Abstract
Background
Soft ticks (Ixodida: Argasidae) are medically important ectoparasites that mainly feed on birds and mammals, which play a key role in their geographic distribution and dispersion. Despite their importance, studies on soft ticks are scarce for many regions and countries of the world, including Pakistan.
Methods
In this study, 2330 soft ticks—179 larvae (7.7%), 850 nymphs (36.4%), 711 males (30.5%) and 590 females (25.3%)—were collected from animal shelters in 18 locations within five districts of Khyber Pakhtunkhwa, Pakistan. A subset of the collected ticks was processed for DNA extraction and polymerase chain reaction (PCR) for the amplification of tick 12S ribosomal DNA (rDNA), 16S rDNA and cytochrome c oxidase subunit I (cox1), and rickettsial 16S rDNA gene fragments. The obtained sequences were used for the construction of a phylogenetic tree.
Results
All the specimens were morphologically identified as Ornithodoros, and were morphologically similar to Ornithodoros tholozani. The genus was confirmed by sequencing partial 12S rDNA, 16S rDNA and cox1 gene fragments. Additionally, a Rickettsia sp. was detected in some of the collected ticks by PCR targeting 16S rDNA. The morphological relatedness of the tick specimens with O. tholozani was confirmed by phylogenetic analysis, in which the Ornithodoros sp. clustered with Ornithodoros tholozani and Ornithodoros verrucosus, both of which belong to the subgenus Pavlovskyella and have been previously reported from Israel, Ukraine and Iran. The phylogenetic tree also indicated that the Ornithodoros sp. from Pakistan corresponds to an undetermined species. Furthermore, the associated Rickettsia sp. grouped with the limoniae group of Rickettsia species previously reported from Argas japonicus ticks from China.
Conclusions
This is the first molecular study of an Ornithodoros species from Pakistan. Further studies are essential to confirm its identity and possible pathogenicity with regard to its associated microorganisms in the studied region.
Graphical abstract

Similar content being viewed by others
Background
Soft ticks (Ixodida: Argasidae) are medically important hematophagous ectoparasites that feed on terrestrial vertebrates, mostly birds and mammals [1, 2]. Among the Argasidae, some species are host specific, while others are generalists and feed on a variety of hosts when encountered in their microhabitats [3]. They are distributed throughout the world and spread to new geographic regions through the movement of their hosts, such as bats and migratory birds [4, 5]. Soft ticks have an endophilic or nidicolous lifestyle and are found in sheltered habitats such as burrows, cracks, crevices, nests and loose soil [3, 6, 7]. Fewer studies have been conducted on soft ticks than on hard ticks (Ixodidae), likely because of the former’s nidicolous lifestyle and short feeding duration, which make these parasites difficult to observe in the field [6].
The family Argasidae comprises 218 species, of which approximately 133 have not been assigned accurately to genera [8, 9]. Ornithodoros is the most diverse genus within the Argasidae, with approximately 130 known species [2, 10,11,12,13]. The phylogeny and taxonomy of argasid ticks are still controversial, and many species have been assigned to more than one genus [14]. In this study, we follow the nomenclature proposed by Mans et al. [15] who, among others, showed that phylogenetic reconstructions using tick mitogenomes indicate that the genus Ornithodoros (Pavlovskyella) is paraphyletic [15]. Ornithodoros tholozani has been described from specimens collected in Persia [16], a geographical region that does not include Pakistan. Only two Ornithodoros species, Ornithodoros tholozani [17, 18] and Ornithodoros papillipes [19], which were identified morphologically, have been reported in Pakistan thus far. Ornithodoros tholozani mainly infests livestock [17], birds and pigs [4]. A similar tick, Alveonasus lahorensis, also parasitizes cattle, camels and sheep, although the Asiatic mouflon is its primary host [4]. In Pakistan, O. tholozani is widely distributed, whereas the distribution of A. lahorensis is limited to the western part of the country [20]. Currently, no genetic data are available for Ornithodoros spp. from Pakistan.
Soft ticks are considered to be reservoirs for several arboviruses, including African swine fever virus, as well as relapsing fever, which is caused by species of the genus Borrelia, and several Rickettsia species [21,22,23]. Some Rickettsia species are endosymbionts, whereas others are pathogens of vertebrate hosts, including humans [24]. Depending on their genotypical and phenotypical characteristics, the rickettsiae have been divided into four major groups, namely the spotted fever, typhus, bellii, and limoniae groups [25]. Further, several Rickettsia species have been reported from Ornithodoros species, such as “Candidatus Rickettsia africaseptentrionalis” and “Candidatus Rickettsia mauretanica” [26], “Candidatus Rickettsia wissemanii” [27], Rickettsia hoogstraalii [28], Rickettsia lusitaniae [28], “Candidatus Rickettsia nicoyana” [29], and many others that have yet to be identified to species level [24].
The traditional method of tick identification is based on morphological traits and requires a specialist’s expertise, particularly for soft ticks. The morphological identification of argasid ticks is particularly difficult in the case of closely related species, immature stages, engorged stages and damaged specimens [30]. The identification of argasid ticks can be facilitated by the use of genetic markers, such as 12S ribosomal DNA (rDNA), 16S rDNA and cytochrome c oxidase subunit I (cox1) mitochondrial genes [31,32,33]. Moreover, there is a lack of data on the Rickettsia species harbored by soft ticks in Pakistan [17, 31, 34,35,36]. Therefore, the aims of this study were to genetically identify soft ticks collected in the mountainous region of Khyber Pakhtunkhwa (KP) in Pakistan, and to detect the presence of Rickettsia DNA in these ticks.
Methods
Study area
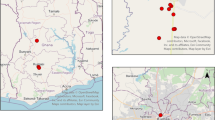
Khyber Pakhtunkhwa (KP) province is in northwestern Pakistan, adjacent to Afghanistan, with Baluchistan province to the south, Punjab province to the east and Azad Jammu Kashmir and Gilgit provinces to the northeast. The present study was conducted in the following five districts of KP: Shangla (34°52'50.6"N, 72°35'27.0"E), Bajaur (34°43'56.1"N, 71°30'33.1"E), Lower Dir (34°52'12.1"N, 71°49'00.8"E), Upper Dir (35°20'28.7"N, 72°03'40.0"E), and Orakzai (33°41'21.3"N, 70°57'26.6"E). These districts are located in mountainous territory at high elevation. The highest elevation, annual average temperature, relative humidity and precipitation of these districts are as follows: Shangla—3440 m, 14 °C, 67% and 850 mm; Bajaur—1150 m, 21 °C, 64% and 700 mm; Lower Dir—1340 m, 17 °C, 68% and 730 mm; Upper Dir—1860 m, 15 °C, 65% and 770 mm; and Orakzai—2500 m, 25 °C, 65% and 600 mm. Google Maps was used to find the geographical coordinates of each location, and the study map (Fig. 1) was designed using ArcGIS 10.3.1.
Tick collection and preservation
Ticks were collected manually from crevices, cracks, burrows and debris in 60 different animal shelters (housing cows, buffaloes, goats, sheep, and equids) in a total of 18 sampling locations in the selected districts (Fig. 1). Tick collection was performed during the day and at night between June and October 2019. The tick samples were stored in Eppendorf tubes and transported to the Department of Zoology, Abdul Wali Khan University, Mardan. Some of the collected ticks were subjected to low intensity ultrasonic vibration for 5 min with a Qsonica sonicator Q700, LLC (Newtown, CT), and washed with distilled water and 70% ethanol to remove contaminants. The specimens were subjected to morphological identification and individually preserved in 100% ethanol in 2-ml Eppendorf tubes for further molecular examination.
Morphological identification
The collected soft ticks were morphologically identified to genus level using a standard identification key [37]. Representative specimens were photographed at 50-200× magnification using a Keyence VHX 900F microscope (Keyence, Itasca, IL).
DNA extraction and PCR
A total of 54 specimens, i.e. two nymphs and one female from each of the 18 sampling locations, were subjected to DNA extraction for subsequent genetic identification. Each specimen was cut in half using a sterile blade and the DNA of one half was extracted using the Nucleospin Tissue Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s guidelines. Extracted DNA was stored at −20 °C until further analysis.
Extracted DNA samples were examined by PCR amplification of partial fragments of the 12S rDNA, 16S rDNA and cox1 tick mitochondrial genes. A 25-µl PCR reaction mixture was prepared for 16S rDNA and cox1 genes; the mixture consisted of 12.75 µl water, 5 µl of 5X Fusion HF Buffer (Mobidiag), 2.5 µl of 2 mM deoxyribose nucleoside triphosphates (dNTPs), 1 µl of 10 µM primers, 0.25 S7 Fusion polymerase and 2.5 µl template DNA. The primers 16S+1 (5’-CCGGTCTGAACTCAGATCAAGT-3’) and 16S-1 (5’-GCTCAATGATTTTTTAAATTGCTGT-3’) were used for the amplification of 16S rDNA fragments [38]. Universal cox1 gene primers HC02198 (5’-TAAACTTCAGGGTGACCAAAAAATCA-3’) and LCO1490 (5’-GGTCAACAAATCATAAAGATATTGG-3’) were used for the amplification of fragments of the cox1 gene [39]. Cycling conditions were as follows: 98 °C for 30 s, followed by 40 cycles of 98 °C for 10 s, 55 °C (16S rDNA) or 63 °C (cox1) for 20 s, 72 °C for 15 s (16S rDNA) or 25 s (cox1), and a final extension step at 72 °C for 5 min. For the amplification of 12S rDNA fragments, PCR was performed in a 25-µl reaction mixture containing 2.5 µl 10X DreamTaq Buffer (Thermo Fisher Scientific), 2.5 µl 2 mM dNTPs, 1 µl of 10 µM primers SR-J-14199 (5’-TACTATGTTACGACTTAT-3’) and SR-N-14594 (5’-AAACTAGGATTAGATACCC-3’) [40], 15.375 µl water, 0.125 (5 U/μl) of DreamTaq polymerase (Thermo Fisher Scientific) and 2.5 µl DNA template. Cycling conditions were as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 30 s, 50 °C for 30 s, 72 °C for 25 s, and finally 72 °C for 5 min.
PCR reaction mixtures (25 µl) for the amplification of fragments of the 16S rDNA gene of Rickettsia species consisted of 12.75 µl water, 5 µl of 5X Fusion HF buffer (Mobidiag), 2.5 µl of 2 mM dNTPs, 1 µl of 10 µM primers Eh-out1 (5’-TTGAGAGTTTGATCCTGGCTCAGAACG-3’) and Ehr3-17 (5’-TAAGGTGGTAATCCAGC-3’) [41], 2.5 µl DNA template and 0.25 µl S7 Fusion polymerase. Cycling conditions were as follows: 98 °C for 30 s, followed by 35 cycles of 98 °C for 10 s, 55.9 °C for 15 s, 72 °C for 10 s, with a final extension at 72 °C for 10 min. The PCR reactions contained Argas persicus DNA as the positive control in the case of tick species, and Rickettsia massiliae DNA as the positive control for Rickettsia species, while PCR-grade water was used as the negative control. The PCR amplified products were run on a 2% agarose gel and observed using gel documentation (BioDoc-It Imaging Systems). Amplicons were purified using the DNA Clean & Concentrator Kit (Zymo Research, Irvine, CA) and sequenced in both directions by LGC Genomics (Berlin, Germany).
Sequencing and phylogenetic analysis
The obtained sequences were trimmed using SeqMan version 5.0 (DNASTAR) to remove primers and poor-quality sequence reads, and subjected to a Basic Local Alignment Search Tool [BLASTn; National Center for Biotechnology Information (NCBI)] search. Trimmed sequences were imported together with sequences from related species in BioEdit alignment editor v 7.0.5 [42] and subjected to ClustalW Multiple alignment [43]. The phylogenetic trees for tick mitochondrial 12S rDNA, 16S rDNA, and cox1 sequences, and for Rickettsia 16S rDNA sequences, were created separately, in accordance with the neighbor-joining method in Molecular evolutionary genetics analysis (MEGA-X) software [44], using align by MUSCLE [45] with a bootstrapping value of 1000 [44].
Results
Identified ticks
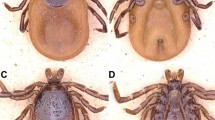
In total, 2330 soft ticks were collected (Table 1) and morphologically identified as Ornithodoros (Pavlovskyella) sp. from the following traits: nymphs and adults had small cheeks; the dorsal idiosome had faint dorsal disks and small mammillae; a dorsoventral groove was present; a pre-anal groove, which completely intersected the transverse postanal groove, was present (Fig. 2).
Abundance and percentage of the Ornithodoros sp. collected from various locations in the examined districts are summarized in Table 2.
Sequences and phylogenetic analysis
The BLAST (NCBI) results for the 12S rDNA [336 base pairs (bp)] gene showed maximum percentage identity of 88.9% with O. tholozani; the 16S rDNA (383 bp) and cox1 (639 bp) genes showed 93.5% and 90.2% identity with Ornithodoros verrucosus (synonym Ornithodoros asperus), respectively. A total of 20, 27 and 21 homologous sequences were downloaded in FASTA format for 12S rDNA, 16S rDNA and cox1 partial genes, respectively, from NCBI. In the phylogenetic analyses (Figs. 3, 4, 5), the obtained sequences clustered with O. tholozani from Israel for the 12S rDNA gene; with O. verrucosus species reported from Iran and Ukraine for the 16S rDNA gene; and with O. tholozani and O. verrucosus reported from Israel and Iran in the case of the cox1 gene.
Neighbor-joining (NJ) phylogenetic tree of Ornithodoros spp. based on the partial mitochondrial 12S ribosomal DNA (rDNA) gene. A group formed by 12S rDNA sequences of Argas giganteus, Proknekalia peringueyi and Alveonasus lahorensis was employed as an outgroup. In the tree, accession numbers are followed by the species name and collection sites. The bootstrap values (1000) are shown at each node. The sequence generated in this study (MW182426) is highlighted by a black circle
NJ phylogenetic tree inferred from the partial mitochondrial 16S rDNA gene of the Ornithodoros spp. A group formed by 16S rDNA sequences of Argas giganteus, Argas persicus and Alveonasus lahorensis was employed as an outgroup. The accession numbers are followed by the species name and collection sites. The bootstrap values (1000) are shown at each node. The sequence generated in this study (MW181620) is highlighted by a black circle. For abbreviations, see Fig. 3
NJ phylogenetic tree of Ornithodoros spp. was constructed based on the partial cytochrome c oxidase subunit I (cox1) gene. A group formed by the cox1 sequences of Argas persicus and Alveonasus lahorensis was employed as an outgroup. GenBank accession numbers are followed by the species name and collection point. The bootstrap values (1000) are shown at each node. The sequence generated in this study (MW182436) is highlighted by a black circle
Ticks collected from seven of the 18 sampling locations (five locations in Shangla and two in Lower Dir) were positive for Rickettsia sp., and had an overall infection rate of 20.4% (11/54) (Table 1). The BLAST results of the obtained Rickettsia 16S rDNA (702 bp) partial gene showed a maximum identity of 100% with a Rickettsia sp. detected in Argas japonicus ticks from China. In the phylogenetic tree, the 16S rDNA gene sequence of Rickettsia sp. (MW308520) from the present study clustered with rickettsial sequences of the limoniae group, including those from Rickettsia limoniae (AF322442) and uncultured Rickettsia species (MG827265, MG827266 and MG827267) detected in A. japonicus (Fig. 6).
NJ phylogenetic tree of Rickettsia spp. based on the partial 16S rDNA gene sequence. The sequence of the 16S rDNA gene of Orientia chuto was used as an outgroup. The bootstrapping values (1000) are shown at each node. The position of the Rickettsia sp. detected in the current study (MW308520) is highlighted by a black circle. For abbreviations, see Fig. 3
Discussion
Like in many other countries, and regions of the world, the diversity of soft ticks in Pakistan has barely been explored. To the best of our knowledge, only three species of soft ticks from Pakistan have been molecularly identified, namely Argas persicus [33], Argas sp. “rousetti” and Carios vespertilionis [46]. In addition, five other species of soft ticks have been reported from Pakistan based on morphological identification: Argas abdussalami [4], Argas reflexus [4], O. tholozani [17, 18], Argas lahorensis [18, 19] and O. papillipes [19]. There is no genetic information on Ornithodoros spp. from Pakistan, and this lack of information needs to be addressed. Therefore, we aimed to molecularly characterize Ornithodoros ticks by using three genetic markers to delineate the species of this genus collected in the KP region of Pakistan. The next objective was to molecularly detect Rickettsia spp. in the collected ticks. The phylogenetic analysis revealed that the collected ticks are related to O. verrucosus and O. tholozani reported from Ukraine, Iran and Israel [21, 47,48,49]. The phylogenetic analysis of the Rickettsia sp. showed that it is closely related to the limoniae group, a basal group of Rickettsia species.
Many argasids, including Ornithodoros species, are nidicolous and live in sheltered habitats such as burrows, crevices, nests and old man-made shelters [6, 48, 50], and have a long lifespan of up to 20 years [51]. Ornithodoros tholozani, a member of the subgenus Pavlovskyella within the genus Ornithodoros, has been reported from stables, barns, stone walls, recording studios and human dwellings [4], whereas O. verrucosus has been reported to mainly inhabit burrows and/or crevices inhabited by reptiles and small mammals [3, 4]. These tick species are found in deserts, semi-deserts and dry areas of the Palearctic zoogeographical region, and chiefly feed on rodents of the family Cricetidae [4, 47]. In the present study, Ornithodoros ticks were collected from burrows, crevices, and cracks in the walls and from the ground debris of domestic animal shelters in northern and southern districts of KP. These districts generally have cold winters, mild summers and rainfall in all the seasons.
The collected ticks were morphologically comparable to Ornithodoros (Pavlovskyella), and their genetic identity confirmed this similarity since the sequences of their loci clustered with those of Ornithodoros (Pavlovskyella) specimens from Iran, Israel, and Ukraine in the phylogenetic tree (Figs. 3, 4, 5). However, it is important to note that, on the basis of the topology of the phylogenetic tree and genetic comparisons with other Ornithodoros ticks, the collected ticks might represent separate species to O. tholozani or O. verrucosus. Ornithodoros tholozani was originally described from Persia [16], an antique territory that does not include Pakistan, and the sequences presently available for this species are from Israel. Ornithodoros verrucosus occurs mainly in the Caucasus and eastern Europe, thus the available sequences from Ukraine are likely representative of this species [21]. In the 16S rDNA phylogenetic tree, both O. tholozani from Israel and O. verrucosus from Ukraine grouped into separate clades (Fig. 4). However, it is noteworthy that the sequences of the ticks identified as O. verrucosus from Iran did not cluster with those of O. verrucosus from Ukraine, and thus are thought to correspond to another Pavlovskyella species. These findings highlight the need for the accurate morphological identification of Ornithodoros (Pavlovskyella) spp. with a Palearctic distribution. Remarkably, there are only a limited number of sequences available in NCBI for O. tholozani and O. verrucosus, and our obtained 12S rDNA, 16S rDNA and cox1 sequences showed maximum percentage identity with O. tholozani (88.9%), O. verrucosus (93.5%) and O. verrucosus (90.2%) reported from Israel, Ukraine and Iran, respectively. As the study of larvae has proved useful for the differentiation of Ornithodoros spp. [4], a taxonomic approach that considers this life stage should be used to elucidate the status of O. verrucosus from Iran, and also that of the Ornithodoros sp. collected in this study.
The nidicolous lifestyle of Ornithodoros species makes it challenging to develop potential mathematical distribution models for these soft ticks using correlative niche modeling [52]. It is also difficult to judge the actual relationships between their life history traits and external environment [3]. However, it has been observed that the geographical distribution of species of the subgenus Pavlovskyella within the genus Ornithodoros, which include O. tholozani and O. verrucosus, is influenced by humidity and precipitation [3, 53]. These ticks have been mostly reported from areas with microclimatic conditions marked by temperature and relative humidity ranges of 17–20 °C and 70–80%, respectively [48, 54]. Similarly, in the present study, the Ornithodoros specimens were collected from regions with high relative humidity, high precipitation, and high temperatures. The monthly mean temperature, relative humidity and annual precipitation in the studied districts in the summer were as follows: Bajaur—25 °C, 68%, and 700 mm; Lower and Upper Dir—22 °C, 64% and 750 mm; Shangla—20 °C, 72% and 850 mm; and Orakzai—25 °C, 61%, and 600 mm (climate-data.org). Interestingly, favorable conditions for the survival and growth of most argasid ticks, including Ornithodoros species, were reported to be provided by the heat and respiration generated by their host animals inside the caves in which they were found [3, 48]. Additionally, temperature has a profound effect on the activity of argasids [3], including Ornithodoros species [55]. We collected Ornithodoros ticks in the summer, from June to September, and even in this season, the specimens were never found out of their microhabitats during the daytime, which agrees with previous findings that these soft ticks are mainly nocturnal [50]. According to Hopla et al. [56], few Ornithodoros species that feed on livestock and poultry also feed on wild animals. Accordingly, the inspected districts in the present study were mostly rural, with an abundance of livestock (buffaloes, other cattle, equids and small ruminants).
Being an agricultural country, inhabitants of several regions of Pakistan rear livestock and poultry in animal shelters, and within or outside their houses. Soft ticks have been documented as vectors and reservoirs for several disease-causing agents throughout the world [57]. Thus, Ornithodoros species might pose a threat to livestock, poultry, and to people who rear livestock, in Pakistan. To the best of our knowledge, to date, there is no published report on the molecular detection of Rickettsia spp. associated with Ornithodoros species in Pakistan. Despite the fact that soft ticks are not considered natural vectors of Rickettsia spp., several rickettsiae have been detected in them [10, 51, 58,59,60]. In several regions of the world, Ornithodoros species are vectors and reservoirs of causal agents of tick-borne relapsing fever (TBRF) [54, 57]. In the present study, a Rickettsia sp. was recorded for the first time in Ornithodoros specimens from Pakistan. The pathogenicity of the Rickettsia sp. detected in the present study needs to be investigated, given the relevance of the rickettsiae bacterial group as causal agents of emerging infectious tick-borne diseases [24].
Conclusions
The present study reports on an Ornithodoros (Pavlovskyella) sp. tick and an associated Rickettsia sp. of the limoniae group for the first time from Pakistan. The morphological and phylogenetic analyses revealed that the Ornithodoros sp. is an undetermined sister species of O. verrucosus and O. tholozani; formal identification of this Ornithodoros sp. is pending. The Rickettsia sp. detected in the Ornithodoros specimens clustered within the limoniae group of Rickettsia species reported from China. Further studies on soft ticks, especially Ornithodoros species, are essential to explore their diversity, associated pathogens, and any economic losses that they may cause.
Availability of data and materials
The datasets that support the conclusions are given in the article.
Abbreviations
- BLAST:
-
Basic local alignment search tool
- cox1 :
-
Cytochrome c oxidase subunit I
- dNTP:
-
Deoxyribose nucleoside triphosphate
- KP:
-
Khyber Pakhtunkhwa
- NCBI:
-
National Center for Biotechnology Information
- NJ:
-
Neighbor-joining
- PCR:
-
Polymerase chain reaction
- rDNA:
-
Ribosomal DNA
References
Ali A, Khan MA, Zahid H, Yaseen PM, Khan MQ, Nawab J, et al. Seasonal dynamics, record of ticks infesting humans, wild and domestic animals, and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front Physiol. 2019;10:793.
Muñoz-Leal S, Toledo LF, Venzal JM, Marcili A, Martins TF, Acosta IC, et al. Description of a new soft tick species (Acari: Argasidae: Ornithodoros) associated with stream-breeding frogs (Anura: Cycloramphidae: Cycloramphus) in Brazil. Ticks Tick Borne Dis. 2017;8:682–92.
Vial L. Biological and ecological characteristics of soft ticks (Ixodida: Argasidae) and their impact for predicting tick and associated disease distribution. Parasite. 2009;16:191–202.
Hoogstraal H. Argasid and nuttalliellid ticks as parasites and vectors. Adv Parasit. 1985;24:135–238.
Müller MA, Devignot S, Lattwein E, Corman VM, Maganga GD, Gloza-Rausch F, et al. Evidence for widespread infection of African bats with Crimean-Congo hemorrhagic fever-like viruses. Sci Rep. 2016;6:26637.
Gray J, Estrada-Peña A, Vial L, Sonenshine D, Roe R. Ecology of nidicolous ticks. Biol Ticks. 2013;2:39.
Ravaomanana J, Michaud V, Jori F, Andriatsimahavandy A, Roger F, Albina E, et al. First detection of African swine fever virus in Ornithodoros porcinus in Madagascar and new insights into tick distribution and taxonomy. Parasit Vectors. 2010;3:115.
Guglielmone AA, Robbins RG, Apanaskevich DA, Petney TN, Estrada-Pena A, Horak IG, et al. The Argasidae, Ixodidae and Nuttalliellidae (Acari: Ixodida) of the world: a list of valid species names. Zootaxa. 2010;2528:1.
Karataş A. A systematic investigation on the ticks (Acari: Ixodida) of the domestic sheep in Niğde Province, Turkey. J Wildl Biodivers. 2020;4:31.
Bakkes DK, De Klerk D, Latif AA, Mans BJ. Integrative taxonomy of Afrotropical Ornithodoros (Ornithodoros) (Acari: Ixodida: Argasidae). Ticks Tick Borne Dis. 2018;9:1006–37.
Luz HR, Bezerra BB, Flausino W, Marcili A, Muñoz-Leal S, Faccini JL. First record of Ornithodoros faccinii (Acari: Argasidae) on toads of genus Rhinella (Anura: Bufonidae) in Brazil. Rev Bras Parasitol Vet. 2018;27:390–5.
Muñoz-Leal S, Domínguez L, Armstrong BA, Labruna MB, Bermúdez S. Ornithodoros capensis sensu stricto (Ixodida: Argasidae) in Coiba National Park: first report for Panama, with notes on the O. capensis group in Panamanian shores and Costa Rica. Exp Appl Acarol. 2020;81:469–81.
Muñoz-Leal S, Venzal JM, Nava S, Marcili A, González-Acuña D, Martins TF, et al. Description of a new soft tick species (Acari: Argasidae: Ornithodoros) parasite of Octodon degus (Rodentia: Octodontidae) in northern Chile. Ticks Tick Borne Dis. 2020;11:101385.
Estrada-Peña A, Mangold AJ, Nava S, Venzal JM, Labruna M, Guglielmone AA. A review of the systematics of the tick family Argasidae (Ixodida). Acarologia. 2010;50:317–33.
Mans BJ, Kelava S, Pienaar R, Featherston J, de Castro MH, Quetglas J, et al. Nuclear (18S-28S rRNA) and mitochondrial genome markers of Carios (Carios) vespertilionis (Argasidae) support Carios Latreille, 1796 as a lineage embedded in the Ornithodorinae: re-classification of the Carios sensu Klompen and Oliver (1993) clade into its respective subgenera. Ticks Tick Borne Dis. 2021;12:101688.
Nuttall GHF, Warburton C, Cooper WF, Robinson LE. Ticks, a monograph of the Ixodoidea. Part I. Argasidae. London: Cambridge University Press, London; 1908.
Karim S, Budachetri K, Mukherjee N, Williams J, Kausar A, Hassan MJ, et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLOS Negl Trop Dis. 2017;11:e0005681.
Rao KNA, Kalra SL. Tick-borne relapsing fever in Kashmir. Curr Sci. 1949;18:249.
Doss MA. Ticks and tick-borne diseases: geographical distribution of ticks. US Government Printing Office; 1974.
Kleinerman G, Baneth G. Ornithodoros (Alectorobius) lahorensis Neumann, 1908 (Figs. 19 and 20). In: Ticks of Europe and North Africa. Springer, Cham. pp. 63–66; 2017.
Filatov S, Krishnavajhala A, Armstrong BA, Kneubehl AR, Nieto NC, De León PAA, et al. Isolation and molecular characterization of tick-borne relapsing fever Borrelia infecting Ornithodoros (Pavlovskyella) verrucosus ticks collected in Ukraine. J Infect Dis. 2020;221:804–11.
Muñoz-Leal S, Macedo C, Gonçalves TC, Barreira JD, Labruna MB, de Lemos ER, et al. Detected microorganisms and new geographic records of Ornithodoros rietcorreai (Acari: Argasidae) from northern Brazil. Ticks Tick Borne Dis. 2019;10:853–61.
Ndlovu S, Williamson AL, Malesa R, Van Heerden J, Boshoff CI, Bastos AD, et al. Genome sequences of three African swine fever viruses of genotypes I, III, and XXII from South Africa and Zambia, isolated from Ornithodoros soft ticks. Microbiol Resour Announc. 2020;9:e01376-19.
Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702.
Yan P, Qiu Z, Zhang T, Li Y, Wang W, Li M, et al. Microbial diversity in the tick Argas japonicus (Acari: Argasidae) with a focus on Rickettsia pathogens. Med Vet Entomol. 2019;33:327–35.
Buysse M, Duron O. Two novel Rickettsia species of soft ticks in North Africa: ‘Candidatus Rickettsia africaseptentrionalis’ and ‘Candidatus Rickettsia mauretanica’. Ticks Tick Borne Dis. 2020;11:101376.
Tahir D, Socolovschi C, Marié JL, Ganay G, Berenger JM, Bompar JM, et al. New Rickettsia species in soft ticks Ornithodoros hasei collected from bats in French Guiana. Ticks Tick Borne Dis. 2016;7:1089–96.
Qiu Y, Simuunza M, Kajihara M, Chambaro H, Harima H, Eto Y, et al. Screening of tick-borne pathogens in argasid ticks in Zambia: expansion of the geographic distribution of Rickettsia lusitaniae and Rickettsia hoogstraalii and detection of putative novel Anaplasma species. Ticks Tick Borne Dis. 2021;12:101720.
Moreira-Soto RD, Moreira-Soto A, Corrales-Aguilar E, Calderón-Arguedas Ó, Troyo A. ‘Candidatus Rickettsia nicoyana’: a novel Rickettsia species isolated from Ornithodoros knoxjonesi in Costa Rica. Ticks Tick Borne Dis. 2017;8:532–6.
Nava S, Guglielmone AA, Mangold AJ. An overview of systematics and evolution of ticks. Front Biosci. 2009;14:2857–77.
Ali A, Zahid H, Zeb I, Tufail M, Khan S, Haroon M, et al. Risk factors associated with tick infestations on equids in Khyber Pakhtunkhwa, Pakistan, with notes on Rickettsia massiliae detection. Parasit Vectors. 2021;14:363.
Ali A, Mulenga A, Vaz IS Jr. Tick and tick-borne pathogens: molecular and immune targets for control strategies. Front Physiol. 2020;11:744.
Zahid H, Muñoz-Leal S, Khan MQ, Alouffi AS, Labruna MB, Ali A. Life cycle and genetic identification of Argas persicus infesting domestic fowl in Khyber Pakhtunkhwa. Pakistan Front Vet Sci. 2021;8:302.
Ali A, Shehla S, Zahid H, Ullah F, Zeb I, Ahmed H, et al. Molecular survey and spatial distribution of Rickettsia spp. in ticks infesting free-ranging wild animals in Pakistan (2017–2021). Pathogens. 2022;11:162.
Kamran K, Ali A, Villagra CA, Siddiqui S, Alouffi AS, Iqbal A. A cross-sectional study of hard ticks (Acari: Ixodidae) on horse farms to assess the risk factors associated with tick-borne diseases. Zoonoses Public Health. 2021;68:247–62.
Kamran K, Ali A, Villagra CA, Bazai ZA, Iqbal A, Sajid MS. Hyalomma anatolicum resistance against ivermectin and fipronil is associated with indiscriminate use of acaricides in southwestern Balochistan, Pakistan. Parasitol Res. 2021;120:15–25.
Estrada-Peña A, Mihalca AD, Petney TN, editors. Ticks of Europe and North Africa: a guide to species identification. Springer; 2018.
Mangold AJ, Bargues MD, Mas-Coma S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol Res. 1998;84:478–84.
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9.
Kambhampati S, Smith PT. PCR primers for the amplification of four insect mitochondrial gene fragments. Insect Mol Biol. 1995;4:233–6.
Wen B, Cao W, Pan H. Ehrlichiae and ehrlichial diseases in China. Ann N Y Acad Sci. 2003;990:45–53.
Hall T, Biosciences I, Carlsbad C. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2011;2:60–1.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9.
Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113.
Ullah H, Kontschán J, Takács N, Wijnveld M, Schötta AM, Boldogh SA, et al. A new Rickettsia honei-related genotype, two novel soft tick haplotypes and first records of three mite species associated with bats in Pakistan. Syst Appl Acarol. 2019;24:2106–18.
Hosseini-Chegeni A, Tavakoli M, Koshki H, Khedri J. A new record of Ornithodoros (Pavlovskyella) verrucosus (Acari: Argasidae) from a porcupine burrow in western Iran. Syst Appl Acarol. 2019;24:771–81.
Kleinerman G, King R, Nachum-Biala Y, Baneth G. Borrelia persica infection in rock hyraxes. Ticks Tick Borne Dis. 2018;9:382–8.
Mans BJ, Featherston J, Kvas M, Pillay KA, de Klerk DG, Pienaar R, et al. Argasid and ixodid systematics: implications for soft tick evolution and systematics, with a new argasid species list. Ticks Tick Borne Dis. 2019;10:219–40.
Kleinerman G, Baneth G. Ornithodoros (Pavlovskyella) tholozani (Laboulbène and Mégnin, 1882) (Figs. 21 and 22). Ticks of Europe and North Africa. Springer, Cham. 2017; pp. 67–70.
Muñoz-Leal S, Barbier E, Soares FA, Bernard E, Labruna MB, Dantas-Torres F. New records of ticks infesting bats in Brazil, with observations on the first nymphal stage of Ornithodoros hasei. Exp Appl Acarol. 2018;76:537–49.
Elith J, Leathwick JR. Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst. 2009;40:677–97.
Lidror R, Shulov A. Preliminary observations on the reactions of Ornithodoros tholozani (Laboulbene & Megnin, 1882) to some environmental factors. Isr J Zool. 1972;21:29–39.
Assous MV, Wilamowski A. Relapsing fever borreliosis in Eurasia-forgotten, but certainly not gone. CMI. 2009;15:407–14.
Moradi-Asl Jafari. The habitat suitability model for the potential distribution of Ornithodoros tholozani (Laboulbène et Mégnin, 1882) and Ornithodoros lahorensis (Neumann, 1908) (Acari: Argasidae): the main vectors of tick-borne relapsing fever in Iran. Ann Parasitol. 2020;66:357–63.
Hopla CE, Durden LA, Keirans JE. Ectoparasites and classification. Rev Sci Tech. 1994;13:985–1034.
Rafinejad J, Choubdar N, Oshaghi MA, Piazak N, Satvat T, Mohtarami F, et al. Detection of Borrelia persica infection in Ornithodoros tholozani using PCR targeting rrs gene and xenodiagnosis. Iran J Public Health. 2011;40:138.
Colombo VC, Montani ME, Pavé R, Antoniazzi LR, Gamboa MD, Fasano AA, et al. First detection of “Candidatus Rickettsia wissemanii” in Ornithodoros hasei (Schulze, 1935) (Acari: Argasidae) from Argentina. Ticks Tick Borne Dis. 2020;11:101442.
Hornok S, Szőke K, Meli ML, Sándor AD, Görföl T, Estók P, et al. Molecular detection of vector-borne bacteria in bat ticks (Acari: Ixodidae, Argasidae) from eight countries of the Old and New Worlds. Parasit Vectors. 2019;12:50.
Kooshki H, Goudarzi G, Faghihi F, Telmadarraiy Z, Edalat H, Hosseini-Chegeni A. The first record of Rickettsia hoogstraalii (Rickettsiales: Rickettsiaceae) from Argas persicus (Acari: Argasidae) in Iran. Syst Appl Acarol. 2020;25:1611–7.
Acknowledgements
We are grateful for the financial support provided by the Pakistan Science Foundation and the Higher Education Commission of Pakistan.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
AA, MN, and MK designed the study. AA acquired the financing. AA, MN and MK collected the samples. AA, MN, MK and SML carried out the statistical data analysis and designed the map of the study locations. AA, MN, MBL, SML, AMN, OA, LCD and MK did the experimental work. LCD took the photos. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The design of this study was approved by the Advanced Study and Research Board of the Faculty of Chemical and Life Sciences (Dir/A&R/AWKUM/2018/1410), Abdul Wali Khan University Mardan. Written consent was obtained from the owners of the animal shelters for the collection of ticks used for the screening.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ali, A., Numan, M., Khan, M. et al. Ornithodoros (Pavlovskyella) ticks associated with a Rickettsia sp. in Pakistan. Parasites Vectors 15, 138 (2022). https://doi.org/10.1186/s13071-022-05248-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05248-0