Abstract
Background
Salmonella enterica serovar Enteritidis, the most common cause of human gastroenteritis, employs several virulence factors including lipopolysaccharide (LPS) for infection and establishment of disease inside the host. The LPS of S. enterica serovar Enteritidis consists of lipid A, core oligosaccharide and O-antigen (OAg). The OAg consists of repeating units containing different sugars. The sugars of OAg are synthesized and assembled by a set of enzymes encoded by genes organized into clusters. Present study focuses on the effect of deletion of genes involved in biosynthesis of OAg repeating units on resistance to antimicrobial peptides and virulence in mice.
Methods
In the present study, the OAg biosynthesis was impaired by deleting tyv, prt and wbaV genes involved in tyvelose biosynthesis and its transfer to OAg. The virulence phenotype of resulting mutants was evaluated by assessing resistance to antimicrobial peptides, serum complement, adhesion, invasion and in vivo colonization.
Results
Deletion of the above three genes resulted in the production of OAg-negative LPS. All the OAg-negative mutants showed phenotype reported for rough strains. Interestingly, ΔwbaV mutant showed increased resistance against antimicrobial peptides and normal human serum. In addition, the ΔwbaV mutant also showed increased adhesion and invasion as compared to the other two O-Ag negative mutants Δtyv and Δprt. In vivo experiments also confirmed the increased virulent phenotype of ΔwbaV mutant as compared to Δprt mutant.
Conclusion
OAg-negative mutants are known to be avirulent; however, this study demonstrates that certain OAg negative mutants e.g. ∆wbaV may also show resistance to antimicrobial peptides and cause colitis in Streptomyces pretreated mouse model.
Similar content being viewed by others
Background
Salmonella are Gram-negative, intracellular bacteria that cause diseases ranging from acute gastroenteritis to typhoid fever, posing a significant threat to public health globally. Infections with non typhoidal serovars of Salmonella enterica, predominantly S. enterica serovar Enteritidis (S. Enteritidis) and S. enterica serovar Typhimurium (S. Typhimurium) are more frequent and occur in both developing and industrialized nations [1]. Salmonella employs a number of virulence factors to successfully colonize and replicate inside the host. The most critical virulence determinants of Salmonella infections are Salmonella pathogenicity islands (SPI), of which SPI-1 and SPI-2 play crucial role in invasion and intracellular replication respectively. Apart from these, numerous additional virulence factors such as pilli or fimbria [2], flagella [3], lipopolysaccharide (LPS) [4] etc. are required for a successful infection. Among these factors, role of LPS in Salmonella virulence has been established by several studies [5–7]. LPS is a major structural component of the outer membrane of all the Gram-negative bacteria and plays a critical role in the bacterial pathogenesis. It consists of three distinct domains: lipid A, core oligosaccharide (OS) and O-antigen polysaccharide (OAg). The different components of LPS interact with different parts of host cells and contribute to bacterial pathogenesis. For example, different sugars of outer core interact with epithelial cells, whereas, lipid A interacts with the TLR4, a surface receptor of immune cells. While lipid A and core OS structures are fairly conserved, the OAg is highly variable, leading to serological specificity among Gram-negative bacteria. OAg is a modular assembly of oligosaccharide units that varies with respect to the sugar composition and number of their modal repeats [8]. Typically, in all the Gram-negative bacteria, this region comprises of 16 to more than 100 repeats of oligosaccharide units containing 4–6 monosaccharides each. Bacteria lacking OAg are called rough. The repeating unit of OAg of Salmonella mainly consists of three hexose sugars namely mannose, rhamnose, galactose and one dideoxy hexose as the fourth component [9]. Mannose, rhamnose and galactose form the backbone of the OAg and are conserved across different serovars of Salmonella. The dideoxy sugar linked (α-1, 3) to mannose residue varies among different serovars. Based on the agglutination by antibodies against different O-antigens, Salmonella have been grouped into six serogroups namely A, B, C1, C2, D and E. The group A (e.g. S. Typhi) contain paratose (3,6-dideoxy-d-ribo-hexose), group B (S. Typhimurium) has abequose, whereas group D (e.g. S. Enteritidis) contain tyvelose (3,6-dideoxy-d-arabino-hexose) as fourth component of OAg repeating unit [10]. A schematic representation of the biosynthetic pathway of these sugars is shown in Fig. 1a. In S. Enteritidis, enzyme paratose synthase (Prt, formerly known as RfbS), synthesizes paratose, which is further epimerized by the enzyme CDP-tyvelose-2-epimerase (Tyv, formerly known as RfbE), to tyvelose which is then transferred to OAg repeating unit by tyvelosyl transferase (WbaV, formerly known as rfbV) (Fig. 1a). The genes responsible for OAg biosynthesis are generally found on the chromosome as an OAg gene cluster and genetic variation in this cluster reflects the structural variations of OAg across Gram-negative bacteria. This gene cluster encodes proteins that can be further categorized into three groups [11]. First group involve proteins that synthesize nucleotide sugar precursors. Proteins of the second group are mainly glycosyl transferases (GTase) that build sequentially, the OAg-repeating unit on the carrier lipid, undecaprenyl phosphate (UndP). The third group of enzyme is mainly OAg processing enzyme involved in the polymerization and translocation of OAg across the membrane. The enzymes, Tyv and Prt are involved in biosynthesis of CDP-sugars whereas WbaV is a GTase. The location of the above genes on chromosome has been depicted in Fig. 1b.
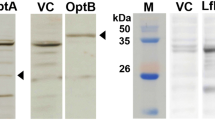
Schematic presentation of final steps of dideoxy sugar biosynthesis, location of genes on chromosome and LPS profile of wild-type S. Enteritidis and its isogenic mutants. a CDP-paratose is synthesized by CDP-4-keto-3,6-dideoxy-d-glucose by the action of prt which is epimerized by Tyv to tyvelose. After synthesis, tyvelose is transferred to OAg backbone of mannose (Man), rhamnose (Rha) and galactose (Gal) by WbaV. CDP-abequose is also synthesized from same CDP sugar intermediate as CDP-paratose. b Location of genes on chromosome. The regions enclosed by the black bar were deleted with the help of lambda red recombinase system. White arrow indicate that genes are located on negative strand. c LPS from wild-type Salmonella as well as the mutants were isolated using the protocol described in materials and methods and separated on polyacrylamide gel electrophoresis (PAGE) gel using tricine-SDS buffer system. LPS were visualized by silver staining. c∆ indicates the mutants are complemented with the corresponding genes
Deletion of the genes involved in nucleotide diphosphate (NDP) sugar biosynthesis renders bacteria unable to synthesize complete OAg [12]. For example, deletion of galE or pmi genes involved in NDP sugar biosynthesis, results in the truncation of LPS which can be further restored via trans-complementation [12–14]. Since, both prt and tyv are involved in CDP-sugar biosynthesis [15, 16], their deletion could possibly affect OAg biosynthesis. OAg biosynthesis and assembly is a highly co-ordinated process and is accomplished step by step. Interference at any step may not only affect the OAg biosynthesis but also other cellular processes as sugar intermediates are synthesized from precursors obtained from common metabolic pathways [16]. In the present study, we have blocked tyvelose biosynthesis and its transfer to OAg repeating units by gene deletion (tyv, prt and wbaV) and investigated their effect on S. Enteritidis virulence. Interestingly, we found that deletion of tyvelosyl transferase gene (wbaV) resulted in increased resistance against antimicrobial peptides (AMPs) and decreased degree of attenuation in the rough strain of S. Enteritidis as compared to other OAg-negative strains.
Results
Deletion of genes involved in OAg biosynthesis from the genome of S. Enteritidis results in the production of OAg-negative LPS
In order to investigate the effect of deletion of genes involved in biosynthesis of OAg, the ΔwbaV, Δtyv, and Δprt gene were deleted from the genome of S. Enteritidis and LPS from the respective deletion mutant was analyzed by SDS PAGE and silver staining. It was observed that the strains were unable to synthesize OAg repeating units (Fig. 1b). However, the synthesis of lipid A and core region remained unaffected. To prove that the deletion of genes had no polar effect on downstream genes, mutant strains were complemented with their respective genes with the help of plasmid listed in Table 1. Upon complementation, all the strains synthesized complete OAg repeating unit (Fig. 1c).
OAg-negative mutants showed significant impairment in the motility
Bacterial strains lacking OAg are known to be less motile as compared to those with long OAg repeating units. To confirm if the OAg-negative mutants used in this study were also less motile, the motilities of the mutants were analyzed by soft agar assay. All the OAg-negative mutants were found to be less motile as compared to the wild-type and showed almost two fold reduction in their motility (Fig. 2a).
Assessment of motility and sensitivity to antimicrobial peptides and serum. a Wild-type Salmonella and its isogenic mutants were placed at the center of the agar plate containing 0.3 % agar and incubated for 8 h. Diameter of growth zone was measured in centimeter (cm). Experiments were performed in triplicates at three independent occasions. b For AMP sensitivity assay, approximately 1 × 107 CFU/ml bacteria in PBS were incubated with or without antimicrobial peptides (1 µg/ml polymyxin B, 5 µg/ml protamine, 10 µg/ml LL37, and 2 µg/ml cecropin) for 1 h at 37 °C. The number of surviving bacteria was determined by plating serial dilutions on LB agar plates and survival percentage was calculated. Statistical analysis was performed using two-way ANOVA. c For analysis of serum resistance 1 × 107 CFU/ml of bacteria was incubated with or without 50 % of normal human serum and serial dilutions were plated to enumerate the number of surviving bacteria. Statistical analysis was performed using student t-test. The survival of Δprt and Δtyv mutants was compared with that of ΔwbaV. Level of significance is indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001), ns not significant. Error bar indicate the standard deviation of three independent experiments
OAg-negative mutants showed differential sensitivity to AMPs and complement mediated killing
The structure and composition of LPS is known to affect the susceptibility of bacterial strains to various cationic AMPs [17, 18]. Therefore, we evaluated the sensitivity of these mutants to different cationic AMPs. The survival efficiency of the mutants was compared to that of the wild-type (normalized to 100 percent). All the mutants showed increased susceptibility to AMPs tested in this study as compared to the wild-type. Interestingly, the ΔwbaV mutant showed significantly increased resistance to all the AMPs as compared to other OAg-negative mutants (P < 0.001), which was reduced up to the level of wild-type upon complementation (Fig. 2b).
To further characterize the mutants, the sensitivities of mutants to complement mediated killing was determined. All the OAg-negative mutants were found to be more susceptible to killing by serum as compared to wild-type strain. The OAg-negative strains showed 50 % or less survival in serum when compared to the wild-type (normalized to 100 %). The ΔwbaV mutant, however, showed significantly increased survival (P < 0.05–0.01) as compared to other two OAg-negative mutants (Δprt and Δtyv) (Fig. 2c).
The ΔwbaV mutant showed increased adhesion and invasion efficiencies
To study the effect of deletion of above genes in adhesion and invasion, the ability of different OAg mutants to adhere and invade HCT-116 cells was compared. OAg- negative mutants attached and invaded HCT-116 cells more efficiently as compared to the parental strain. Among all the three mutants, ΔwbaV mutant showed the highest rate of adhesion (~2.5-fold) and invasion (~4.5-fold) when compared with the wild-type (Fig. 3a, b) and the complemented strain showed adhesion and invasion similar to wild type. However, variations in the efficiency of adhesion and invasion among mutants were observed. The ΔwbaV mutant showed significantly increased adhesion and invasion when compared with other two OAg- negative mutants (P < 0.05–0.001).
Attachment and invasion of S. Enteritidis wild-type and its isogenic mutants to HCT116 cell line. a HCT116 cells were seeded at the density of 2 × 105 cells per well and bacterial strains were infected at the MOI of 10. Infection was performed on ice followed by incubation at 4 °C for 30 min. Non adherent bacteria were removed by washing and number of adhering bacteria was enumerated by plating serial dilution. b For invasion, HCT116 cells were seeded at the density of 2 × 105 cells per well and bacterial strains were infected at the MOI of 10. Number of invading bacteria was determined through standard gentamicin protection assay. SPI1 negative strain ΔinvC served as an experimental control. Experiments were performed in triplicates. Percentage of adhesion, invasion and uptake of mutant was compared to wild-type (normalized to 100). Error bar indicate the standard deviation of three independent experiments. Statistical analysis was performed using t test. The percentage of adhesion and invasion replication of Δprt and Δtyv was compared with that of ΔwbaV and level of significance is indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001), ns not significant
S. Enteritidis ΔwbaV mutant showed increased systemic load as compared to Δprt mutant in C57BL/6 mice
To understand the effect of deletion of above genes on virulence of ΔwbaV mutant in vivo, C57BL/6 mice were infected with wild-type, ΔwbaV and Δprt mutants. In a 3 days p. i. experiment, the cecal colonization of ΔwbaV mutant was comparable to wild-type, whereas the Δprt mutant colonized less efficiently than ΔwbaV mutant and the wild-type (Fig. 4a), however, the differences were not statistically significant. On the contrary, the pathogen load in the mesenteric lymph node (mLN) varied between both the mutants. The CFU of ΔwbaV mutant recovered from mLN was significantly (P = 0.0085) higher than Δprt mutant (Fig. 4b). Similarly, the bacterial load in spleen and liver was also found to be significantly higher in case of ΔwbaV mutant as compared to Δprt mutant (P < 0.05) (Fig. 4c, d). Notably, the ΔwbaV mutant showed reduced colonization in mLN, spleen and liver as compared to the wild-type. Further, histopathological evaluation was carried out to compare the cecal inflammation between the mutants and wild-type. The ΔwbaV mutant was found to induce moderate cecal inflammation whereas Δprt mutant was unable to induce inflammation in the C57BL/6 mice (Fig. 5a). The cecal pathology was assessed by a semi-quantitative scoring method as described in materials and methods. The day 3 p. i. experiment showed a significant difference in the cecal pathology of the mice infected with Δprt mutant (pathoscore: 8) and ΔwbaV mutant (pathoscore: 2) as compared to the wild-type (P < 0.01) (Fig. 5b). The phenotype of complemented ΔwbaV strain was similar to wild type.
Colonization of cecum, mLN, spleen and liver of C57BL/6 mouse by S. Enteritidis and OAg-negative mutants: Streptomycin pretreated mice were infected with respective bacterial strains through oral route of administration. Mice were sacrificed on 3 days p. i. and cecum (a), mLN (b), spleen (c) and liver (d) were collected and CFU in each organ were determined. Statistical significance is indicated by asterisks (*P < 0.05, **P < 0.01, ***P < 0.001), ns not significant
Histopathological evaluation of cecum from mouse infected with S. Enteritidis and OAg-negative mutants. HE stained sections of cryoembeded cecal tissue showing cecal inflammation (a) and respective pathoscore (b) was determined by semi quantitative method as described in “Methods”. S submucosal edema, LP lamina propria, L lumen. Bars 200 μm
Discussion
LPS forms the major component of the outer membrane of Gram-negative bacteria, though ubiquitously present, its structure and composition varies among species. The most variable part of LPS is the OAg chain, consisting of repeating unit of four to six sugars which forms the basis of serological typing. The serovars of Salmonella mainly differ in the presence of dideoxy sugars in OAg repeating unit. In the present study, we have investigated the effect of deletion of three OAg biosynthetic genes on virulence of S. Enteritidis.
Deletion of each gene targeted in this study resulted in the production of OAg-negative LPS. Deletion of prt gene blocks the synthesis of CDP-paratose which cannot be further epimerized to tyvelose by tyv gene product; as a result Δprt mutant cannot synthesize OAg. However, in Δtyv mutant, epimerization of CDP-paratose into tyvelose was hampered due to deletion of tyv gene but the synthesis of CDP-paratose remains unaffected. It has been reported earlier, that tyvelosyl transferase of S. durban has an affinity for CDP-paratose and can transfer it to OAg [10]. Similarly, the group A, B and D glycosyl transferases are able to transfer paratose, abequose and tyvelose to OA [19, 20]. However, Osborn and Weiner, 1968 reported that glycosyl transferase of B1 has more specificity for its natural substrate [21]. In this study, the ability to synthesize complete OAg was restored after complementation indicating that rough phenotype of Δtyv mutant was not due to polar effect. Therefore, the inability of Δtyv mutant to synthesize complete OAg suggests that the tyvelosyl transferase (wbaV) of S. Enteritidis is unable to transfer paratose to OAg repeating unit. Further, deletion of wbaV gene also resulted in the synthesis of OAg-negative LPS, suggesting that, the addition of dideoxy sugar to the OAg sugar backbone is crucial for the complete biosynthesis of OAg. Similar finding was reported by Hong et al. [22].
It is already established in previous studies, that OAg-negative mutants are more susceptible to killing by serum [23, 24], and cationic AMPs such as polymyxin B [23–25]. We also reported the similar findings, and showed that all the mutants were more susceptible to killing by serum and AMPs as compared to wild-type. Earlier reports also demonstrated that the OAg-negative strains have increased efficiency of adhesion and invasion into non phagocytic cells [25, 26]. Our data correlates well with these findings where all the OAg-negative mutants were adhering and invading more efficiently into HCT-116 cells as compared to the wild-type. It was reported that the outer core sugars of LPS are crucial for the interaction of the bacteria with the epithelial cells, [27, 28] therefore, in the absence of OAg, early interaction of core sugars with epithelial cells is facilitated contributing to increased adhesion. Further, increased invasion of O-Ag negative mutants can be explained on the basis of the model proposed by Holzer et al. [25]. The model states that long and very long OAg interferes with the interaction of type III secretion complex with the host cell membrane and impairs the translocation of effector proteins from bacteria to host cell [25]. Thus, in the absence of OAg, this interference is abolished resulting in the increased efficiency of invasion. In this study, mutants showing increased invasion also showed increased adhesion. Therefore, we propose that increased adhesion efficiency along with the enhanced type III secretion system functionality contributed to the significantly increased invasion efficiency to OAg-negative mutants. Notably, in spite of increased invasiveness, the O-Ag negative mutants did not show increased colonization of cecum suggesting, invasiveness alone does not determine the colonization capacity of Salmonella.
In the present study, the ΔwbaV mutant showed increased resistance to AMPs and serum as compared to other OAg-negative mutants. It also showed increased adhesion and invasion as compared to other OAg-negative mutants. To support the in vitro data, mouse experiments were carried out with wild-type, ΔwbaV and Δprt mutants of S. Enteritidis. In, in vivo experiments, no significant difference was observed in cecal colonization of both the mutants, however, the bacterial count of ΔwbaV mutant in mLN was significantly higher than the Δprt mutant. Similarly, ΔwbaV mutant showed increased colonization in spleen and liver as compared to Δprt mutant. Further, ΔwbaV mutant elicited colitis while Δprt mutant was unable to cause colitis. These findings suggest that ΔwbaV mutant is less attenuated as compared to Δprt mutant, which can be explained on the basis of its increased resistance to AMPs and serum.
All the mutants in the present study showed phenotypes similar to an OAg negative strains which are generally avirulent [26]. However, our in vitro and in vivo data demonstrates that the ΔwbaV mutant, in spite of being OAg negative, showed a virulent phenotype. Similar finding was reported by Ilg et al. [26] where mutant of S. Typhimurium lacking O-Ag side chains elicited acute intestinal inflammation. The ability to cause colitis is determined by several factors including stimulation of host immune response which depends on the interaction of LPS and other pathogenesis related patterns with host cell receptors. Modification of LPS is one of the mechanism through which bacteria attain resistance to antimicrobial peptides. Further, LPS modification also affects the interaction of bacteria with TLRs affecting the signaling pathways which are responsible for production of inflammatory cytokines. Recently, Spahich et al. [29] highlighted an important relation between LPS biosynthesis and outer membrane composition [29]. They found that deletion of specific LPS biosynthetic genes downregulates the transcription of an outer membrane protein, Hap and block its insertion into outer membrane [29]. Further, their study also demonstrated that glycosyltransferase activity but not LPS structure affected the insertion of Hap into outer membrane. Their finding suggests that deletion of LPS biosynthetic genes may affect gene expression through unknown mechanism. According to Tam and Missiaka [30], the intermediate compounds of LPS biosynthesis may exert feedback control on LPS biosynthetic pathway which is regulated by σE response. σE belongs to the family of extracytoplasmic factors and serves as an alternative sigma factor encoded by rpoE gene. Earlier studies have reported that changes in outer membrane protein as well as lipopolysaccharide structure induce σE dependent response [30, 31]. In Escherichia coli, σE factor is also involved in transcription of several genes of LPS biosynthetic pathway (for example, rfaD, lpxD, lpxA and ecfA) [32]. rpoE gene is also known to control the alteration in the structure of both lipid A and core oligosaccharide [33]. LPS remodeling has been depicted as a survival strategy employed by bacteria for adaptation to various environmental changes [34]. In present study, the gene wbaV is a glycosyltransferase and deletion of which is expected to result in the accumulation of CDP-tyvelose (intermediate sugar) in the cytoplasm. Also, the ∆wbaV mutant showed increased resistance to antimicrobial peptides and serum which most commonly is observed in case of LPS modifications particularly in lipidA modification [18, 35–37]. Based on the phenotype observed in ∆wbaV mutant, it can be hypothesized that deletion of wbaV gene not only affects the OAg biosynthesis but also certain other unknown cellular process which contributes to the increased resistance to antimicrobial peptides and decreased attenuation in mice.
Conclusion
To conclude, we found that the deletion of wbaV gene conferred an adaptive resistance to antimicrobial peptides resulting in less attenuation in mice as compared to other OAg-negative mutants. Present study demonstrates that certain OAg negative strains can trigger colitis in mouse. Thus, increased resistance to AMPs might be one of the mechanisms employed by S. Enteritidis to compensate the loss of OAg side chains for retaining its pathogenicity.
Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Wild-type S. Enteritidis P125109 strain and its isogenic mutants were grown in Luria–Bertani (LB) medium (10 g/l Bacto tryptone, 5 g/l Bacto yeast extract, 5 g/l NaCl, HiMedia, India), at 37 °C. For invasion experiments, bacterial strains were grown in SPI1 inducing environment (LB containing 0.3 M NaCl) at 120 rpm. Antibiotics in growth media were used at the following concentrations: ampicillin, 100 µg/ml; kanamycin, 50 µg/ml; and streptomycin, 50 µg/ml.
Generation of deletion mutant and complementation
Gene deletion was performed using one-step inactivation method as described previously [38]. Briefly, the Tn5 neomycin phosphotransferase gene (aphT) (conferring resistance to kanamycin) in template plasmid pKD4 was PCR amplified using primers listed in Table 2. The primers were designed such that the overlapping coding sequences of adjacent genes do not get deleted and thus the expression of adjacent genes was not affected. The deleted regions have been shown in Fig. 1c. S. Enteritidis wild-type strain harboring plasmid pKD46 was transformed with PCR amplified product and transformants were screened on LB agar plates supplemented with kanamycin. Gene knock-out was confirmed by PCR using kanamycin internal primer and gene specific primers listed in Table 2. For complementation, the respective genes along with 600-bp upstream sequence were cloned between NcoI and XbaI site of plasmid pCH112 and transformed into the corresponding mutant strain [39]. Primers used for complementation have been listed in Table 2.
LPS analysis
LPS was prepared as described previously [40]. Briefly, bacterial cultures collected from LB agar plate were suspended in phosphate-buffered saline (PBS) with the help of cotton swab and optical density at 600 nm was adjusted to 2.0. From this 1.5 ml suspension was centrifuged and the pellet was resuspended in lysis buffer and boiled at 100 °C for 10 min followed by treatment with proteinase K enzyme and extraction in hot phenol. Final extraction of the aqueous phase was carried out with Tris-saturated ether. LPS preparations were resolved on 14 % acrylamide gels using Tricine-SDS buffer system and visualized by silver staining method as described earlier [40].
Motility assay
Motility of wild-type S. Enteritidis and its isogenic mutants was analyzed by soft agar assay. Briefly, 1 µl of overnight grown culture of S. Enteritidis wild-type and its isogenic mutants was placed at the centre of the soft agar (0.3 % agar) plates and incubated at 37 °C. The diameter of the halo zone was measured after 4 h and 8 h. Plates were photographed with Nikon camera COOLPIXL810. Experiments were performed in triplicates and repeated thrice.
Analysis of resistance to antimicrobial peptides
Overnight grown culture of wild-type and mutant strains was subcultured for 4 h. Cells were washed with PBS and diluted to approximately 108 CFU/ml. A 50 µl aliquot of bacterial suspension was incubated at 37 °C for 1 h with or without AMPs, polymyxin B; 1 µg/ml (Sigma), protamine; 5 µg/ml (Himedia, India), LL37; 10 µg/ml (Sigma), cecropin; 2 µg/ml (Sigma) in PBS. Subsequently, samples were serially diluted and plated on LB agar plates to obtain the number of surviving bacteria. The survival efficiency was calculated as percent ratio of the CFU obtained from AMP treated culture versus untreated culture. Experiment was repeated thrice in triplicates.
Analysis of serum resistance
Human serum samples, devoid of antibody against Salmonella, were obtained from healthy volunteers and pooled, divided into small aliquots and stored at −80 °C. Bacterial cultures grown overnight in LB medium were subcultured for 4 h. Cells were washed with PBS and diluted to approximately 108 CFU/ml. 50 µl of diluted cell suspension was incubated with 50 µl of serum at 37 °C for 1 h. To determine the survival percentage, equal bacterial densities were incubated without serum under similar conditions. Number of surviving bacteria was evaluated by plating serial dilutions on LB agar plate. Survival percentage was calculated by dividing the number of bacteria survived with serum by the number of bacteria obtained from untreated culture. Experiment was performed thrice in triplicates.
Attachment and invasion assay
For attachment and invasion assay, human colonic epithelial cell line (HCT-116) was cultured in Roswell Park Memorial Institute Medium (RPMI 1640) (HiMedia, India) supplemented with 10 % fetal bovine serum (FBS) (HiMedia, India). Cells were seeded on 24 well plates at the density of 2 × 105 cells 16 h prior to infection. For infection, bacterial cultures were grown overnight under SPI1 inducing environment at 120 rpm. Overnight grown culture was diluted to 1:20 in fresh LB and subcultured without antibiotics for 4 h in SPI1 inducing condition. Bacterial cells were washed with PBS and inoculum was prepared in RPMI without antibiotics such that each well received 1 × 106 bacterial cells to achieve a multiplication of infection (MOI) equal to 10. For the attachment assay, both inoculum and cells were prechilled on ice for 15 min to prevent the invasion. Infection was carried out on ice and further incubated for 30 min at 4 °C. Invasion assay was performed as described earlier [41]. Briefly, after 50 min of infection at room temperature, media was replaced with fresh infection media containing 100 µg/ml gentamicin to kill extracellular bacteria and incubated at 37 °C for 2 h in CO2 incubator (5 %). For both the assays, cells were lysed with 0.1 % sodium deoxycholate and appropriate dilutions were plated on LB agar plates supplemented with appropriate antibiotic/s to obtain the adherent and invasive bacteria. The percent of adherent/invasive bacteria was calculated by dividing the number of adherent/invasive bacteria by the number of infecting bacteria and multiplying by 100. Both, adhesion and invasion assays were performed thrice in triplicates.
Mice infection experiments
All animals (C57BL/6 mice) used for the experiments were specific pathogen free (SPF). This animal model is used because of its susceptibility to many broad and narrow host range Salmonella species [42]. Mice were purchased and maintained at the animal house facility of KIIT School of Biotechnology, Odisha, India. 6–8 weeks old specific pathogen free mice were used. All experiments were performed in according to the guidelines of the Institutional Animal Ethics Committee (IAEC), KIIT University, under approval number: KSBT/IAEC/2013/MEET1/A6. During the experiments, all mice were maintained in individually ventilated cages. All the animal experiments were performed in strict accordance with guidelines laid by the Institutional Animal Ethics Committee (IAEC). The infection experiments were carried out in streptomycin pre-treated mice models as established previously [43]. Briefly, a group of five mice were infected (by gavage) with approximately 107 CFU of wild-type, ΔwbaV and Δprt mutants of S. Enteritidis. The bacterial loads in the cecum content, mesenteric lymph node (mLN), spleen and liver were determined at 3 days p. i. by plating suitable dilutions of tissue homogenates on MacConkey agar supplemented with the appropriate antibiotics. For statistical analysis, samples without bacterial counts were adjusted to the minimal detection level (20 CFU/organ in the spleen and 60 CFU/organ in liver). Cecal inflammation of the infected mice was scored to analyze the degree of inflammation.
Histopathological evaluation
Segments of the cecum, colon and ileum were embedded in O.C.T. (Sakura Finetek Inc., USA), snap-frozen in liquid nitrogen, and stored at −80 °C. The 5 μm thick tissue sections were obtained on glass slides and stained with hematoxylin and eosin (H&E) after drying for at least 2 h at room temperature. The stained cryosections were evaluated on the basis of a previously described scoring system for the quantitative analysis of cecal inflammation [43, 44]. The stained sections were scored on the basis of the pathological changes that include sub-mucosal edema, polymorphonuclear leukocyte (PMN) infiltration, loss of goblet cells and epithelial ulceration. The collective pathological score ranges from 0 to 13 with arbitrary units covering the inflammation levels that included intact intestine without any sign of inflammation (pathoscore 0); least sign of inflammation (pathoscore 1–2), slight inflammation as a minimal sign of tissue pathology (pathoscore 3–4); moderate inflammation (pathoscore 5–8); and significant inflammation (pathoscore 9–13).
Statistical analysis
All the in vitro data represent mean ± standard deviation of three independent experiments. Two-way analysis of variance (ANOVA) was employed to determine significant differences in case of sensitivity to different antimicrobial peptides whereas in remaining experiments t test was used. All the statistical calculations were performed with the help of GraphPad Prism software version 5.
Abbreviations
- S. Enteritidis:
-
Salmonella enterica serovar Enteritidis
- PBS:
-
phosphate buffered saline
- LPS:
-
lipopolysaccharide
- OAg:
-
O-antigen
- mLN:
-
mesenteric lymph node
- AMP:
-
antimicrobial peptide
References
Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ et al (2010) The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis Off Publ Infect Dis Soc Am 50(6):882–889. doi:10.1086/650733
Allen-Vercoe E, Woodward MJ (1999) The role of flagella, but not fimbriae, in the adherence of Salmonella enterica serotype Enteritidis to chick gut explant. J Med Microbiol 48(8):771–780
Allen-Vercoe E, Woodward MJ (1999) Colonisation of the chicken caecum by afimbriate and aflagellate derivatives of Salmonella enterica serotype Enteritidis. Vet Microbiol 69(4):265–275
Nevola JJ, Laux DC, Cohen PS (1987) In vivo colonization of the mouse large intestine and in vitro penetration of intestinal mucus by an avirulent smooth strain of Salmonella typhimurium and its lipopolysaccharide-deficient mutant. Infect Immun 55(12):2884–2890
Nevola JJ, Stocker BA, Laux DC, Cohen PS (1985) Colonization of the mouse intestine by an avirulent Salmonella typhimurium strain and its lipopolysaccharide-defective mutants. Infect Immun 50(1):152–159
Freudenberg MA, Merlin T, Gumenscheimer M, Kalis C, Landmann R, Galanos C (2001) Role of lipopolysaccharide susceptibility in the innate immune response to Salmonella typhimurium infection: LPS, a primary target for recognition of Gram-negative bacteria. Microbes Infect/Inst Pasteur 3(14–15):1213–1222
Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ et al (2004) Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol 54(4):994–1010. doi:10.1111/j.1365-2958.2004.04323.x
Raetz CR, Whitfield C (2002) Lipopolysaccharide endotoxins. Ann Rev Biochem 71:635–700. doi:10.1146/annurev.biochem.71.110601.135414
Kita H, Nikaido H (1973) Structure of cell wall lipopolysaccharide from Salmonella typhimurium. IV. Anomeric configuration of l-rhamnose residues and its taxonomic implications. J Bacteriol 113(2):672–679
Sasaki T, Uchida T (1974) Mutants of group D1 Salmonella carrying the somatic antigen of group A organisms: evidence for the lack of cytidine diphosphate paratose-2-epimerase activity. J Bacteriol 117(1):13–18
Reeves PR, Hobbs M, Valvano MA, Skurnik M, Whitfield C, Coplin D et al (1996) Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol 4(12):495–503
Collins LV, Attridge S, Hackett J (1991) Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect Immun 59(3):1079–1085
Hone DM, Attridge SR, Forrest B, Morona R, Daniels D, LaBrooy JT et al (1988) A galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect Immun 56(5):1326–1333
Li Y, Wang S, Scarpellini G, Gunn B, Xin W, Wanda SY et al (2009) Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc Natl Acad Sci USA 106(2):593–598. doi:10.1073/pnas.0811697106
Hallis TM, Lei Y, Que NL, Liu H (1998) Mechanistic studies of the biosynthesis of paratose: purification and characterization of CDP-paratose synthase. Biochemistry 37(14):4935–4945. doi:10.1021/bi9725529
Samuel G, Reeves P (2003) Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res 338(23):2503–2519
Gunn JS (2001) Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res 7(1):57–62
Nummila K, Kilpelainen I, Zahringer U, Vaara M, Helander IM (1995) Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol Microbiol 16(2):271–278
Verma N, Reeves P (1989) Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol 171(10):5694–5701
Wyk P, Reeves P (1989) Identification and sequence of the gene for abequose synthase, which confers antigenic specificity on group B salmonellae: homology with galactose epimerase. J Bacteriol 171(10):5687–5693
Osborn MJ, Weiner IM (1968) Biosynthesis of a bacterial lipopolysaccharide. VI. Mechanism of incorporation of abequose into the O-antigen of Salmonella typhimurium. J Biol Chem 243(10):2631–2639
Hong Y, Cunneen MM, Reeves PR (2012) The Wzx translocases for Salmonella enterica O-antigen processing have unexpected serotype specificity. Mol Microbiol 84(4):620–630. doi:10.1111/j.1365-2958.2012.08048.x
Grossman N, Schmetz MA, Foulds J, Klima EN, Jimenez-Lucho VE, Leive LL et al (1987) Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J Bacteriol 169(2):856–863
Liang-Takasaki CJ, Saxen H, Makela PH, Leive L (1983) Complement activation by polysaccharide of lipopolysaccharide: an important virulence determinant of salmonellae. Infect Immun 41(2):563–569
Holzer SU, Schlumberger MC, Jackel D, Hensel M (2009) Effect of the O-antigen length of lipopolysaccharide on the functions of Type III secretion systems in Salmonella enterica. Infect Immun 77(12):5458–5470. doi:10.1128/IAI.00871-09
Ilg K, Endt K, Misselwitz B, Stecher B, Aebi M, Hardt WD (2009) O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect Immun 77(6):2568–2575. doi:10.1128/IAI.01537-08
Bravo D, Hoare A, Silipo A, Valenzuela C, Salinas C, Alvarez SA et al (2011) Different sugar residues of the lipopolysaccharide outer core are required for early interactions of Salmonella enterica serovars Typhi and Typhimurium with epithelial cells. Microb Pathog 50(2):70–80. doi:10.1016/j.micpath.2010.11.001
Hoare A, Bittner M, Carter J, Alvarez S, Zaldivar M, Bravo D et al (2006) The outer core lipopolysaccharide of Salmonella enterica serovar Typhi is required for bacterial entry into epithelial cells. Infect Immun 74(3):1555–1564. doi:10.1128/IAI.74.3.1555-1564.2006
Spahich NA, Hood DW, Moxon ER, St Geme JW 3rd (2012) Inactivation of Haemophilus influenzae lipopolysaccharide biosynthesis genes interferes with outer membrane localization of the hap autotransporter. J Bacteriol 194(7):1815–1822. doi:10.1128/JB.06316-11
Tam C, Missiakas D (2005) Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol Microbiol 55(5):1403–1412. doi:10.1111/j.1365-2958.2005.04497.x
Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA (1993) The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev 7(12B):2618–2628
Dartigalongue C, Missiakas D, Raina S (2001) Characterization of the Escherichia coli sigma E regulon. J Biol Chem 276(24):20866–20875. doi:10.1074/jbc.M100464200
Klein G, Lindner B, Brade H, Raina S (2011) Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-alpha-d-manno-oct-2-ulosonic acid and rhamnose. J Biol Chem 286(50):42787–42807. doi:10.1074/jbc.M111.291799
Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD et al (2012) LPS remodeling is an evolved survival strategy for bacteria. Proc Natl Acad Sci USA 109(22):8716–8721. doi:10.1073/pnas.1202908109
Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M et al (1998) PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol Microbiol 27(6):1171–1182
Helander IM, Kilpelainen I, Vaara M (1994) Increased substitution of phosphate groups in lipopolysaccharides and lipid A of the polymyxin-resistant pmrA mutants of Salmonella typhimurium: a 31P-NMR study. Mol Microbiol 11(3):481–487
Helander IM, Kato Y, Kilpelainen I, Kostiainen R, Lindner B, Nummila K et al (1996) Characterization of lipopolysaccharides of polymyxin-resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur J Biochem/FEBS 237(1):272–278
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97(12):6640–6645. doi:10.1073/pnas.120163297
Vishwakarma V, Periaswamy B, Bhusan Pati N, Slack E, Hardt WD, Suar M (2012) A novel phage element of Salmonella enterica serovar Enteritidis P125109 contributes to accelerated type III secretion system 2-dependent early inflammation kinetics in a mouse colitis model. Infect Immun 80(9):3236–3246. doi:10.1128/IAI.00180-12
Marolda CL, Lahiry P, Vines E, Saldias S, Valvano MA (2006) Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol Biol 347:237–252. doi:10.1385/1-59745-167-3:237
Pati NB, Vishwakarma V, Jaiswal S, Periaswamy B, Hardt WD, Suar M (2013) Deletion of invH gene in Salmonella enterica serovar Typhimurium limits the secretion of Sip effector proteins. Microbes Infect/Inst Pasteur 15(1):66–73. doi:10.1016/j.micinf.2012.10.014
Suar M, Jantsch J, Hapfelmeier S, Kremer M, Stallmach T, Barrow PA et al (2006) Virulence of broad- and narrow-host-range Salmonella enterica serovars in the streptomycin-pretreated mouse model. Infect Immun 74(1):632–644. doi:10.1128/IAI.74.1.632-644.2006
Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M et al (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71(5):2839–2858
Vishwakarma V, Pati NB, Ray S, Das S, Suar M (2014) TTSS2-deficient hha mutant of Salmonella typhimurium exhibits significant systemic attenuation in immunocompromised hosts. Virulence 5(2):311–320. doi:10.4161/viru.27605
Authors’ contributions
MS and SJ designed the experiment. SJ, MD, CP, PKS, SR, AA and NKM, performed experiment. NBP performed in vivo experiment. SJ and MS wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the grant from Department of Biotechnology, Govt. of India (Project No. BT/PR7515/MED/29/673/2012). The research fellowship awarded by “Council of Scientific and Industrial Research (CSIR), New Delhi, India” to Ms. Sangeeta Jaiswal is greatly acknowledged.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jaiswal, S., Pati, N.B., Dubey, M. et al. The O-antigen negative ∆wbaV mutant of Salmonella enterica serovar Enteritidis shows adaptive resistance to antimicrobial peptides and elicits colitis in streptomycin pretreated mouse model. Gut Pathog 7, 24 (2015). https://doi.org/10.1186/s13099-015-0070-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-015-0070-4