Abstract
Background
Plasmodium ovale is a neglected malarial parasite that can form latent hypnozoites in the human liver. Over the last decade, molecular surveillance studies of non-falciparum malaria in Africa have highlighted that P. ovale is circulating below the radar, including areas where Plasmodium falciparum is in decline. To eliminate malaria where P. ovale is endemic, a better understanding of its epidemiology, asymptomatic carriage, and transmission biology is needed.
Methods
We performed a pilot study on P. ovale transmission as part of an ongoing study of human-to-mosquito transmission of P. falciparum from asymptomatic carriers. To characterize the malaria asymptomatic reservoir, cross-sectional qPCR surveys were conducted in Bagamoyo, Tanzania, over three transmission seasons. Positive individuals were enrolled in transmission studies of P. falciparum using direct skin feeding assays (DFAs) with Anopheles gambiae s.s. (IFAKARA strain) mosquitoes. For a subset of participants who screened positive for P. ovale on the day of DFA, we incubated blood-fed mosquitoes for 14 days to assess sporozoite development.
Results
Molecular surveillance of asymptomatic individuals revealed a P. ovale prevalence of 11% (300/2718), compared to 29% (780/2718) for P. falciparum. Prevalence for P. ovale was highest at the beginning of the long rainy season (15.5%, 128/826) in contrast to P. falciparum, which peaked later in both the long and short rainy seasons. Considering that these early-season P. ovale infections were low-density mono-infections (127/128), we speculate many were due to hypnozoite-induced relapse. Six of eight P. ovale-infected asymptomatic individuals who underwent DFAs successfully transmitted P. ovale parasites to A. gambiae.
Conclusions
Plasmodium ovale is circulating at 4–15% prevalence among asymptomatic individuals in coastal Tanzania, largely invisible to field diagnostics. A different seasonal peak from co-endemic P. falciparum, the capacity to relapse, and efficient transmission to Anopheles vectors likely contribute to its persistence amid control efforts focused on P. falciparum.
Graphical Abstract

Similar content being viewed by others
Molecular surveillance studies of non-falciparum malaria in Africa over the last decade have highlighted that Plasmodium ovale is circulating below the radar [1, 2] and consists of two distinct species, P. ovale curtisi and P. ovale wallikeri [3, 4], that are rising in prevalence, including in areas where Plasmodium falciparum is in decline [5,6,7,8]. These studies have been primarily country-level surveys conducted intermittently over several years, while fewer studies have examined the local epidemiology driving these trends. We applied P. ovale-specific real-time polymerase chain reaction (qPCR) assays in the context of an ongoing study of the asymptomatic P. falciparum infectious reservoir in Bagamoyo, Tanzania, to examine the seasonality and transmissibility of P. ovale sensu lato (hereafter P. ovale). Herein, we describe the results of cross-sectional surveys across three malaria transmission seasons and of a pilot study that used direct skin feeding assays (DFAs) in subjects with P. falciparum/P. ovale infection to explore their infectivity to mosquitoes.
Bagamoyo district in coastal Tanzania has moderate malaria transmission that occurs throughout the year, with peaks during the long (March to June) and short (October to December) rainy seasons. Anopheles gambiae sensu stricto, Anopheles arabiensis and Anopheles funestus s.s. are the main vectors [9]. Participants aged 6 years and above without fevers or chills in the preceding 3 days were recruited from schools and health centers in the villages of Fukayosi, Mwavi, and Yombo for screening during the long and short rainy seasons from October 2018–December 2019, as well as for 1 month into the dry season following the long rains in 2019.
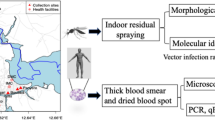
Finger-prick blood was used to make thick and thin blood smears, perform dual-antigen histidine-rich protein 2 (HRP2) and parasite lactate dehydrogenase (pLDH) rapid diagnostic tests (RDT; SD Bioline), and create two to three dried 50 µl blood spots (DBS) on Whatman 3MM filter paper. Species-specific P. falciparum qPCR targeting 18S rRNA was performed on DBS-extracted DNA as described [10]. For P. ovale, primers and probe targeting an 18S rRNA region conserved in P. ovale curtisi and P. ovale wallikeri were used [2]. Midway through the study, qPCR was shifted to Bagamoyo, with the same protocol modified for use with Sahara Hot Start PCR Master Mix (Chai Biotechnologies). For P. falciparum detection, samples were run in duplicate and considered positive if the cycle threshold (Ct) was < 40 cycles in at least one reaction. For P. ovale detection, samples were initially run singly and considered positive if Ct was < 40 cycles. Since low-density parasitemia is common in P. ovale infections, samples were also considered positive if Ct ≥ 40 but < 45 and then confirmed by qPCR in a subsequent independent reaction. All participants positive for malaria by microscopy and/or RDT were treated with artemisinin combination therapy in accordance with Tanzanian national guidelines.
Female A. gambiae s.s. (IFAKARA strain) mosquitoes (4–7 days post-emergence) were used in all mosquito feeding assays. The IFAKARA colony was established in 1996 from mosquitoes collected in southeastern Tanzania [11] and has been maintained in the laboratory since then. To generate experimental mosquitoes, eggs were collected on moist filter paper, rinsed with 1% bleach, and hatched in plastic pans containing deionized water. Larvae were reared in plastic bowls in deionized water and fed daily with TetraMin® fish food. Pupae were transferred to 30 × 30 × 30 cm cages in a climate-controlled room at a constant temperature of 27 °C and relative humidity of 70–80%. Emerged adults were maintained in the same cages on 10% sucrose solution until they were transferred to 500 ml mesh-covered cups for feeding assays. Mosquitoes not used in experiments were fed blood (acquired from a blood bank through Bagamoyo District Hospital) using a Hemotek artificial feeding system to produce eggs for colony maintenance.
Participants positive by RDT or P. falciparum qPCR were enrolled for mosquito direct skin feeding assays (DFAs) on the same day (RDT+) or within 6 days of screening (PCR+). For a subset who were qPCR-positive for P. ovale on the day of DFA, mosquito-dissection procedures were modified to enable molecular evaluation for sporozoite development. Briefly, female mosquitoes were prepared as described above and starved 4–6 h prior to DFAs. For each assay, a cup with 25 mosquitoes was attached to each calf of a participant (50 mosquitoes/participant) and allowed to feed for 15 min. If the participant expressed discomfort, the DFA was stopped immediately. Blood-fed mosquitoes were kept in climate-controlled incubators (27 °C, 70–80% humidity, 12:12 h light/dark) for 24 h without access to sucrose or water to kill unfed mosquitoes and were maintained on 10% sucrose thereafter for the duration of the experiment. On day 8 post-DFA, midguts from a subset of mosquitoes were dissected, stained with 0.1% Mercurochrome, and scored for oocysts. Positive midguts were individually stored in DNA/RNA Shield (Zymo Research) for Plasmodium screening by qPCR. If one or more oocyst-positive midguts were found in the first 10 dissected, the remaining mosquitoes were maintained until day 14 post-DFA, when mosquito abdomens were removed and heads/thoraxes were pooled in groups of up to five for DNA extraction and qPCR.
DNA from mosquito heads/thoraxes stored in DNA/RNA Shield was extracted using the Quick-DNA Tissue/Insect Kit (Zymo Research) following the manufacturer’s instructions. DNA from oocyst-positive midguts stored in DNA/RNA Shield was extracted using DNAzol Reagent (Invitrogen). Prior to homogenization, samples were centrifuged at 3000×g for 15 s, DNA/RNA Shield was removed, and 100 µl of DNAzol was added. Samples were homogenized with a sterilized pestle, brought to 1 ml with DNAzol, and incubated at room temperature for 2 min. After incubation, 0.2 µg of glycogen and 500 µl of −20 °C ethanol was added, and samples were centrifuged at 12,000×g for 10 min at 4 °C. The remaining steps followed the DNAzol manufacturer’s instructions. Species-specific qPCR was performed as described above.
Malaria screening and mosquito-infection data were entered into a RedCAP database. The prevalence of P. falciparum and P. ovale was calculated for each enrollment period. Associations with sex, age, and season were analyzed by log-binomial regression in Stata v.16.1. Rainfall data for the study area were acquired from the Tanzania Meteorological Agency (TMA; https://www.meteo.go.tz). The transmissibility of P. ovale parasite carriers was assessed based on frequency of mosquito midgut and salivary gland/sporozoite infection.
Over three transmission seasons (October 2018 to December 2019), 2718 asymptomatic individuals aged 6 years and older underwent malaria screening. Overall, P. falciparum prevalence by qPCR was 29% (780/2718), while 15% (382/2718) were positive by RDT (HRP2 band), and 12% (316/2718) were smear-positive. Plasmodium ovale prevalence by qPCR was 11% (300/2718), ranging from 4 to 15% (Table 1). Only 7.7% (23/300) of P. ovale qPCR-positive infections had any parasites (P. falciparum or P. ovale) detected by microscopy, while 9.7% (29/300) were diagnosed with malaria based on a positive RDT (HRP2 band present indicating presence of P. falciparum antigen). Thus, approximately 90% (271/300) of asymptomatic P. ovale infections were invisible to routine malaria diagnostic testing, compared to roughly half (54%, 422/780) of asymptomatic P. falciparum infections.
Parasite prevalence varied significantly by season with P. falciparum and P. ovale infections peaking at different times of the year (Fig. 1). While P. falciparum prevalence was highest during the short rainy seasons in October–November (32% and 39% for both months combined, respectively, in 2018 and 2019), P. ovale prevalence peaked early in the long rainy season in April–May 2019 (16%), reaching comparable numbers to P. falciparum in April (21% vs. 18% for P. ovale and P. falciparum, respectively). Plasmodium ovale prevalence subsequently dropped three- to fourfold during the October–November short rainy season when P. falciparum cases were highest.
Plasmodium falciparum/P. ovale co-infections were not common overall (33 total, or 4.2% of P. falciparum [33/780] and 11% of P. ovale [33/300] infections) (Table 1), but they exhibited striking seasonality (Fig. 1). The majority of P. falciparum/P. ovale co-infections occurred during the short rainy season in October–November, when 71.4% (15/21) and 38.7% (12/31) of P. ovale infections were mixed in 2018 and 2019, respectively. In contrast, all but one of the 128 P. ovale infections detected during April–May 2019 were mono-infections. These had higher qPCR Ct values on average compared to P. ovale infections detected in all other seasons combined (Ct = 41.0 vs. 39.8, t(298) = 3.79, P < 0.001), suggesting lower parasite densities.
Male sex and younger age were associated with increased risk of infection with P. falciparum but not P. ovale. For P. falciparum, males were 1.5 times more likely to be infected compared to females (RR = 1.5, P < 0.001), and the risk of infection decreased slightly with each year in age (RR = 0.99, P < 0.001). In contrast, sex and age were not associated with P. ovale infection risk when examined as continuous variables, either in univariate analysis or when the model included sex, age, and season. However, the median age of P. ovale-positive versus negative persons was greater by 4 years (Table 1), and the point prevalence of P. ovale was slightly higher in adults aged 18 years and older (12.6%, 188/1496) compared to children and adolescents 6–17 years of age (9.2%, 112/1222) (Z = −2.82, P = 0.005). For P. falciparum/P. ovale co-infections, each yearly increase in age reduced the relative risk of co-infection by 4.3% (RR = 0.96, P = 0.04), with males being more likely to be co-infected than females (RR = 3.3, P = 0.001).
Over 4 weeks in October–November 2019, qPCR screening of participants undergoing DFA detected P. ovale in eight participants, aged 9–52 years (median 23.5 years). Mosquitoes fed on these participants who harbored P. ovale on the day of DFA were evaluated for P. falciparum and P. ovale infection. Five participants were PCR-positive for mixed P. falciparum/P. ovale infection, while three had P. ovale mono-infection by qPCR (Table 2). The majority of DFAs (6 of 8) yielded P. ovale infections in mosquitoes at either day 8 or day 14 post-DFA, regardless of the mixed infection status observed at screening or enrollment (Table 2). In all four experiments where pooled mosquitoes (head and thoraxes) were available for testing at day 14 post-DFA, the presence of P. ovale sporozoites was detected by qPCR, with all but one experiment yielding mosquitoes infected solely or predominantly with P. ovale.
We report the comparative prevalence of P. falciparum and P. ovale among asymptomatic carriers in coastal Tanzania over a 14-month period and demonstrate human-to-mosquito transmission of P. ovale parasites to A. gambiae s.s. mosquitoes. Plasmodium ovale prevalence was substantial, ranging from 4 to 15%, with 90% of this burden invisible to routine diagnostics (RDT and microscopy).
Our longitudinal analysis reveals a seasonality to P. ovale cases that contrasts with co-endemic P. falciparum, peaking near the beginning of the long rainy season in April, when P. falciparum prevalence is relatively lower. This seasonal pattern could be due to variability in transmission of the two parasite species driven by seasonal differences in Anopheles spp. abundance combined with varying transmission efficiency for certain vector–parasite combinations. Nothing is known about which anopheline species efficiently transmit P. ovale, but our pilot study suggests that P. ovale readily infects A. gambiae, historically the main vector for P. falciparum in the region. These observations, along with the relatively high proportion of P. falciparum/P. ovale co-infections in October–November and reported in other studies [2, 5, 6], suggest that the two parasite species are frequently co-transmitted in the same mosquitoes, a phenomenon we also observed in two DFA participants.
Notably, the early-season P. ovale cases in April–May were virtually all mono-infections and exhibited lower parasite densities than co-infections occurring in October–November. This raises the question of whether the influx of low-density mono-infections during these months arose from hypnozoite-induced relapse [12]. Seasonal patterns of P. vivax versus P. falciparum have previously been used to infer the proportion of vivax cases due to relapse [13, 14]. While there is still much to be learned about the relapse patterns of P. ovale, relapses occurring in April–June, 4 to 6 months following mosquito bites in October–November, could be consistent with the relapse intervals (a median of 15 weeks) observed in a longitudinal cohort study of P. ovale-infected persons in Gabon [15]. Interestingly, the seasonal pattern of 77 imported cases of P. ovale in China from 2010 to 2017 resembles our observations, with cases peaking slightly higher in April than October [16]. We speculate that relapses may have evolved to occur when anopheline vectors are increasing in abundance at the onset of the rainy season.
The use of skin feeding assays combined with on-site PCR detection allowed us to highlight what appears to be highly efficient human-to-mosquito transmission of P. ovale from asymptomatic carriers invisible to routine diagnostics, including those with mixed-species infection. There is much more to be learned about whether this holds true during different seasons, with different anopheline vectors, and for different P. ovale species (P. ovale curtisi vs. P. ovale wallikeri).
Limitations to this study include its cross-sectional nature, small sample of subjects in the DFA study, lack of data in the months between rainy seasons, and the use of insectary-reared mosquitoes to assess infectivity. We have not yet determined the species of these P. ovale isolates (as P. ovale curtisi or P. ovale wallikeri), many of which are low-density samples.
We provide the first direct evidence of efficient transmission from asymptomatic P. ovale carriers to A. gambiae and seasonal patterns of P. ovale infection in coastal Tanzania that may provide clues to how this species, which remains largely undetected, continues to circulate amid heightened efforts to control and eliminate malaria.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon request.
Abbreviations
- A. gambiae :
-
Anopheles gambiae
- C t :
-
Cycle threshold
- DBS:
-
Dried blood spot
- DFA:
-
Direct skin feeding assay
- IQR:
-
Interquartile range
- P. falciparum :
-
Plasmodium falciparum
- P. ovale :
-
Plasmodium ovale
- qPCR:
-
Quantitative polymerase chain reaction
- RDT:
-
Rapid diagnostic test
- RR:
-
Risk ratio
- rRNA:
-
Ribosomal RNA
References
Sutherland CJ. Persistent parasitism: the adaptive biology of malariae and ovale malaria. Trends Parasitol. 2016;32:808–19.
Mitchell CL, Brazeau NF, Keeler C, Mwandagalirwa MK, Tshefu AK, Juliano JJ, et al. Under the radar: epidemiology of Plasmodium ovale in the Democratic Republic of the Congo. J Infect Dis. 2021;223:1005–14.
Sutherland CJ, Tanomsing N, Nolder D, Oguike M, Jennison C, Pukrittayakamee S, et al. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J Infect Dis. 2010;201:1544–50.
Rutledge GG, Böhme U, Sanders M, Reid AJ, Cotton JA, Maiga-Ascofare O, et al. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature. 2017;542:101–4.
Yman V, Wandell G, Mutemi DD, Miglar A, Asghar M, Hammar U, et al. Persistent transmission of Plasmodium malariae and Plasmodium ovale species in an area of declining Plasmodium falciparum transmission in eastern Tanzania. PLoS Negl Trop Dis. 2019;13: e0007414.
Akala HM, Watson OJ, Mitei KK, Juma DW, Verity R, Ingasia LA, et al. Plasmodium interspecies interactions during a period of increasing prevalence of Plasmodium ovale in symptomatic individuals seeking treatment: an observational study. Lancet Microbe. 2021;2:e141–50.
Betson M, Clifford S, Stanton M, Kabatereine NB, Stothard JR. Emergence of nonfalciparum Plasmodium infection despite regular artemisinin combination therapy in an 18-month longitudinal study of Ugandan children and their mothers. J Infect Dis. 2018;217:1099–109.
Cook J, Xu W, Msellem M, Vonk M, Bergström B, Gosling R, et al. Mass screening and treatment on the basis of results of a Plasmodium falciparum-specific rapid diagnostic test did not reduce malaria incidence in Zanzibar. J Infect Dis. 2015;211:1476–83.
Temu EA, Minjas JN, Coetzee M, Hunt RH, Shiff CJ. The role of four anopheline species (Diptera: Culicidae) in malaria transmission in coastal Tanzania. Trans R Soc Trop Med Hyg. 1998;92:152–8.
Markwalter CF, Ngasala B, Mowatt T, Basham C, Zackary Park Z, Loya M, et al. Direct comparison of standard and ultrasensitive PCR for the detection of Plasmodium falciparum from dried blood spots in Bagamoyo, Tanzania. Am J Trop Med Hyg. 2021;104:1371–4.
Knols BG, Njiru BN, Mathenge EM, Mukabana WR, Beier JC, Killeen GF. MalariaSphere: a greenhouse-enclosed simulation of a natural Anopheles gambiae (Diptera: Culicidae) ecosystem in western Kenya. Malar J. 2002;1:19.
Joyner C, Moreno A, Meyer EVS, Cabrera-Mora M, The MaHPIC Consortium, Kissinger JC, et al. Plasmodium cynomolgi infections in rhesus macaques display clinical and parasitological features pertinent to modelling vivax malaria pathology and relapse infections. Malar J. 2016;15:451.
Golassa L, White MT. Population-level estimates of the proportion of Plasmodium vivax blood-stage infections attributable to relapses among febrile patients attending Adama Malaria Diagnostic Centre, East Shoa Zone, Oromia, Ethiopia. Malar J. 2017;16:301.
Ross A, Koepfli C, Schoepflin S, Timinao L, Siba P, Smith T, et al. The incidence and differential seasonal patterns of Plasmodium vivax primary infections and relapses in a cohort of children in Papua New Guinea. PLoS Negl Trop Dis. 2016;10: e0004582.
Groger M, Veletzky L, Lalremruata A, Cattaneo C, Mischlinger J, Zoleko RM, et al. Prospective clinical and molecular evaluation of potential Plasmodium ovale curtisi and wallikeri relapses in a high-transmission setting. Clin Infect Dis. 2019;69:2119–26.
Zhou R, Li S, Zhao Y, Yang C, Liu Y, Qian D, et al. Characterization of Plasmodium ovale spp. imported from Africa to Henan Province, China. Sci Rep. 2019;9:2191.
Acknowledgements
We thank the study participants as well as the staff at the schools and health centers in Bagamoyo for their support. We thank the study teams at Muhimbili University of Health and Allied Sciences (MUHAS) for coordinating the field activities and the Ifakara Health Institute for coordinating the mosquito experimental infections.
Funding
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health through grant R01AI137395 to JTL. The funders had no role in the study design, data collection, or interpretation.
Author information
Authors and Affiliations
Contributions
JL, DM, RD, and JJ conceptualized and designed the study. BT, DM, IR, and VN led the mosquito feeding studies, including acquisition and analysis of data. CB, MM, SC, and RR carried out the molecular work, including acquisition and interpretation of data. ML and BN led the field team for subject enrollment, including acquisition and interpretation of data. DM and BT carried out data analysis. DM, JL, BT, and VN drafted the manuscript. RD and JJ helped revise the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by institutional review boards at the University of North Carolina (ID 276606), Tanzania National Institute for Medical Research (NIMR/HQ/R.8a/Vol.IX/3150), Ifakara Health Institute (IHI/IRB/33–2018), and Muhimbili University of Health and Allied Sciences (MUHAS/DA.282/298/01/C). Informed consent in Kiswahili was obtained prior to screening for malaria and for enrollment into the mosquito feeding portion of the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tarimo, B.B., Nyasembe, V.O., Ngasala, B. et al. Seasonality and transmissibility of Plasmodium ovale in Bagamoyo District, Tanzania. Parasites Vectors 15, 56 (2022). https://doi.org/10.1186/s13071-022-05181-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05181-2