from different parts of the world.
Conclusions
A weakened immune system of the patient due to diabetes mellitus and the additional virulence genes of the isolate may have contributed to the unusual presentation of carbuncle. The possibility of S. Enteritidis to cause carbuncle should be considered.
Similar content being viewed by others
Background
Non-typhoid Salmonella infections lead to not only self-limited acute gastrointestinal infections, but also, bacteremia with or without extra-intestinal focal infections [1, 2]. Extra-intestinal focal infections include septic arthritis, osteomyelitis, cholangitis, aortitis, endocarditis, pneumonia, urinary tract infection, and meningitis [1, 2]. Non-typhoid salmonellae do not produce cutaneous infections in immunocompetent individuals, but a single case of leg abscesses has been reported in an immunocompromised patient [3]. Here we report carbuncle in a patient with type 2 diabetes mellitus. Carbuncle is an aggregate of connected furuncles (hair follicle infections) with multiple pustular openings. To our knowledge, this is the first case of carbuncle due to non-typhoid salmonellosis. Clinical manifestation is the result of interaction between the host and the pathogen. In this report, we also investigated whether there are unique genetic properties of the Salmonella isolate that could have contributed to this unusual clinical manifestation. This was achieved by whole genome sequencing of the isolate.
Case presentation
A 55-year-old Bangladeshi male with a history of type 2 diabetes mellitus and hypertension presented to the Emergency Department of Farwaniya Hospital, Kuwait in December 2015 with a left-sided neck swelling (8 cm × 6 cm) discharging pus through multiple sinuses. He was suffering from this condition for the past 2 weeks. On examination on alert, voice, pain and unresponsive (AVPU) scale, he was conscious and oriented with stable signs and without fever (oral temperature of 36.7 °C). Examinations covering the respiratory, cardiovascular, gastrointestinal and respiratory systems were unremarkable. His body mass index (BMI) was 27.4 (classified as overweight [4] and a risk factor for type 2 diabetes [5]). Blood examination by flow cytometry (Sysmex 9000, Bornbarch, Norderstedt, Germany) showed leukocytosis (18 × 109/L, mainly neutrophils, normal range is 3.7 × 109/L)) and by oxygen rate method (Beckman Coulter DxC 800, Brea, CA, USA) hyperglycemia (random blood sugar of 20 mmol/L; normal range is 3.9–6.1 mmol/L). A diagnosis of carbuncle with hyperglycemia was made. He was given parenteral insulin on a sliding scale (1–14 units with increments of 1–2 units during a 24 h period). Incision and drainage of the carbuncle were done and pus was sent for microbiological studies. An empirical therapy with clindamycin (300 mg intravenously every 8 h) was started suspecting infection with Staphylococcus aureus which is a common cause of carbuncle [6]. This therapy was continued for 3 days until culture and susceptibility report became available (see below).
Pus was cultured on blood agar, MacConkey agar, chocolate agar and gentamicin blood agar. Blood agar and MacConkey agar were incubated aerobically, chocolate agar microaerobically and gentamicin blood agar anaerobically. Incubation was done at 37 °C for 24–48 h. All plates except the gentamicin plate grew a pure culture (organism was susceptible to gentamicin, see below) which was identified as a Salmonella species using Phoenix method (Becton–Dickinson, Franklin Lakes, NJ, USA) and Vitek 2 and Vitek-MS methods (Biomerieux, Marcy l’Etoile, France). Blood, urine and stool were sent for culture. Blood culture was done using BD BACTEC system (Becton–Dickinson) and there was no growth of any organism. Urine culture was done on blood agar and cysteine-, lactose-, and electrolyte-deficient (CLED) agar. There was no bacteriuria and Salmonella species was not isolated. Stool was cultured on MacConkey agar, Campy agar and Salmonella–Shigella agar (SSA) and enriched in selenite F broth with subsequent subculture on SSA. No bacterial diarrheal pathogen including Salmonella species was isolated. In vitro susceptibility of the Salmonella isolated from carbuncle (designated as CSE76F) to antibiotics was done by Vitek II system (bioMerieux) and E test (AB Biodisk, Solna, Sweden) and interpreted by Clinical and Laboratory Standards Institute (CLSI) guidelines [7]. It was susceptible to amikacin, amoxicillin–clavulanic acid, chloramphenicol, tetracycline, ceftazidime, ceftriaxone, cefuroxime, cephalothin, imipenem, meropenem, ciprofloxacin, gentamicin, piperacillin–tazobactam, tigecycline, and trimethoprim–sulfamethoxazole, but resistant to ampicillin and clindamycin. The antibiotic therapy was changed to ciprofloxacin (400 mg intravenously every 12 h) for 14 days at which time he recovered. This prolonged intravenous therapy was necessitated to avoid relapse because the patient was immunocompromised (due to diabetes mellitus) and the wound was large. After debridement of the wound (Fig. 1), the patient was referred to plastic surgery for skin grafting.
The Salmonella organism was typed as Enteritidis species by multilocus sequence typing (MLST) [8] and a sefA (S almonella Enteritidis fimbria)A gene-specific PCR [9]. A draft genome sequence for isolate CSE76F was determined using the whole genome shotgun method. The sequencing library was prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA) and the sequence read data were produced on the Illumina NextSeq instrument (paired end, 150 base reads). A total of 3,213,288 reads yielded 477,626,349 bases of usable sequence data after filtering to remove low quality sequence data and adapter sequences (approximately 100- fold read coverage of the genome) (Sequence Read Archive [SRA] Accession Number, SRR5198927). De novo assembly of the read data with MegaHit [10] yielded a draft genome sequence comprising a total of 4,743,613 bases in 61 contigs (minimum contig size 500 bases). The MLST type was confirmed from the draft genome sequence (Senterica scheme, ST 11) from the genome sequence [11] and the antimicrobial resistance gene profile of the isolate was determined using Abricate [12] and the ResFinder database [13]. This showed that the isolate carried a bla TEM-1b gene (encoding a class A beta-lactamase). This fits with the observed resistance of the isolate to ampicillin [14]. The most related closed genome sequence was that from Salmonella enterica subsp. enterica serovar Enteritidis strain P125109 isolated from an outbreak of human food-poisoning (RefSeq: NC_011294) with 55 SNPs identified across 98.82% of the strain P125109 genome sequence. Aligning the CSE76F contigs to the P125109 genome sequence revealed no apparent major genomic deletions, inversions or rearrangements (Fig. 2). Analysis of the virulence gene profile was performed using Abricate with the VFDB database of virulence genes [15]. CSE76F strain possessed three additional virulence genes-pef (p lasmid-e ncoded f imbriae with all four subunits-pefA, pefB, pefC, pefD), spv (s almonella p lasmid v irulence with all three subunits- spvA, spvB, spvC) and rck ( r esistance to c omplement k illing)-that were absent in strain P125109. Phylogenetic relationship of CSE76F strain based on single nucleotide polymorphism (SNP) with that of 60 closed genomes of S. Enteritidis strains available in GenBank (shown in Additional file 1: Table S1) was constructed using FastTree [16]. Core SNP differences were called using Nullarbor [17]. Strain CSE76F did not occupy a unique position, but clustered with several other strains from the United Kingdom, South Korea, Canada and the United States of America (Fig. 3).
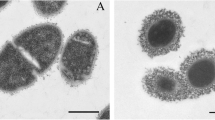
A graphical overview of comparison of genome sequence of reference strain P125109 (a) with draft genome sequence of strain CSE76F (b). No evidence of insertions/deletions or genomic rearrangements was seen in CSE76F. The figure shows 61 contigs in CSE76F aligned to the P125109 genome. Vertical red lines in B show the edges of contigs
Core genome comparison shows a cluster of strains with core genome sequences that are less than 100 SNPs different from each other (indicated as “Node A”). Strain P125109 was used as the reference genome sequence for the core genome comparison. All pairwise genome comparisons covered more than 97% of the reference genome sequence. Among the 60 sequences compared in the tree, there were 2452 aligned positions for which there was a sequence difference in one or more of the genome sequences of the strains
Discussion and conclusions
pef gene mediates adhesion and biofilm formation [18], spv gene cytotoxicity [19] and rck gene serum-resistance and survival inside the macrophage [20]. All the three genes are located in a 60 MDa virulence plasmid that enhances the growth rate during the systemic phase of the disease [21]. It is not surprising that the strain CSE76F possessed the additional virulence genes which would have contributed to its ability to produce the unusual manifestation of carbuncle in the immunologically weakened diabetes mellitus patient. However, the strain clustered with several other S. Enteritidis strains from different parts of the world.
Since there was no history of diarrhea in this patient or in his close family contacts, he would have had an asymptomatic intestinal infection with S. Enteritidis. The organism would have entered the blood stream from the intestinal tract and then the skin. Another possibility that the patient might have introduced the organism into his skin by scratching it by fecally contaminated fingers. This case shows that the possibility of non-typhoid Salmonella (S. Enteritidis) causing carbuncle should be taken into consideration.
Abbreviations
- AVPU:
-
alert voice pain unresponsive scale
- CLED:
-
cysteine lysine electrolyte deficient agar
- SSA:
-
Salmonella-Shigella agar
- CLSI:
-
Clinical and laboratory Standards Institute
- sef :
-
Salmonella Enteritidis fimbriaA
- pef :
-
plasmid-encided fimbriae
- spv :
-
Salmonella plasmid virulence
- rck :
-
resistance to complement killing
- SRA:
-
equenced read archive
- SNPs:
-
single nucleotide polymorphisms
- BMI:
-
body mass index
References
Hohman EL. Nontyphoidal salmonellosis. Clin Infect Dis. 2001;32:263–9.
Chen P-L, Chang C-M, Wu C-J, Ko N-Y, Lee N-Y, Lee H-C, et al. Extraintestinal focal infections in adults with nontyphoid Salmonella bacteraemia: predisposing factors and clinical outcome. J Intern Med. 2007;261:91–100.
Shamiss A, Thaler M, Nussinovitch N, Zissin R, Rosenthal T. Multiple Salmonella enteritidis leg abscess in a patient with systemic lupus erythematosus. Postgrad Med J. 1990;66:486–8.
Defining adult overweight and obesity. https://www.cdc.gov/obesity/adults/defining.html. Accessed 28 Aug 2017.
Huelgas RG. Weight and type 2 diabetes: new recommendations. Med Clin (Barc). 2016;147(Suppl 1):17–21.
Sharma S, Verma KK. Skin and soft tissue infection. Indian J Pediatr. 2001;3:S46–50.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 18th informal supplement. CLSI/NCCLS M100-S18. Wayne: Clinical and Laboratory Standards Institute; 2008.
Achtman M, Wain J, Weill F-X, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enteritica. PLoS Pathog. 2012;8(6):e1002776.
Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G, et al. Identification by a multiplex PCR-based assay of Salmonella Typhimurium and Salmonella Enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol. 1999;29:1–6.
Li D, Liu CM, Luo R, Sadakane K, Lam TW. An ultrafast single node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6.
Scan contig files against PubMLST typing schemes. https://github.com/tseemann/mlst. Accessed 8 Aug 2017.
Mass screening of contigs for antimicrobial and virulence genes. https://github.com/tseemann/abricate. Accessed 8 Aug 2017.
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired resistance genes. J Antimicrob Chemother. 2012;67:2640–4.
Shaikh S, Fatima J, Shakil S, Rizvi SMD, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J Biol Sci. 2015;22:90–101.
Chen L, Zheng D, Liu B, Yang J, Jin Q. Hierarchical and refined dataset for big data analysis-10 years on. Nucleic Acids Res. 2016;44:D694–7.
Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5(3):e9490.
“Reads to report” for public health and clinical microbiology. https://github.com/tseemann/nullarbor. Accessed 29 Aug 2017.
Ledeboer NA, Frye JG, McClelland M, Jones BD. Salmonella enterica serovar Typhimurium requires Lpf, Pef and Tafi fimbriae for biofilm formation on Hep-2 tissue culture cells and chicken intestinal epithelium. Infect Immun. 2006;74:3156–9.
Guiney DG, Fierer J. The role of the spv genes in Salmonella pathogenesis. Front Microbiol. 2011;2:129.
Hefferman EJ, Harwood J, Fierer J, Guiney D. The Salmonella typhimurium virulence plasmid complement resistance gene rck is homologous to a family of virulence-related outer membrane protein genes including pagC and ail. J Bacteriol. 1992;174:84–91.
Gulig PA, Doyle TJ. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61:504–11.
Authors’ contributions
WA, SS and NA did clinical microbiology work, DB did whole genome analysis, HI typed the isolate, KA did surgical debridement of the wound, and MJA supervised collection of data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Available.
Consent for publication
Available from the patient.
Ethics approval and consent to participate
Not needed as this case is a routine case admitted in the hospital for treatment. Therefore, ethics approval was waived by Farwaniya Hospital Ethics Committee. Local, national and international guidelines were followed in the conduct of this study.
Funding
Not funded by any agency.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Alfouzan, W., Bulach, D., Izumiya, H. et al. Carbuncle due to Salmonella Enteritidis: a novel presentation. Gut Pathog 9, 51 (2017). https://doi.org/10.1186/s13099-017-0200-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-017-0200-2