Abstract
Introduction
Gene expression profiling is rapidly becoming a useful and informative tool in a much needed area of research. Identifying patients as to whether they will respond or not to a given treatment before prescription is not only essential to optimise treatment outcome but also to lessen the economic burden that such drugs can have on healthcare resources. In rheumatoid arthritis (RA), there is of yet no genetic/genomic biomarker which can accurately predict response to TNF inhibitor biologics prior to treatment, despite much interest in this area. Multiple studies have reported findings on potential candidate genes; however, due to relatively small sample sizes or lack of sufficient validation, results have been disappointingly inconsistent. The aim of this research was to further explore the predictive value of a previously reported association between CD11c expression and response to the TNF inhibitor biologics, adalimumab and etanercept.
Methods
Real-time qPCR was performed using whole blood RNA samples obtained from seventy-five rheumatoid arthritis patients about to commence treatment with a TNF inhibitor biologic drug, whose response status was determined at 3-month follow-up using the EULAR classification criteria. Relative quantification of CD11c using the comparative CT method outputted differential expression between good-responders and non-responders as a fold-change.
Results
Relative expression of CD11c in patients receiving TNF inhibitor biologics yielded a decrease of 1.025 fold in good-responders as compared to non-responders (p-value = 0.36). Upon stratification of patients dependent upon the specific drug administered, adalimumab or etanercept, similar findings to the full cohort were observed, decreases of 1.015 (p-value = 0.33) and 1.032 fold (p-value = 0.13) in good-responders compared to non-responders, respectively.
Conclusion
The results from this study reveal that CD11c expression does not correlate with response to TNF inhibitor biologics when tested for within pre-treatment whole blood samples of rheumatoid arthritis patients.
Similar content being viewed by others
Introduction
Rheumatoid arthritis (RA) is a systemic autoimmune disorder of unknown aetiology affecting around 400,000 adults in the UK [1] and is characterised by chronic inflammation, joint swelling and fatigue. Early diagnosis and aggressive treatment are known to correlate with improved long-term outcome, ultimately resulting in a better quality of life [2–5]. Within the last decade, the treatment strategy for RA has progressed considerably, particularly since the introduction of biologic therapies, which greatly benefit patients who do not respond to traditional disease-modifying anti-rheumatic drugs (DMARDs) [6]. Although effective, biologic agents are costly (approximately £10,000 per patient per year) and are reserved in many countries for those patients with moderate to severe RA, who have previously failed to respond to more than one DMARD, including methotrexate (MTX) [1]. However, up to 40 % of RA patients receiving biologic agents fail to respond satisfactorily.
Clinical characteristics such as age, gender and concurrent DMARD use only contribute 15–17 % of the variance observed in treatment response [7]. Hence, identifying predictive biomarkers to help clinicians select an effective drug early in the disease course has been highlighted as a research priority. In the field of oncology, transcriptional profiling is routinely utilized to stratify treatment regimens; for example, over-expression of the HER2/neu gene in breast cancer patients is used to inform the prescription of trastuzumab (Herceptin), whilst oestrogen receptor expression is associated with a better response to tamoxifen [8–10]. It is possible, therefore, that expression biomarkers of response could be identified in other fields of medicine. Indeed, expression of CD11c (ITGAX), an integrin molecule involved in cell adhesion and chemotaxis [11] has shown evidence for association with response to treatment with a TNF inhibitor drug (TNFi) in RA patients. It was reported that increased expression of CD11c prior to treatment significantly correlates with good response to adalimumab monotherapy (p value <0.0001); with sensitivity of 100 % and specificity of 91.7 % [12].
However, the previously observed association was conducted using RA monocytes and due to the impracticality of using such a sample source in clinical settings, it is likely to hamper the adoption of this biomarker for routine clinical use. Ideally, for a predictive biomarker to be useful in a clinical setting, it should be possible to test the biomarker in a biological sample that is readily available, acquired through a minimally invasive procedure (resulting in the least possible distress to the patient) and requiring minimal processing following collection. Hence, whole blood would be an ideal source for a biomarker. We therefore set out with the aim not to replicate the previously reported observation, but to (1) establish whether expression of CD11c is detectable in whole blood samples collected from RA patients about to commence treatment with a TNFi biologic drug and (2) correlate pre-treatment whole blood expression levels with response to TNFi biologic drugs both as a whole and sub-grouped by the drug received (both adalimumab and etanercept were tested).
Methods
Patients
Seventy-five patients with RA were selected for analysis from the Biologics in Rheumatoid Arthritis Genetics and Genomics Syndicate (BRAGGSS), which recruits patients from over 50 contributing centres across the UK, who are about to commence treatment with biologic drugs for the first time, as described in detail previously [7]. Inclusion criteria were as follows: participants were Caucasian, over the age of 18 years, fulfilled the 1987 American College of Rheumatology (ACR) criteria for RA and gave written informed consent. Upon recruitment, patients were asked to provide blood samples for the collection of laboratory and serological data and provide psychological and clinical information. As part of the study, patients are prospectively followed for 12 months and provide further samples/information at months 3, 6 and 12. As such, the 28-joint count disease activity score (DAS28) using four variables (the number of tender and swollen joints, erythrocyte sedimentation rate (ESR)/C-reactive protein (CRP) and patient global assessment score) could be recorded prior to and at 3, 6 and 12 months [13]. The BRAGGSS study was approved by NRES Committee North West - Greater Manchester South (REC Ref: 04/Q1403/37).
For this study, patients were selected if they were about to commence treatment with the TNFi biologic drugs, adalimumab or etanercept, and if they showed either a good response or no response to therapy at 3-month follow up. Clinical efficacy of the TNFi biologic drug was assessed at 3-month follow-up using the established European League Against Rheumatism (EULAR) classification criteria. Good-responder patients were defined by having a DAS28 ≤ 3.2 but also having decreased from the pre-treatment DAS28 by >1.2. A non-responder was defined as having a DAS28 score that did not decrease by >0.6 or was between 0.6–1.2 but also with an end score >5.1. Moderate responders were excluded from this study.
Blood collection
Pre-treatment whole blood samples were collected into either Tempus ™ Blood RNA Tubes (3 ml) (Applied Biosystems, Foster City, CA, USA) or PAXgene Blood RNA collection tubes (2.5 ml) (Qiagen, Valencia, CA, USA). Following collection, samples were shipped to the central processing laboratory at the Arthritis Research UK Centre for Genetics and Genomics (ARUK-CoGG) where they were logged onto the laboratory information management system (LIMS) and stored in an allocated freezer location at –80 °C until RNA extraction.
RNA extraction
RNA extraction kits were used to extract total RNA from whole blood samples following the manufacturer’s instructions: i.e., Tempus™ Spin RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) and PAXgene RNA Isolation Kit (Qiagen, Valencia, CA, USA). In both cases an on-column DNase digestion step was performed to remove any potential genomic DNA contamination. Quantification was performed using the Nanodrop ND-1000 (Thermo Scientific, Waltham, MA, USA) and assessment of RNA integrity was assessed using the 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Total RNA with 260/280 and 260/230 ratios in the range of 1.8–2.2 and RNA integrity number (RIN) >5 were taken forward for analysis.
cDNA Synthesis and RT-qPCR
Total RNA was reverse transcribed into high-quality single stranded cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. Briefly, 10 μl total RNA was reverse transcribed in a 10-μl master-mix solution containing MultiScribe™ Reverse Transcriptase and placed in a thermal cycler programmed according to the manufacturer’s instructions.
Quantitative real-time PCR (RT-qPCR) was performed using the QuantStudio™ 12 K Flex real-time qPCR system (Applied Biosystems, Foster City, CA, USA) which employs TaqMan based technology. Relative expression levels of CD11c were determined by normalising CD11c mRNA transcripts against GAPDH and ACTB mRNA transcripts (housekeeping genes (HKGs)). All TaqMan® gene expression assays used were assays-by-demand supplied by Applied Biosystems: ACTB Hs99999903_m1, GAPDH Hs99999905_m1 and ITGAX (CD11c) Hs00174217_m1. All experiments were performed in triplicate, under PCR amplification conditions in accordance with the manufacturer’s instructions.
Statistical analysis
All statistical analysis was performed using the data analysis and statistical software programme STATA/SE 11.2 (StataCorp LP, College Station, TX, USA) [14]. The comparative cycle threshold (CT) method was used to relatively quantify CD11c expression against an average of the HKGs resulting in CD11c expression being presented as a fold-change in good responders as compared to non-responders [15]. Subsequently, normalised CD11c expression values were used in a logistic regression model; both univariate and multivariate models were tested (the multivariate model including baseline characteristics with a p value <0.05 for comparison between the response phenotypes as covariates). The analysis was performed in two stages. First, the analysis was performed on the full cohort and second, the analysis was repeated following stratification of patients into sub-groups based on the TNFi biologic drugs they received.
Results
Seventy-five patients receiving TNFi biological therapy were included in this study. The summarised cohort characteristics of these patients can be seen in Table 1 and are typical of patients with active and severe disease. Similar baseline characteristics were observed for good responders and non-responders except that good responders were slightly younger (p value = 0.023) and presented with a lower health assessment questionnaire (HAQ) score (p value = 0.026). Using the EULAR classification criteria, 41 patients were classified as TNFi responders and 34 patients as TNFi non-responders at 3 months following commencement of treatment (Table 1).
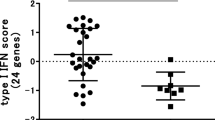
Good-responder and non-responder patients receiving TNFi biologic drugs demonstrated similar baseline CD11c expression profiles when normalised against the average HKGs profile using the comparative CT method (see Fig. 1), equating to a 1.025-fold decrease in good responders as compared to non-responders (multivariate p value = 0.36; using age at baseline and baseline HAQ score as covariates).
Relative CD11c expression stratified by response phenotype. Pre-treatment CD11c expression normalised to the average housekeeping gene (HKG) profile and grouped according to treatment response (presented as 2-ΔC t). Measured in 75 total RNA samples from patients about to receive anti-TNF treatment (a), in 50 total RNA samples from patients about to receive the anti-TNF etanercept (b) and in 25 total RNA samples from patients about to receive the anti-TNF adalimumab (c)
As the previously reported association was reported in patients receiving adalimumab, the analysis was repeated after stratifying patients based on the type of treatment received; this equated to 25 adalimumab patients (16 good responders and 9 non-responders) and 50 etanercept patients (25 good responders and 25 non-responders). Stratified cohort characteristics are listed in Table 1. No significant associations were observed in the stratified analysis. Adalimumab patients yielded a 1.015-fold decrease in good-responders as compared to non-responders (multivariate p value = 0.33, using concurrent DMARD use as a covariate) and etanercept patients yielded a 1.032-fold decrease in good-responders as compared to non-responders (multivariate p value = 0.13, using baseline HAQ score as a covariate).
The previous studies initial exploratory analysis of seven RA patients treated with adalimumab showed that higher CD11c expression correlated with good response. However further analysis revealed that the association was only observed in patients receiving monotherapy and was not observed in adalimumab-treated RA patients when adalimumab was prescribed with MTX, as is common in clinical practice. Further analysis restricted to patients on monotherapy was undertaken in the current study where possible; however, this did not lead to any significant findings at the 5 % significance threshold, though the small group numbers (n = 7) included in the analysis limited the conclusions that could be drawn.
Discussion
This study has employed a candidate gene approach to further explore the potential predictive capability of expression of a previously reported treatment response gene, CD11c, using a clinically applicable sample source (as opposed to the original sample source of monocytes). To achieve this, expression of CD11c was measured in whole blood samples collected at baseline from RA patients about to undergo treatment with TNFi biologic drugs. However, no evidence for association was detected for TNFi drug therapy as a class or after sub-grouping by specific TNFi agents.
A strength of this study was the large sample size tested relative to previous treatment response gene expression profiling studies and the use of samples from extreme ends of the response spectrum, which resulted in greater power to detect the same effect size as previously reported in the study of adalimumab-treated patients [12]. For example, in the original study, after initial identification of CD11c expression associating with treatment response in 7 patients by microarray analysis, validation was conducted in 27 patients from an independent cohort by RT-PCR [12]. By contrast, the current study of 75 RA samples is, to the best of our knowledge, one of the largest and most well-powered treatment response studies of gene expression conducted to date in RA.
A major difference between this study and the original study was the testing of CD11c expression within whole blood rather than in monocytes; therefore, as previously stated, the previous association has not been faithfully tested. The discovery of the correlation between CD11c expression and adalimumab response may be wholly cell-type-specific and due to the heterogeneous nature of whole blood any signal could be confounded by saturation from other cells. However, a recent study [16] demonstrated that CD11c expression is quantifiable in whole blood and we therefore elected to test whole blood, as this sample type would be most readily translated to clinical benefit, whereas isolation of specific cell populations is currently impractical in a clinical setting. Further experiments in an enriched monocyte cell population and whole blood samples will be needed to explore this further.
Another reason for the lack of association could be due to the use of the EULAR classification criteria to define response to treatment within this study, whereas in the original study patients were classified according to the ACR response criteria. Both outcome measures are well-validated and therefore commonly used to assess response to treatment; however, there is debate as to the concordance between them, with the use of which measure being at the discretion of the researcher. For example, one study reported that the concordance between classifying an ACR20 and EULAR response was high (with all 94 ACR20 patients found in the EULAR response group), however, in contrast, concordance between the ACR50 and EULAR response groups was poor (with only 34 ACR50 patients (out of 53) found within the EULAR group) [17]. Of note, ACR scores could not be calculated within the current study.
Another consideration to take into account is that different TaqMan® primers were used within this study, compared to the original. However, both primer sets detected the same transcript so this is not the reason for the lack of replication. Furthermore, it could be that the association between CD11c expression and response is only applicable to patients receiving adalimumab monotherapy, as the signal was lost upon administration of concomitant MTX. However, according to the National Institute for Health and Care Excellence (NICE) guidelines for the treatment of patients with RA in the UK, biologic drugs should be given in conjunction with MTX and only in patients who have proven intolerant or developed an adverse reaction should a biologic be given as monotherapy [18]. This would therefore limit the usefulness of CD11c as a treatment response biomarker, as those receiving adalimumab monotherapy will only represent a small minority of the overall number of patients receiving biologic drugs.
Given the success of gene expression profiling in guiding therapy decisions in the field of oncology, expression profiling is currently being explored as a biomarker predictive of treatment response to other therapies, including biologic treatments used for RA [19–23]. However, results so far have been disappointing as inconsistencies in the reported findings have been observed. This is due to a number of reasons, including differences in study design (type of tissue used, treatment/dose received, classification criteria applied or the platform used to generate the results) or lack of statistical power. Future well-powered studies of treatment response are therefore needed in RA and whole blood remains an attractive tissue to test for biomarkers in the clinical setting. Considering this, it would be ideal to agree upon a standardised method of generating data in treatment response studies before a clinical biomarker can be identified and sufficiently validated for use in the clinic.
Conclusion
We hypothesised that the expression profile of CD11c, previously reported to be associated with response to adalimumab therapy when tested in monocytes, would also correlate with response to TNFi agents with enough discriminatory power to be detectable in whole blood samples. However, our study shows that expression of CD11c in baseline whole blood samples does not correlate with treatment outcome in etanercept-treated or adalimumab-treated RA patients.
Abbreviations
- ACR:
-
American College of Rheumatology
- ACTB:
-
beta-actin
- BRAGGSS:
-
Biologics in Rheumatoid Arthritis Genetics and Genomics Study Syndicate
- CT :
-
cycle threshold
- DAS28:
-
disease activity score in 28 joints
- DMARD:
-
disease modifying anti-rheumatic drug
- ESR:
-
erythrocyte sedimentation rate
- EULAR:
-
the European League Against Rheumatism
- GAPDH:
-
glyceraldehyde 3-phosphate dehydrogenase
- gDNA:
-
genomic DNA
- HAQ:
-
health assessment questionnaire
- HKG:
-
housekeeping gene
- ITGAX:
-
integrin alpha X (complement component 3 receptor 4 subunit)
- MTX:
-
methotrexate
- NICE:
-
National Institute for Health Care Excellence
- qPCR:
-
quantitative polymerase chain reaction
- RA:
-
rheumatoid arthritis
- RIN:
-
RNA integrity number
- SJC:
-
swollen joint count
- TJC:
-
tender joint count
- TNF:
-
tumour necrosis factor
- TNFi:
-
tumour necrosis factor inhibitor
References
National Institute for Health and Care Excellence. CG79 Rheumatoid Arthritis: The Management of rheumatoid arthrits in adults. 2009. http://www.nice.org.uk/guidance/cg79/chapter/Introduction.
Lard LR, Visser H, Speyer I, vander Horst-Bruinsma IE, Zwinderman AH, Breedveld FC, et al. Early versus delayed treatment in patients with recent-onset rheumatoid arthritis: comparison of two cohorts who received different treatment strategies. Am J Med. 2001;111:446–51.
Machold KP, Stamm TA, Nell VP, Pflugbeil S, Aletaha D, Steiner G, et al. Very recent onset rheumatoid arthritis: clinical and serological patient characteristics associated with radiographic progression over the first years of disease. Rheumatology (Oxford). 2007;46:342–9.
Nell VP, Machold KP, Eberl G, Stamm TA, Uffmann M, Smolen JS. Benefit of very early referral and very early therapy with disease-modifying anti-rheumatic drugs in patients with early rheumatoid arthritis. Rheumatology (Oxford). 2004;43:906–14.
van der Linden MP, le Cessie S, Raza K, van der Woude D, Knevel R, Huizinga TW, et al. Long-term impact of delay in assessment of patients with early arthritis. Arthritis Rheum. 2010;62:3537–46.
Upchurch KS, Kay J. Evolution of treatment for rheumatoid arthritis. Rheumatology (Oxford). 2012;51 Suppl 6:vi28–36.
Potter C, Hyrich KL, Tracey A, Lunt M, Plant D, Symmons DP, et al. Association of rheumatoid factor and anti-cyclic citrullinated peptide positivity, but not carriage of shared epitope or PTPN22 susceptibility variants, with anti-tumour necrosis factor response in rheumatoid arthritis. Ann Rheum Dis. 2009;68:69–74.
Bast Jr RC, Ravdin P, Hayes DF, Bates S, Fritsche Jr H, Jessup JM, et al. 2000 update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol. 2001;19:1865–78.
Nahta R, Esteva FJ. HER-2-targeted therapy: lessons learned and future directions. Clin Cancer Res. 2003;9:5078–84.
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43.
Lowin T, Straub RH. Integrins and their ligands in rheumatoid arthritis. Arthritis Res Ther. 2011;13:244.
Stuhlmüller B, Häupl T, Hernandez MM, Grützkau A, Kuban RJ, Tandon N, et al. CD11c as a transcriptional biomarker to predict response to anti-TNF monotherapy with adalimumab in patients with rheumatoid arthritis. Clin Pharmacol Ther. 2010;87:311–21.
Prevoo ML, van’ t Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–8.
StataCorp. Stata Statisitical Software: Release 11. College Station, TX: StataCorp LP; 2009. [computer program]. 2009.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8.
Toonen EJ, Gilissen C, Franke B, Kievit W, Eijsbouts AM, den Broeder AA, et al. Validation study of existing gene expression signatures for anti-TNF treatment in patients with rheumatoid arthritis. PLoS One. 2012;7, e33199.
Gülfe A, Geborek P, Saxne T. Response criteria for rheumatoid arthritis in clinical practice: how useful are they? Ann Rheum Dis. 2005;64:1186–9.
National Institute for Health and Clinical Excellence. TA130 Adalimumab, etanercept and infliximab for the treatment of rheumatoid arthritis. 2007. http://www.nice.org.uk/guidance/ta130.
Julià A, Barceló M, Erra A, Palacio C, Marsal S. Identification of candidate genes for rituximab response in rheumatoid arthritis patients by microarray expression profiling in blood cells. Pharmacogenomics. 2009;10:1697–708.
Julià A, Erra A, Palacio C, Tomas C, Sans X, Barceló P, et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009;4, e7556.
Lequerré T, Gauthier-Jauneau AC, Bansard C, Derambure C, Hiron M, Vittecoq O, et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8:R105.
Koczan D, Drynda S, Hecker M, Drynda A, Guthke R, Kekow J, et al. Molecular discrimination of responders and nonresponders to anti-TNF alpha therapy in rheumatoid arthritis by etanercept. Arthritis Res Ther. 2008;10:R50.
van Baarsen LG, Wijbrandts CA, Rustenburg F, Cantaert T, van der Pouw Kraan TC, Baeten DL, et al. Regulation of IFN response gene activity during infliximab treatment in rheumatoid arthritis is associated with clinical response to treatment. Arthritis Res Ther. 2010;12:R11.
Acknowledgements
We are very grateful to all the patients who consented to participate in this study and to all the nurses who helped collect the samples for this important study. SLS was a PhD student funded by an investigator-initiated award to AB from Pfizer (award number WS1940162). AY is funded by the innovative medicines initiative joint undertaking (IMI JU) funded project BeTheCure, (contract number 115142-2). The work is supported by the National Institute for Health Research (NIHR)’s Manchester Musculoskeletal Biomedical Research Unit, Leeds Musculoskeletal Biomedical Research Unit and Newcastle’s Biomedical Research Centre. The views expressed in this publication are those of the author(s) and not necessarily of the NHS; the National Institute for Health research or the Department of Health. We also thank Arthritis Research for their support (grant number 20385).
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SLS devised the study concept and design, performed the experiments, participated in the data analysis and drafted and revised the manuscript. DP devised the study concept and design, participated in the data analysis, interpretation of the findings and helped draft and revised the manuscript. AY devised the study concept and design, participated in the data analysis, interpretation of the findings and critically appraised the manuscript for intellectual content. SE devised the study concept, was involved in the interpretation of the findings and critically appraised the manuscript for intellectual content. AB devised the study concept and design, was critically involved in sample acquisition, interpretation of the findings and appraising the manuscript for intellectual content. KH was critically involved in sample acquisition, interpretation of the findings and critically appraising the manuscript for intellectual content. AWM was critically involved in sample acquisition, interpretation of the findings and critically appraising the manuscript for intellectual content. AGW was critically involved in sample acquisition, interpretation of the findings and critically appraising the manuscript for intellectual content. JI was critically involved in sample acquisition, interpretation of the findings and critically appraising the manuscript for intellectual content. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Smith, S.L., Eyre, S., Yarwood, A. et al. Investigating CD11c expression as a potential genomic biomarker of response to TNF inhibitor biologics in whole blood rheumatoid arthritis samples. Arthritis Res Ther 17, 359 (2015). https://doi.org/10.1186/s13075-015-0868-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13075-015-0868-y