Abstract
Background
Helicobacter pylori infection is associated with risk for chronic gastritis (CG), gastric ulcer (GU), duodenal ulcer (DU), and gastric cancer (GC). The H. pylori Cag type IV secretion system (TFSS) translocates the virulence factor cytotoxin-associated gene A protein into host cells and plays an important role in initiating gastric carcinogenesis. The CagL and CagI proteins are components of the TFSS. The Arg-Gly-Asp (RGD) motif of CagL, and the six most distal C-terminal amino acids (Ser-Lys-Ile-Ile-Val-Lys, and Ser-Lys-Val-Ile-Val-Lys) of CagL and CagI are essential for TFSS adhesion to host cells. Additionally, the CagL variant Tyr58Glu59 was previously shown to be associated with GC patients.
Results
We isolated 43 H. pylori isolates from 17 CG, 8 GU, 8 DU, and 10 GC patients in Southeast Asia. Total DNAs were extracted and sequenced with MiSeq. H. pylori strain ATCC 26695, which was isolated from CG patients, was used as a reference. We examined the full sequences of H. pylori cagL and cagI using whole-genome sequencing (WGS), and analyzed whether single nucleotide variants and amino acid changes (AACs) correlated with adverse clinical outcomes. Three isolates were excluded from the analysis due to cagPAI rearrangements. CagL RGD motifs were conserved in 39 isolates (97.5%). CagL-Glu59 and Ile234 in the C-terminal motif were more common in 10 H. pylori isolates from GC patients (p < 0.001 and p < 0.05, respectively). When 5 Vietnamese isolates from GC patients were excluded, CagL-Glu59 still remains significant (p < 0.05), but not Ile234. CagL-Tyr58 was seen in only one isolate. The CagI C-terminal motif was completely conserved across all 40 isolates, and there were no significant AACs in CagI.
Conclusions
Using WGS, we analyzed genetic variants in clinical H. pylori isolates and identified putative novel and candidate variants in uncharacterized CagL and CagI sequences that are related to gastric carcinogenesis. In particular, CagL-Glu59 has the possible association with GC.
Similar content being viewed by others
Background
The infection rate for the Gram-negative bacterium Helicobacter pylori is around 50% worldwide [1, 2]. H. pylori infection increases the risk of chronic gastritis (CG), gastric ulcer (GU), duodenal ulcer (DU), and gastric cancer (GC). Nevertheless, the exact molecular action to the development of these adverse clinical outcomes remains not well-defined. Especially, in the East Asia, since the infection of cytotoxin-associated gene A (cagA) positive H. pylori is nearly 100%, their correlation to the different clinical outcomes could not be fully assessed [3–5].
Most H. pylori strains (so-called type I strains) contain the cag pathogenicity island (cagPAI), a chromosomal region that includes about 37,000 bp and 28 genes [3, 4]. Genes encoded in the cagPAI allow H. pylori bacteria to translocate its major virulence protein cytotoxin associated gene A (CagA) into host gastric epithelial cells using a type-IV secretion system (TFSS) [5, 6]. The role of the H. pylori TFSS and CagA translocation was examined in previous sequential studies that showed Src-mediated phosphorylation of CagA tyrosines is important for H. pylori virulence [5, 7, 8]. In East Asia in particular, nearly all H. pylori infections are CagA positive, which complicates assessment of how clinical H. pylori isolates are associated with disease outcomes [9–11]. Moreover, the mechanisms by which H. pylori expresses and regulates its TFSS injection apparatus when adapting to human epithelial cell receptors are unclear.
A recent study identified integrin α5β1 expressed on gastric epithelial cells as the putative host receptor for H. pylori TFSS [12]. The H. pylori CagL protein was found to be an adhesion target on the injected pilus surface for binding to host integrin α5β1 through the CagL Arg-Gly-Asp (RGD) motif [13]. Initial CagL-integrin binding properly induced to locate the bacterial TFSS prior to CagA translocation as well as to activate host tyrosine kinase [12, 14]. This interaction between the H. pylori TFSS and host integrin α5β1 can activate the NF-kB proteins and several important pro-inflammatory cytokines that resulted in more adverse clinical outcomes, such as gastric carcinogenesis.
CagI is another H. pylori protein, but its function is less clear [12, 15]. CagI has no sequence similarities to any other TFSS components, or to other known proteins [16, 17]. Although an isogenic cagI mutant has been examined, there were conflicting reports about whether CagI is required for TFSS function [3, 18]. Based on H. pylori transcriptome evidence [19], cagI is certainly part of an operon containing cagPAI genes involved in the TFSS, but the actual contribution of CagI to clinical phenotypes is unknown.
Here, we used whole-genome sequencing (WGS) to analyze genetic variants of 43 H. pylori isolates from patients in Southeast Asia who had different clinical disease. Using the WGS data, we examined whether CagL and/or CagI amino acid changes (AACs) correlated with adverse clinical outcomes such as GC.
Results
Characteristics of clinical H. pylori isolates
We previously performed WGS on 19 H. pylori clinical isolates that we deposited under accession number DRA001250 (see “Methods”). Here we undertook WGS of 24 new clinical H. pylori isolates, and analyzed a total of 43 H. pylori whole genome sequences (Table 1). The 43 isolates were from 17 chronic gastritis (CG), 8 gastric ulcer (GU), 8 duodenal ulcer (DU) and 10 gastric cancer (GC) patients whose diagnosis was based on endoscopy results. The 43 H. pylori isolates we analyzed also had different geographic origins in that 31, 7, and 5 isolates were isolated from Japanese, Chinese, and Vietnamese patients, respectively.
Sequence reads mapping to ATCC 26695 and quality check
The total reads for the 43 H. pylori isolates ranged from 1.99 to 10.87 million (Table 1). Sequencing data were mapped to the genome of the H. pylori strain ATCC 26695, which was isolated from CG patients, as a reference. Total consensus length (bp) ranged from 1,503,522 to 1,664,897, and total consensus coverage (%) ranged from 90.15 to 99.82%. Average coverage (fold) ranged from 79.5 to 669.4-fold.
Following the initial quality check, we focused on the 28 genes in the cagPAI region (Additional file 1: Table S1). Among the 43 isolates, strain ID 189 had lower coverage (under 100-fold) in the cagPAI region, strain ID 194 had no genes in the cagPAI region, and strain ID F51 carried the cagA gene alone. Due to these major sequence differences in the cagPAI region, we excluded data for these three isolates, which were all from Japanese patients, such that 40 clinical H. pylori isolates were subjected to further analysis. Of these 40 isolates, 15, 8, 7, and 10 were from CG, GU, DU, and GC patients, respectively, and 28, 7, and 5 isolates were derived from Japanese, Chinese, and Vietnamese patients, respectively. CagA motifs of 40 clinical isolates were different (Additional file 2: Table S2).
After the quality check, the average coverage of the remaining 40 isolates ranged from 99.6- to 361.4-fold for cagL, and from 105.4- to 416.3-fold for cagI (nearly over 100-fold). Consistent with our earlier report, the WGS data in this study had high sequencing coverage, and were of sufficiently high quality to allow detection of SNVs in the H. pylori genome [20].
CagL variants in patients with different clinical disease outcomes
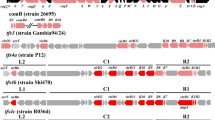
We translated the CagL nucleotide sequences into amino acid sequences (residues 1–237) with Genomics Workbench 8.5.1, and analyzed CagL variants based on clinical disease outcomes. Table 2 lists CagL variants, and the partial alignments of CagL amino acid changes (AACs) and their locations are shown in Fig. 1. In particular, we characterized AACs present in 10 clinical H. pylori isolates derived from GC patients.
Partial alignment of CagL sequences from 40 isolates from patients with different clinical outcomes. A partial alignment of CagL sequences (aa 1–150 and 201–237) is shown. The 40 clinical isolates included 15 from chronic gastritis (CG), 8 from gastric ulcer (GU), 7 from duodenal ulcer (DU) and 10 from gastric cancer (GC) patients. The amino acid sequence of the H. pylori reference strain ATCC 26695 is shown on the top line. Tyr58, Glu59, RGD motifs (76–78), Ala141, Glu142, Asn201, and C-terminal motifs of Ser-Lys-Ile-Ile-Val-Lys (232–237) are marked in grey blocks. Sequences of 10 isolates from GC patients are indicated in red
More recently, the CagL variants Tyr58 and/or Glu59 (CagL-Y58E59) was found to occur at significantly higher rates in H. pylori isolates from Taiwanese GC patients. CagL-Tyr58Glu59 can induce higher integrin α5β1 expression levels in the upper stomach and increase inflammation in the corpus [21]. Consistent with this report, we found that CagL-Glu59 occurred at a significantly (p < 0.001) higher rate (7/10, 70.0%) in H. pylori isolates from GC patients compared to that for 30 H. pylori isolates from non-GC patients (4/30, 13.3%). Shown in Table 3, this association between CagL-Glu59 and clinical outcome was still significant with the exception of 5 Vietnamese isolates from GC patients (p < 0.05). The remaining 26 isolates from non-GC patients had Lys59 (K59), and all 15 isolates from DU and GU patients had the CagL-Lys59 variant. In contrast, the reference H. pylori strain ATCC 26695 carried CagL-Glu59.
Meanwhile, CagL-Tyr58 was present in only one isolate (HZ67) from a GC patient, and its frequency was not significant. Aspartic acid was the most commonly present amino acid at position 58 (Asp58), and occurred in 38 of 40 isolates (95.0%). The remaining isolate (F32) had CagL-Asn58, as did the reference strain ATCC 26695. Only one isolate (HZ67) among the 43 tested had a CagL sequence with both Tyr58 and Glu59.
The C-terminal motifs that include the most distal amino acids of both CagL and CagI are functionally important for the TFSS [22]. In CagL, the sequence of this motif is Ser-Lys-Ile-Ile-Val-Lys (232–237). In this study, we found that Ile234 occurred at a significantly (p = 0.018) higher rate in GC patients (7/10 isolates, 70.0%) relative to that for non-GC patients (23.3%, 7/30). However, Ile234 lost significance with the exception of 5 Vietnamese isolates from GC patients (Table 3). The other five residues showed no significant amino acid sequence differences among the disease outcomes tested.
For CagL, Ala141 and Glu142 variants occurred in all 5 isolates isolated from Vietnamese GC patients. Asp201 had a significantly (p = 0.006) lower frequency in GC patients (3/10 isolates, 30.0%) compared to that for isolates from non-GC patients (24/30, 80.0%). However, Ala141, Glu142, and Asp201 variants lost significance when 5 Vietnamese isolates were excluded (Table 3). Notably, the Arg-Gly-Asp (RGD) motif was well conserved in 39 of 40 isolates (97.5%), but there were no significant differences among disease outcomes.
CagI variants in patients with different clinical disease outcomes
We also translated CagI nucleotide sequences into amino acid sequences (1–381), and analyzed rates and locations of CagI variants based on clinical disease outcomes (Table 4; Fig. 2).
Partial alignment of CagI sequences from 40 isolates from patients with different clinical outcomes. A partial alignment of CagI sequences (aa 101–120, 221–270, and 361–380) is shown. The 40 clinical isolates included 15 from chronic gastritis (CG), 8 from gastric ulcer (GU), 7 from duodenal ulcer (DU), and 10 from gastric cancer (GC) patients. The amino acid sequence of the H. pylori reference strain ATCC 26695 is shown on the top line. Val109, Ile262, and the Ser-Lys-Val-Ile-Val-Lys (376–381) C-terminal motif are marked by grey blocks. The results for 10 isolates from GC patients are indicated in red
As with CagL, the C-terminal motif of Ser-Lys-Val-Ile-Val-Lys (376–381) in CagI is functionally essential for the TFSS. In our analysis, all 40 H. pylori isolates from both GC and non-GC patients had the same motif, which had a completely conserved sequence.
Valine at CagI amino acid residue 109 (Val109) was frequent in H. pylori isolates from both GC patients (8/10, 80.0%) and non-GC patients (18/30, 60.0%). Isoleucine at position 262 (Ile262) was similarly frequent in GC patients (8/10, 80%) and non-GC patients (16/30, 53.3%), and the difference in rates was not significant. There were no other AACs associated with clinical outcome in the CagI sequence.
Phylogenetic implications of H. pylori CagL and CagI diversity
Phylogenetic trees were conducted using MEGA7 [23]. In general, CagL sequences showed no characteristic clusters around disease outcomes (Fig. 3a), although there was a cluster among the five Vietnamese isolates (Fig. 3b). Meanwhile, CagI sequences had no characteristic clusters for either region or disease outcome (Fig. 3c, d).
Phylogenetic tree of 40 clinical isolates based on CagL and CagI sequences. Neighbor-Joining tree analysis of concatenated CagL (a, b) and CagI (c, d) sequences for 40 isolates is shown. Each Neighbor-Joining method tree was made using MEGA7 software. Open square, open circle, filled circle, and filled triangle symbols correspond to isolates derived from gastric cancer patients, Japanese isolates, Chinese isolates, and Vietnamese isolates, respectively
Discussion
Using the advantages provided by whole-genome sequencing (WGS), we analyzed candidate and novel variants of CagL and CagI proteins in 40 clinical H. pylori isolates from patients in Southeast Asia. We showed that CagL from H. pylori isolates derived from GC patients carried several specific amino acid changes (AACs), but we detected no significant changes in the CagI amino acid sequence.
Whole-genome sequencing technology was recently applied to clarify the pathogenicity and evolution of H. pylori, as well as to identify its virulence factors [24, 25]. Using WGS, we and others detected potential mutations throughout the H. pylori genome and identified variants when sequence changes were present [20, 24, 25]. Here, we used WGS technology to detect novel variants in uncharacterized cagPAI genes associated with H. pylori pathogenicity.
cagPAI is a 37 kb segment of H. pylori DNA that contains 28 genes [3, 4], and is found in about 60% of Western isolates, whereas nearly all East Asian isolates are cagPAI positive [26]. We analyzed cagPAI integrity and showed the rearrangement of this island in three Japanese isolates (189, 194 and F51). Although cagPAI was most intact in Japanese isolates, it was disrupted in isolates isolated throughout the world [27]. Since the pathogenic role of the cagPAI is well defined as a whole or in part, we excluded the three isolates that had cagPAI rearrangements.
Several Cag proteins have been detected as constituents of the H. pylori cag TFSS apparatus and have important roles in CagA translocation [14, 15, 22]. The CagL and CagI proteins have been previously characterized [16, 17], so in this study we used WGS to screen 40 clinical H. pylori isolates for CagL and CagI variants, and analyzed the relationship between amino acid sequence and clinical outcomes. Consistent with a previous report [21], we detected complete RGD motifs in CagL sequences from all isolates. These data highlight the importance of the RGD motif for CagL function in the TFSS. We also checked whether other AACs in CagL and CagI were correlated with clinical outcomes.
We further confirmed that the frequency of the candidate variant CagL-Glu59 in GC patients significantly differed from that seen for isolates from non-GC patients. This association of Glu59 was still significant with the exception of 5 Vietnamese isolates, which was the half of all GC isolates (5/10). However, the frequency of Tyr58 was not significantly different, which is in contrast to a previous study that showed the CagL-Tyr58Glu59 variants were more common in H. pylori isolates from GC patients [21]. CagL-Tyr58Glu59 variants have strong binding affinity for integrin α5β1 and also promote increased expression of this integrin, and significantly enhances CagA translocation and phosphorylation relative to wild type CagL [28]. However, these results contrasted with those shown by Tegtmeyer et al. [29]. Our data support the importance of CagL-Glu59 variant, and imply that Glu59 could be incorporated into strategies to screen clinical H. pylori isolates. However, the current study is rather small and limited to the patients in Southeast Asia. These results require validation with larger isolates in Southeast Asia and the other samples in Western countries.
The C-terminal motif in CagL and CagI consisting of six amino acids (Ser-Lys-Ile-Ile-Val-Lys, and Ser-Lys-Val-Ile-Val-Lys, respectively) is important for TFSS function [22]. However, whether these sequences were conserved among genomes of clinical H. pylori isolates was unclear. Here, we showed that the CagI C-terminal motif was completely conserved. Although the CagL C-terminal motif was also well conserved, we found a significant difference at position 234 of CagL among H. pylori isolates derived from GC and non-GC patients. However, Ile234 lost significance with the exception of 5 Vietnamese isolates from GC patients. Future studies on additional H. pylori isolates could validate whether CagL-Ile234 could serve as a marker that indicates an increased risk for gastric carcinogenesis.
Conclusions
We analyzed genetic variants of H. pylori using WGS, which has significant advantages over other approaches that examine only a fraction of the genome at any one time. WGS identified several putative novel variants of CagL and CagI sequences from previously uncharacterized H. pylori isolates. These variants, particularly in CagL-Glu59, have the possible effect on the TFSS activity and the relevance with clinical outcomes.
Methods
H. pylori samples
Forty-three H. pylori clinical isolates were obtained from gastric epithelium biopsy tissues taken during upper gastroduodenal endoscopy procedures performed at Okinawa Prefectural Chubu Hospital, Kobe University Hospital, and Fukui University Hospital in Japan, as well as Zhejiang University Hospital in China and Cho Ray Hospital in Vietnam. All patients gave written informed consent for use of their samples in this study, which was performed according to the principles of the Declaration of Helsinki. The major reference strain, ATCC 26695 (NC_000915), was isolated from CG patients in the United Kingdom [30], and its sequence served as the reference sequence.
H. pylori culture
Gastric biopsy specimens were first inoculated onto trypticase soy agar II (TSA-II)-5% sheep blood plates (Becton, Dickinson and Company: BD) and cultured under microaerophilic conditions (O2 5%; CO2 5%; N2 90%) at 37 °C for 3–5 days. Then, one colony was picked from each primary culture plate, and seeded onto a Columbia Helicobacter pylori agar plate containing vancomycin (10 mg/l), trimethoprim (5 mg/l), amphotericin B (5 mg/l), and polymyxin B (2500 units/l), and cultured under the same conditions. A colony was picked from this second plate, seeded onto a TSA-II plate, and cultured under the same conditions. Several colonies were picked from the third plate, transferred into Brucella Broth medium (2 ml) containing 10% fetal calf serum, and cultured for 18 h under the same conditions.
A portion of each culture was stored at −80 °C in 0.01 M phosphate-buffered saline (PBS), pH 7.4, containing 20% glycerol. H. pylori DNA was extracted from bacterial pellets prepared from liquid cultures using the protease–phenol–chloroform method. The extracted DNA was suspended in 100 μl distilled water and stored at 4 °C.
Whole-genome sequencing (WGS)
Total DNA of H. pylori isolated from patients and the reference strain ATCC 26695 were sequenced. The bacterial DNA concentration of each sample was measured with a Qubit dsDNA HS assay kit (Q32851; Invitrogen, Carlsbad, CA) and the concentration of each sample was between 250 and 320 pg/μl.
A DNA library of H. pylori isolates was prepared using a Nextera XT DNA Sample Prep Kit (Illumina, Carlsbad, CA), which was used according to the manufacturer’s instructions to uniformly shear the DNA into 500 bp fragments and add unique adapter sequences to the fragments. The resulting DNA library was run on a MiSeq sequencer (Illumina) with a reagent kit (300 cycle, paired-end). Fluorescence images were analyzed using MiSeq Control Software, and FASTQ-formatted sequence data were generated using MiSeq Reporter Analysis.
Sequence read mapping and single nucleotide variant (SNV) detection
For the analyzed DNA sequence data, read qualities having a Q30 value above 80% were selected according to recommendations by Illumina. After a quality check and data trimming, the sequence reads were assembled with Genomics Workbench 8.5.1 (CLC bio, Aarhus, Denmark). The read mapping module was termed as CLC Assembly Cell 4.0, which was based on an uncompressed Suffix-Array representing the entire reference genome in a single data structure (White paper on CLC read mapper; October 10, 2012). Sequence reads were mapped against the ATCC 26695 genome (NC_000915) as a reference, and single nucleotide variants (SNVs) were identified with Fixed Ploidy Variant Detection modules with default parameters and minor modifications to the mapping algorithm. Variant detection of the software was set to 1.
To exclude false-positive variants that resulted from sequencing errors, we selected variants that were present in >90.0% of mapped reads with a minimum coverage of 100. Insertions, deletions, and successive multi nucleotide variants were also excluded due to the previously reported complexity involved in detecting true variants [18].
Phylogenetic analysis
We constructed a phylogenetic tree from CagL and CagI sequences of H. pylori isolates using Molecular Evolutionary Genetics Analysis version 7.0 (MEGA7) [23]. Evolutionary history was inferred using the Neighbor-Joining tree [31]. Trees were drawn to scale, wherein branch lengths are shown in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The analysis involved 40 isolates, and the CagL and CagI sequences included 237 and 381 amino acids, respectively.
Statistical analysis
Differences in the number of amino acid changes (AAC) in CagL and CagI in clinical outcomes and regions in Southeast Asia were compared using the Fisher’s exact test. A difference associated with a p value <0.05 was considered to be significant. The SPSS statistical software package version 23.0.0.0 (SPSS, Inc., Chicago, IL) was used for all statistical analyses.
Nucleotide sequence accession number
Sequence reads of 19 Japanese clinical isolates and ATCC 26695 were previously deposited in the DNA Data Bank of Japan Sequence Read Archive (http://www.ddbj.nig.ac.jp/index-e.html) under accession number DRA001250. Sequence reads of 5 Vietnamese clinical isolates were deposited under accession number DRA002946, whereas 7 Chinese isolates and an additional 12 Japanese isolates were deposited under DRA004713.
Abbreviations
- H. pylori :
-
Helicobacter pylori
- WGS:
-
whole-genome sequencing
- SNVs:
-
single nucleotide variants
- AACs:
-
amino acid changes
- TFSS:
-
type-IV secretion system
- cag:
-
cytotoxin-associated gene
- CG:
-
chronic gastritis
- GU:
-
gastric ulcer
- DU:
-
duodenal ulcer
- GC:
-
gastric cancer
References
Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–86.
Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–73.
Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42:1337–48.
Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069.
Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683–6.
Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300:1430–4.
Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci U S A. 2002;99:14428–33.
Naito M, Yamazaki T, Tsutsumi R, Higashi H, Onoe K, Yamazaki S, Azuma T, Hatakeyama M. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. Gastroenterology. 2006;130:1181–90.
Sheu BS, Sheu SM, Yang HB, Huang AH, Wu JJ. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection. Gut. 2003;52:927–32.
Mizushima T, Sugiyama T, Komatsu Y, Ishizuka J, Kato M, Asaka M. Clinical relevance of the babA2 genotype of Helicobacter pylori in Japanese clinical isolates. J Clin Microbiol. 2001;39:2463–5.
Yamaoka Y, Souchek J, Odenbreit S, Haas R, Arnqvist A, Boren T, Kodama T, Osato MS, Gutierrez O, Kim JG, Graham DY. Discrimination between cases of duodenal ulcer and gastritis on the basis of putative virulence factors of Helicobacter pylori. J Clin Microbiol. 2002;40:2244–6.
Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, Konig W, Backert S. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449:862–6.
Barden S, Lange S, Tegtmeyer N, Conradi J, Sewald N, Backert S, Niemann HH. A helical RGD motif promoting cell adhesion: crystal structures of the Helicobacter pylori type IV secretion system pilus protein CagL. Structure. 2013;21(11):1931–41.
Bonsor DA, Pham KT, Beadenkopf R, Diederichs K, Haas R, Beckett D, Fischer W, Sundberg EJ. Integrin engagement by the helical RGD motif of the Helicobacter pylori CagL protein is regulated by pH-induced displacement of a neighboring helix. J Biol Chem. 2015;290(20):12929–40.
Jimenez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5:e1000684.
Pham KT, Weiss E, Jimenez Soto LF, Breithaupt U, Haas R, Fischer W. CagI is an essential component of the Helicobacter pylori Cag type IV secretion system and forms a complex with CagL. PLoS ONE. 2012;7:e35341.
Kumar N, Shariq M, Kumari R, Tyagi RK, Mukhopadhyay G. Cag type IV secretion system: CagI independent bacterial surface localization of CagA. PLoS ONE. 2013;8:e74620.
Selbach M, Moese S, Meyer TF, Backert S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. Infect Immun. 2002;70:665–71.
Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5.
Iwamoto A, Tanahashi T, Okada R, Yoshida Y, Kikuchi K, Keida Y, Murakami Y, Yang L, Yamamoto K, Nishiumi S, Yoshida M, Azuma T. Whole-genome sequencing of clarithromycin resistant Helicobacter pylori characterizes unidentified variants of multidrug resistant efflux pump genes. Gut Pathog. 2014;6:27.
Yeh YC, Chang WL, Yang HB, Cheng HC, Wu JJ, Sheu BS. H. pylori cagL amino acid sequence polymorphism Y58E59 induces a corpus shift of gastric integrin alpha5beta1 related with gastric carcinogenesis. Mol Carcinog. 2011;50:751–9.
Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 2011;7:e1002237.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Lu W, Wise MJ, Tay CY, Windsor HM, Marshall BJ, Peacock C, Perkins T. Comparative analysis of the full genome of Helicobacter pylori isolate Sahul64 identifies genes of high divergence. J Bacteriol. 2014;196:1073–83.
Lehours P, Vale FF, Bjursell MK, Melefors O, Advani R, Glavas S, Guegueniat J, Gontier E, Lacomme S, Alves Matos A, Menard A, Mégraud F, Engstrand L, Andersson AF. Genome sequencing reveals a phage in Helicobacter pylori. MBio. 2011;2(6). doi:10.1128/mBio.00239-11.
Sahara S, Sugimoto M, Vilaichone RK, Mahachai V, Miyajima H, Furuta T, Yamaoka Y. Role of Helicobacter pylori cagA EPIYA motif and vacA genotypes for the development of gastrointestinal diseases in Southeast Asian countries: a meta-analysis. BMC Infect Dis. 2012;12:223.
Kauser F, Khan AA, Hussain MA, Carroll IM, Ahmad N, Tiwari S, Shouche Y, Das B, Alam M, Ali SM, Habibullah CM, Sierra R, Megraud F, Sechi LA, Ahmed N. The cag pathogenicity island of Helicobacter pylori is disrupted in the majority of patient isolates from different human populations. J Clin Microbiol. 2004;42:5302–8.
Yeh YC, Cheng HC, Yang HB, Chang WL, Sheu BS. H. pylori CagL-Y58/E59 prime higher integrin alpha5beta1 in adverse pH condition to enhance hypochlorhydria vicious cycle for gastric carcinogenesis. PLoS ONE. 2013;8:e72735.
Tegtmeyer N, Lind J, Schmid B, Backert S. Helicobacter pylori CagL Y58/E59 mutation turns-off type IV secretion-dependent delivery of CagA into host cells. PLoS ONE. 2014;9:e97782.
Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–47.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.
Authors’ contributions
HO, TT, AI, and RO conceived and designed the research. HO, AI, RO, and KY collected samples and performed experiments. HO, TT, AI, and RO analyzed the data and prepared figures, interpreted results of the experiments, and drafted the manuscript. HO and TT edited the manuscript. SN, MY, and TA supervised the study. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files. Sequence reads for the 43 H. pylori clinical isolates and ATCC 26695 were deposited in the DNA Data Bank of Japan Sequence Read Archive (http://www.ddbj.nig.ac.jp/index-e.html).
Funding
This study was supported by a Grant-in-Aid (No. 26460212) to T. T., a Grant-in-Aid (No. 15H06404) to A. I., a Grant-in-Aid for Young Scientists (B) (No. 15K19092) to K. Y., and a Grant-in-Aid (No. 16H05835) to T. A. from the Japan Society for the Promotion of Science (JSPS).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
13099_2017_165_MOESM1_ESM.docx
Additional file 1: Table S1. Average coverage (fold) of 28 cagPAI genes in 43 clinical H. pylori isolates mapped to the ATCC26695 sequence.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ogawa, H., Iwamoto, A., Tanahashi, T. et al. Genetic variants of Helicobacter pylori type IV secretion system components CagL and CagI and their association with clinical outcomes. Gut Pathog 9, 21 (2017). https://doi.org/10.1186/s13099-017-0165-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-017-0165-1