Abstract
Background
We previously showed that zonula occludens toxin (Zot) encoded by Campylobacter concisus zot 808T gene has the potential to initiate inflammatory bowel disease. This Zot protein caused prolonged intestinal epithelial barrier damage, induced intestinal epithelial and macrophage production of tumor necrosis factor-α and enhanced the responses of macrophages to other microbes. In order to understand the potential virulence of Zot proteins in other Campylobacter species, in this study we examined their presence, similarities, motifs and prophages.
Methods
The presence of Zot proteins in Campylobacter species was examined by searching for the Zot family domain in multiple protein databases. Walker A and Walker B motifs in Zot proteins were identified using protein sequence alignment. A phylogenetic tree based on Campylobacter zot genes was constructed using maximum-likelihood method. Campylobacter Zot proteins were compared using protein sequence alignment. The zot-containing prophages in Campylobacter species were identified and compared with known prophage proteins and other viral proteins using protein sequence alignment and protein BLAST.
Results
Twelve Zot proteins were found in nine Campylobacter species/subspecies. Among these Campylobacter species, three species had two Zot proteins and the remaining six species/subspecies had one Zot protein. Walker A and Walker B motifs and a transmembrane domain were found in all identified Campylobacter Zot proteins. The twelve Campylobacter zot genes from the nine Campylobacter species/subspecies formed two clusters. The ZotCampyType_1 proteins encoded by Cluster 1 Campylobacter zot genes showed high similarities to each other. However, ZotCampyType_2 proteins encoded by Cluster 2 Campylobacter zot genes were more diverse. Furthermore, the zot-containing Campylobacter prophages were identified.
Conclusion
This study reports the identification of two types of Campylobacter Zot proteins. The high similarities of ZotCampyType_1 proteins suggest that they are likely to have similar virulence. ZotCampyType_2 proteins are less similar to each other and their virulent properties, if any, remain to be examined individually.
Similar content being viewed by others
Background
Campylobacter concisus is a Gram-negative spiral shaped motile bacterium [1]. Their growth under both anaerobic and microaerobic conditions is largely determined by the presence of H2 [2]. This bacterium usually colonizes the human oral cavity [3, 4]. However, it may also colonize the intestinal tract of some individuals and its prevalence in intestinal biopsies has been associated with human inflammatory bowel disease (IBD) [5–9]. Translocation of C. concisus from the oral cavity to intestinal tract has been suggested to be a cause of a subgroup of IBD [10]. Campylobacter concisus has also been suggested to be involved in diarrheal disease due to the frequent isolation of this bacterium from diarrheal stool samples [8, 11–13].
Campylobacter concisus zonula occludens toxin (Zot) is encoded by the zot gene located in CON_phi2 prophage [14, 15]. The zot genes in different C. concisus strains have polymorphisms [14]. In a recent study, we examined the effects of C. concisus Zot encoded by zot 808T gene on human intestinal epithelial cells and macrophages using cell line models. In that study, we found that C. concisus Zot caused prolonged intestinal epithelial barrier damage, induced intestinal epithelial and macrophage production of proinflammatory cytokines such as tumor necrosis factor-α, and enhanced the responses of macrophages to Escherichia coli [16]. These data suggest that C. concisus Zot may play a role in initiating chronic intestinal inflammatory conditions such as IBD through damaging the intestinal barrier and enhancing the immune responses to luminal microbes.
In addition to C. concisus, four additional Campylobacter species including Campylobacter ureolyticus, Campylobacter corcagiensis, Campylobacter gracilis and Campylobacter iguaniorum were recently reported to possess the zot gene [17–21]. The similarities of Zot proteins in different Campylobacter species have not been examined. In this study, we examined Zot proteins and their genes in Campylobacter species, which revealed two clusters of Campylobacter zot genes and two types of Campylobacter Zot proteins. Furthermore, we identified the prophages that contain zot genes in Campylobacter species.
Methods
Examination of the presence of Zot proteins in Campylobacter species
The presence of Zot proteins in Campylobacter species was examined by searching for proteins in Campylobacter species that have the Zot family domain in multiple protein databases including NCBI protein database, InterPro and Pfam [22–24]. The Zot proteins in these databases were annotated based on the presence of Zot family domain.
Examination of the presence of Walker A and Walker B in Campylobacter Zot proteins
It is shown in the InterPro database that the Zot family proteins (InterPro entry identity: IPR008900) belong to p-loop containing nucleoside triphosphate hydrolase (p-loop NTPase) superfamily. The proteins of p-loop NTPase superfamily have Walker A and Walker B motifs [25]. In this study, we examined the presence of Walker A and Walker B motifs in Campylobacter Zot proteins by protein alignment using Clustal Omega software [26]. The Walker A motif has a sequence of GxxxxGK[S/T], where x is any residue and the Walker B motif has a sequence of hhhh[D/E], where h is a hydrophobic residue [25].
Generation of a phylogenetic tree based on zot genes in Campylobacter species
To examine the genetic relationship between the zot genes in different Campylobacter species, the nucleotide sequences of the zot genes in Campylobacter species identified above were obtained from NCBI genome database. These sequences were used to generate a phylogenetic tree using the maximum likelihood method implemented in molecular evolutionary genetics analysis software version 6.0 [27]. In order to differentiate the zot genes in different Campylobacter species, the zot genes found in different Campylobacter species were indicated by the last four digits of their locus tag numbers.
Comparison of zot-containing prophages in Campylobacter species
To examine whether the zot genes in different Campylobacter species are carried by prophages similar to that in C. concisus, the zot-containing prophages in these Campylobacter species were identified by examination of the genes adjacent to their zot genes. The prophages were defined based on the presence of integrase, hypothetical proteins and attachment sites [28]. The attachment sites were identified by the presence of repetitive sequences located at both ends of the prophages [28].
The proteins in the identified Campylobacter prophages in this study were compared with the corresponding proteins in C. concisus prophages CON_phi2 and CON_phi3. The comparison was performed using Clustal Omega [26]. Proteins sharing more than 40 % sequence identity were considered to have high similarities and were recorded [29].
Comparison of the proteins of zot-containing prophages in Campylobacter species with other viral proteins
To examine whether the zot containing prophages identified in Campylobacter species are similar to previously reported prophages, proteins of Campylobacter prophages were compared with viral proteins in the NCBI non-redundant protein sequence database (taxonomy identity for viruses: 10,239) using protein BLAST with default settings [22]. The identified viral proteins with E-values lower than 10 were noted. For prophage proteins which shared sequence similarities with multiple viral proteins, the viral proteins with the lowest E-values were noted. The full length sequences of the identified viral proteins were then aligned with the Campylobacter prophage proteins using Clustal Omega [26]. The protein identities were calculated by dividing the number of identical amino acids by the total number of amino acids in proteins from Campylobacter prophages. The Campylobacter prophage proteins were also compared with viral proteins in the virus pathogen database and analysis resource (ViPR) using BLAST with a cut-off E-value of 10. In addition to C. concisus, the zot gene was also found in other bacterial species such as Vibrio cholerae and Neisseria meningitis [30, 31]. The identities between Campylobacter Zot, V. cholerae Zot and N. meningitidis Zot proteins were also compared in this study.
Prediction of secreted proteins and transmembrane proteins in Campylobacter prophages
Secreted proteins in zot-containing prophages in different Campylobacter species were predicted using SignalP 4.1 and SecretomeP 2.0. The software SignalP 4.1 predicts secreted proteins based on the presence of the signal peptide at the N-terminus [32]. The software SecretomeP 2.0 predicts non-classical (not signal peptide triggered) protein secretion based on analysis of post-translational and localizational aspects of the proteins [33]. Transmembrane proteins were predicted using the software Phobius [34].
Results
The Zot proteins in different Campylobacter species and their Walker A and Walker B motifs
Twelve Zot proteins were found in nine Campylobacter species/subspecies. Three Campylobacter species including C. concisus, C. ureolyticus and C. corcagiensis had two Zot proteins and the remaining six Campylobacter species/subspecies including C. gracilis, Campylobacter jejuni subsp. doylei, Campylobacter jejuni subsp. jejuni, Campylobacter hyointestinalis subsp. hyointestinalis, C. hyointestinalis subsp. lawsonii and C. iguaniorum had one Zot protein (Table 1).
The Zot family domains were localized at the N-terminal side of the identified Campylobacter Zot proteins, prior to the transmembrane domains. The Zot family proteins had p-loop NTPase domains and the entry identity for p-loop NTPase domain was IPR027417 in InterPro database. We identified the Walker A and Walker B motifs in Campylobacter Zot proteins, which were at the N-terminal side of the Zot proteins (Fig. 1).
Walker A and walker B motifs in Campylobacter Zot proteins. Campylobacter Zot proteins have a transmembrane domain (underlined). The amino acids prior to the transmembrane domains constitute the Zot family domains (approximately 1–210, shaded in grey) in different Campylobacter Zot. The Zot family domains belong to p-loop NTPase superfamily. Walker A and walker B motifs are in the N-terminus of Campylobacter Zot proteins. Walker A has a sequence of GxxxxGK[S/T], where x is any residue. Walker B motif has a sequence of hhhh[D/E], where h is a hydrophobic residue [25]
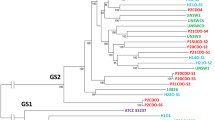
The phylogenetic tree formed based on zot genes in Campylobacter species
The Campylobacter zot genes formed two clusters. Cluster 1 contained three zot genes, including C. concisus zot2276, C. ureolyticus zot3935 and C. corcagiensis zot6485. Cluster 2 contained nine Campylobacter zot genes (Fig. 2).
The phylogenetic tree generated based on zot genes in different Campylobacter species. Maximum likelihood method was used to generate the phylogenetic tree. Bootstrap values were generated from 1000 replicates. Cluster 1 zot genes are shown in bold. The zot gene from Neisseria meningitides (strain 69166) was used as the outgroup
Comparison of ZotCampyType_1 and ZotCampyType_2 proteins
The Zot proteins encoded by Cluster 1 and Cluster 2 Campylobacter zot genes were referred to as ZotCampyType_1 and ZotCampyType_2 proteins respectively. The three ZotCampyType_1 proteins had high similarities; they shared 171 identical amino acids and 77 conservative mutations (Fig. 3). The ZotCampyType_2 proteins were less similar to each other as compared to ZotCampyType_1 proteins; they had 71 identical amino acids and 65 conservative mutations (Fig. 4).
The zot-containing prophages in different Campylobacter species and their similarities to C. concisus prophages CON_phi2 or CON_phi3
We identified the zot-containing prophages in different Campylobacter species. Each of these prophages began with an integrase and had a number of hypothetical proteins (Fig. 5; Additional file 1). The attachment sites were found (Table 2). These prophages were inserted within either tRNA-Met or tRNA-Ser genes, except for URE_phiZB, which was inserted into tRNA-Leu gene (Table 2). For the prophage in C. iguaniorum, two tRNA genes were found after the integrase, suggesting that multiple insertions have occurred.
Schematic illustration of protein similarities in zot-containing Campylobacter prophages. a Campylobacter prophages containing ZotCampyType_1 proteins. b Campylobacter prophages containing ZotCampyType_2 proteins. The prophages and their host Campylobacter strains (in bracket) are listed at the left side of the figure. Proteins with more than 40 % identical amino acids with proteins in CON_phi2 or CON_phi3 were labeled with the same color. The numbers above the proteins are locus tags of the genes in the NCBI database. Int indicates integrase. Asterisk and Hashtag indicate proteins predicted to be secreted via classic secretory pathway or non-classic secretory pathway respectively. Caret indicates transmembrane proteins. Dollar indicates multiple insertion sites for CON_phi prophages in C. concisus 13826 in which only the first attachment site (for CON_phi1) overlapped with tRNA [15]. Filled triangle indicates attachment sites
Prophages containing ZotCampyType_1 proteins had high similarities to CON_phi2. Eight proteins in URE_phiZA and nine proteins in COR_phiZA were found to have more than 40 % identities (41-73 %) with proteins in CON_phi2 (Fig. 5a; Additional file 1). Proteins in prophages containing ZotCampyType_2 proteins were more diverse. However, three to five proteins in these prophages had more than 40 % identities with that in CON_phi3 (40–72 %) (Fig. 5b; Additional file 1).
The similarities between proteins in Campylobacter prophages and viral proteins
The zot-containing Campylobacter were compared with known viral proteins in NCBI non-redundant protein sequence database. The proteins within each individual prophage showed low similarities to multiple phage proteins, except for CCC13826_0188 in CON_phi3 that shared 43 % identity with a phage transferase from an uncultured phage (Additional file 2). The Zot proteins in Campylobacter prophages had low similarities with the Zot proteins in V. cholerae and N. meningitidis (15–21 %) (Additional files 3, 4). None of the Campylobacter prophage proteins shared significant similarities with viral proteins in ViPR database. These data suggest that the zot-containing prophages in Campylobacter species are new prophages that have not been previously characterized.
Secreted and transmembrane proteins in Campylobacter prophages
Proteins secreted via both classical secretory pathway (with signal peptides) and non-classical secretory pathway (without signal peptides), as well as transmembrane proteins were found in all Campylobacter prophages (Fig. 5).
Discussion
In addition to C. concisus, a number of other Campylobacter species were recently reported to have the zot genes [17–21]. In this study, we found that Campylobacter zot genes formed two clusters (Fig. 2). Most of the Campylobacter zot genes were in Cluster 2, and Cluster 1 contained only three zot genes. The three Campylobacter species that had Cluster 1 zot genes also contained Cluster 2 zot genes. The remaining six Campylobacter species/subspecies contained Cluster 2 zot genes only. These data show that Cluster 2 zot gene is more prevalent in Campylobacter species as compared to Cluster 1 zot genes.
ZotCampyType_1 proteins, which were encoded by Cluster 1 zot genes, were highly similar to each other. However they were less similar to ZotCampyType_2 proteins that were encoded by Cluster 2 Campylobacter zot genes. The zot gene in CON_phi2 prophage in C. concisus belongs to ZotCampyType_1. Using cell culture models, we previously showed that ZotCampyType_1 in C. concisus encoded by zot 808T polymorphism damaged intestinal epithelial barrier by induction of epithelial apoptosis and induced production of proinflammatory cytokines such as TNF-α in HT-29 cells and THP-1 macrophage-like cells, supporting its role as a potential virulence factor [16]. The high similarities between ZotCampyType_1 proteins in the three different Campylobacter species suggest that they may have similar effects on human cells. Great variations in protein sequences between ZotCampyType_1 and ZotCampyType_2 proteins as well as within ZotCampyType_2 proteins were observed in this study. Given this, the effects of ZotCampyType_2 proteins on human cells, if any, require to be examined individually.
A transmembrane domain was found in all Zot proteins, showing that Zot proteins are transmembrane proteins. Furthermore, all Zot proteins contained Walker A and Walker B motifs, which are conserved motifs of p-loop NTPase superfamily [25]. P-loop NTPase bind to NTP typically ATP or GTP through the Walker A and B motifs, which are involved in diverse cellular functions [25]. Future studies should be conducted to examine whether Campylobacter Zot proteins have NTPase activities.
In this study, we identified a number of zot-containing prophages in other Campylobacter species in additional to previous reported prophages in C. concisus (Fig. 5). These prophages have an integrase, a number of hypothetical proteins and attachment sites (Fig. 5; Table 2; Additional file 1), which satisfy the previously defined criteria for prophages [28]. The proteins in individual Campylobacter prophages identified in this study have low similarities with multiple viral proteins, suggesting that they are new prophages that have not being characterized previously (Additional file 2).
Campylobacter Zot proteins had very low similarities to V. cholerae Zot and N. meningitis Zot proteins (Additional files 3, 4). These data showed that despite having a common name, the amino acid sequences of Zot proteins in different bacterial species vary greatly. Thus, they may not necessarily exhibit the same effects on human cells.
Conclusions
This study reports the identification of two types of Campylobacter Zot proteins. The high similarities of ZotCampyType_1 proteins suggest that they are likely to have similar virulence. ZotCampyType_2 proteins were less similar to each other and their virulent properties, if any, remain to be examined individually. This study provides useful information for further examination of Campylobacter Zot proteins as potential virulence factors.
Abbreviations
- IBD:
-
inflammatory bowel disease
- Zot:
-
zonula occludens toxin
- ZotCampyType_1 :
-
Campylopbacter Zot protein encoded by Cluster 1 zot gene
- ZotCampyType_2 :
-
Campylopbacter Zot protein encoded by Cluster 2 zot gene
- p-loop NTPase:
-
p-loop containing nucleoside triphosphate hydrolase
References
Lastovica AJ, On SL, Zhang L. The family Campylobacteraceae—the prokaryotes. New York: Springer; 2014. p. 307–35.
Lee H, Ma R, Grimm MC, Riordan SM, Lan R, Zhong L, Raftery M, Zhang L. Examination of the anaerobic growth of Campylobacter concisus strains. Int J Microbiol. 2014;2014:476047.
Zhang L, Budiman V, Day AS, Mitchell H, Lemberg DA, Riordan SM, Grimm M, Leach ST, Ismail Y. Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J Clin Microbiol. 2010;48:2965–7.
Tanner ACR, Badger S, Lai C, Listgarten MA, Visconti RA, Socransky SS. Wolinella gen. nov., WoZinella succinogenes (Vibrio succinogenes Wolin et al.) comb. nov., and Description of bacteroides gracilis sp. nov., Wolinella recta sp. nov., Campylobacter concisus sp. nov., and Eikenella corrodens from humans with periodontal disease. Int J Syst Bacteriol. 1981;31(4):432–45.
Zhang L, Man SM, Day AS, Leach ST, Lemberg DA, Dutt S, Stormon M, Otley A, O’Loughlin EV, Magoffin A, et al. Detection and isolation of Campylobacter species other than C. jejuni from children with Crohn’s disease. J Clin Microbiol. 2009;47:453–5.
Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE. 2011;6:e21490–1.
Mahendran V, Riordan SM, Grimm MC, Tran TA, Major J, Kaakoush NO, Mitchell H, Zhang L. Prevalence of Campylobacter species in adult Crohn’s disease and the preferential colonization sites of Campylobacter species in the human intestine. PLoS ONE. 2011;6:e25417.
Lastovica AJ. Emerging Campylobacter spp.: the tip of the iceberg. Clin Microbiol Newsl. 2006;28:49–56.
Kirk KF, Nielsen HL, Thorlacius-Ussing O, Nielsen H. Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathog. 2016;8:27.
Zhang L. Oral Campylobacter species: initiators of a subgroup of inflammatory bowel disease? World J Gastroenterol. 2015;21:9239–44.
Lindblom G, Sjogren E, Hansson-Westerberg J, Kaijser B. Campylobacter upsaliensis, C. sputorum sputorum and C. concisus as common causes of diarrhoea in Swedish children. Scand J Infect Dis. 1995;27:187–8.
Nielsen H, Ejlertsen T, Engberg J, Nielsen H. High incidence of Campylobacter concisus in gastroenteritis in North Jutland, Denmark: a population-based study. Clin Microbiol Infect. 2013;19:445–50.
Kalischuk L, Inglis G. Comparative genotypic and pathogenic examination of Campylobacter concisus isolates from diarrheic and non-diarrheic humans. BMC Microbiol. 2011;11:53.
Mahendran V, Tan YS, Riordan SM, Grimm MC, Day AS, Lemberg DA, Octavia S, Lan R, Zhang L. The prevalence and polymorphisms of zonula occluden toxin gene in multiple Campylobacter concisus strains isolated from saliva of patients with inflammatory bowel disease and controls. PLoS ONE. 2013;8:e75525.
Zhang L, Lee H, Grimm MC, Riordan SM, Day AS, Lemberg DA. Campylobacter concisus and inflammatory bowel disease. World J Gastroenterol. 2014;20:1259–67.
Mahendran V, Liu F, Riordan S, Grimm M, Tanaka M, Zhang L. Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut Pathog. 2016;8:18.
Bullman S, Lucid A, Corcoran D, Sleator RD, Lucey B. Genomic investigation into strain heterogeneity and pathogenic potential of the emerging gastrointestinal pathogen Campylobacter ureolyticus. PLoS ONE. 2013;8:1.
Koziel M, Lucid A, Bullman S, Corcoran GD, Lucey B, Sleator RD. Draft genome sequence of Campylobacter corcagiensis strain CIT045T, a representative of a novel Campylobacter species isolated from lion-tailed macaques (Macaca silenus). Genome Announc. 2014;2:00248–314.
Miller WG, Yee E, On SL, Andersen LP, Bono JL. Complete genome sequence of the Campylobacter ureolyticus clinical isolate RIGS 9880. Genome Announc. 2015;3(6):01291–315.
Miller WG, Yee E. Complete genome sequence of Campylobacter gracilis ATCC 33236T. Genome Announc. 2015;3(6):e01087–115.
Miller WG, Yee E, Huynh S, Chapman MH, Parker CT. Complete genome sequence of Campylobacter iguaniorum strain RM11343, isolated from an alpaca. Genome Announc. 2016;4(3):646–716.
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9.
Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–21.
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–85.
Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–29.
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Liii WZ, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7(1):539.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol Biol Rev. 2003;67:238–76.
Pearson WR. An introduction to sequence similarity (“homology”) searching. Current Protocols in Bioinformatics. Hoboken: John Wiley & Sons Inc; 2013. p. 311–8.
Kaakoush NO, Man SM, Lamb S, Raftery MJ, Wilkins MR, Kovach Z, Mitchell H. The secretome of Campylobacter concisus. FEBS J. 2010;277:1606–17.
Fasano A, Baudry B, Pumplin DW, Wasserman SS, Tall BD, Ketley JM, Kaper JB. Vibrio cholerae produces a second enterotoxin, which affects intestinal tight junctions. Proc Natl Acad Sci USA. 1991;88:5242–6.
Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6.
Bendtsen JD, Kiemer L, Fausboll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58.
Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007;35:W429–32.
Jackson FL, Goodman YE. Bacteroides ureolyticus, a new species to accommodate strains previously identified as “Bacteroides corrodens, anaerobic”. Int J Syst Bacteriol. 1978;28:197–200.
Koziel M, O’Doherty P, Vandamme P, Corcoran GD, Sleator RD, Lucey B. Campylobacter corcagiensis sp. nov., isolated from faeces of captive lion-tailed macaques (Macaca silenus). Int J Syst Evol Microbiol. 2014;64:2878–83.
Miller WG, Yee E, Chapman MH. Complete genome sequences of Campylobacter hyointestinalis subsp. hyointestinalis strain LMG 9260 and C. hyointestinalis subsp. lawsonii strain LMG 15993. Genome Announc. 2016;4:00665–716.
Authors’ contributions
FL and HL conducted the bioinformatics analysis. LZ conceived the project. RL provided critical feedback on bioinformatics analysis. FL, LZ, HL and RL wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset supporting the conclusions of this article is included within the article (and its additional files).
Funding
This work is supported by a Faculty Research Grant awarded to Dr. Li Zhang from the University of New South Wales (Grant No: PS35329).
Author information
Authors and Affiliations
Corresponding author
Additional files
13099_2016_125_MOESM1_ESM.docx
Additional file 1. Protein identities between prophages in other Campylobacter species and C. concisus CON_phi2 and CON_phi3. Zot proteins are in bold. Integrase proteins are underlined #Identity: Percentage of identical amino acids (number of identical amino acids divided by number of amino acids in proteins from C. concisus 13826).
13099_2016_125_MOESM2_ESM.docx
Additional file 2. Comparison of Campylobacter prophage proteins with known viral proteins. #Identity: Percentage of identical amino acids (number of identical amino acids divided by number of amino acids in proteins from Campylobacter species).
13099_2016_125_MOESM3_ESM.docx
Additional file 3. Comparison of Campylobacter Zot proteins with V. cholerae Zot and N. meningitidis Zot. #Identity: percentage of identical amino acids (number of identical amino acids. divided by number of amino acids of V. cholerae Zot or N. meningitidis Zot). V. cholerae Zot sequence Accession No. is AAF29547. N. meningitidis Zot sequence Accession No. is EJU63554.
13099_2016_125_MOESM4_ESM.docx
Additional file 4. Comparison of Zot proteins from Campylobacter species, N. meningitidis and V. cholerae. * indicates identical amino acids (shaded in red). : indicates conservative mutations (shaded in blue). .indicates semi-conservative mutations. Transmembrane domains are underlined. Walker A and walker B motifs in the N-terminus of Campylobacter Zot proteins were identified and boxed. Walker A has a sequence of GxxxxGK[S/T], where x is any residue. Walker B motif has a sequence of hhhh[D/E], where h is a hydrophobic residue [25].
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, F., Lee, H., Lan, R. et al. Zonula occludens toxins and their prophages in Campylobacter species. Gut Pathog 8, 43 (2016). https://doi.org/10.1186/s13099-016-0125-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-016-0125-1