Abstract
Background
Two species of deer ked (Lipoptena cervi and L. mazamae) have been identified as vectors of Bartonella bacteria in cervids in Europe and the USA. In an earlier study we showed that Japanese sika deer (Cervus nippon) harbor three Bartonella species, namely B. capreoli (lineage A) and two novel Bartonella species (lineages B and C); however, there is currently no information on the vector of Bartonella bacteria in sika deer. The aim of this study was to clarify potential vectors of Bartonella in Japanese sika deer.
Methods
Thirty-eight wingless deer keds (L. fortisetosa) and 36 ticks (Haemaphysalis and Ixodes species) were collected from sika deer. The prevalence of Bartonella in the arthropods was evaluated by real-time PCR targeting the 16S−23S internal transcribed spacer (ITS) and by culture of the organisms. The total number of Bartonella bacteria were quantified using real-time PCR. The distribution of Bartonella bacteria in deer ked organs was examined by immunofluorescence analysis. The relationship of Bartonella strains isolated from sika deer and arthropods were examined by a phylogenetic analysis based on concatenated sequences of the gltA, rpoB, ftsZ, and ribC genes, followed by a BLAST search for gltA and rpoB.
Results
Bartonella prevalence in deer keds was 87.9% by real-time PCR and 51.5% in culture and that in the ticks was 8.3% by real-time PCR and 2.8% in culture. The mean number of Bartonella bacteria per ked was calculated to be 9.2 × 105 cells. Bartonella aggregates were localized in the midgut of the keds. The phylogenetic analysis and BLAST search showed that both the host deer and the keds harbored two Bartonella species (lineages B and C), while B. capreoli (lineage A) was not detected in the keds. Two novel Bartonella species (lineages D and E) were isolated from one ked.
Conclusions
Lipoptena fortisetosa likely serves as a vector of at least two Bartonella species (lineages B and C), whereas ticks do not seem to play a significant role in the transmission of Bartonella between sika deer based on the lower detection rates of Bartonella in ticks compared to keds. Bartonella species in lineages D and E appear to be L. fortisetosa-specific strains.
Graphical Abstract

Similar content being viewed by others
Background
Bartonella species are Gram-negative, facultative intracellular bacteria. To date, more than 30 species and three subspecies have been described in the genus [1]. Several hematophagous arthropods are known to serve as vectors for the transmission of Bartonella bacteria to mammalian hosts, including sand fly (Lutzomyia verrucarum) for B. bacilliformis [2], human body louse (Pediculus humanus humanus) for B. quintana [3, 4] and cat flea (Ctenocephalides felis) for B. henselae [5]. Within the last two decades, it has been shown that Lipoptena, Hippobosca and Melophagus keds in the family Hippoboscidae are involved in the transmission of Bartonella bacteria to ruminants [6].
Deer keds (Lipoptena spp.) are recognized as obligate ectoparasites of ruminants but they can accidentally infest humans and other mammals [7, 8]. The keds have an atypical life-cycle compared with that of other hematophagous arthropods. After a winged ked attaches to its host’s body, it sheds its wings and then loses the ability to fly. Wingless keds are thought to remain on their initial hosts. One possible exception to this general rule is the movement of the Neotropical deer ked (L. mazamae) from a mother deer to its offspring due to close contact [9]. After blood-feeding on the host, a female ked lays a larva on the host body, and the larva pupates immediately. Although the pupa is thought to hatch in the summer or early autumn, precise information on the hatch-timing is lacking.
In Europe, L. cervi is the commonest deer ked found on red deer (Cervus elaphus), roe deer (Capreolus capreolus) and moose (Alces alces) [10]. Bartonella-derived DNA was first detected in wingless L. cervi collected from roe deer in France [11], with the high detection rate (93.8%) indicating that L. cervi may be epidemiologically associated with the transmission of Bartonella in this deer species. Bartonella schoenbuchensis was subsequently isolated from 73.3% of wingless L. cervi collected from roe deer and red deer in Germany [12]. Immunohistochemical analysis has also revealed the presence of large bacterial masses consisting of B. schoenbuchensis in the midguts of deer keds [12]. In addition, B. schoenbuchensis DNA was detected in wingless L. cervi (83.3%) collected from white-tailed deer in Massachusetts, USA [13], and Bartonella DNA was detected not only in developing larvae (71%) but also in winged L. cervi (6.4%) in Hungary [14]. In Finland, B. schoenbuchensis-like DNA was also detected from 13 pupae and one winged L. cervi collected in the field [10]. This epidemiological evidence raises the possibility that B. schoenbuchensis may be transstadially transmitted from the pupal to the winged stages of L. cervi.
The Japanese sika deer (Cervus nippon) is native to Japan and genetically divided into six subspecies, among which Honshu deer (Cervus nippon centralis) is widely distributed throughout the country, with the exception of Hokkaido, Shikoku and Kyushu islands. Yezo deer is also a subspecies (Cervus nippon yesoensis) of sika deer and inhabits only Hokkaido island [15]. In a previous study [16], we isolated Bartonella bacteria from 67.6% of Honshu deer and 50% of Yezo deer examined. A genetic analysis showed that Honshu and Yezo deer harbor at least three Bartonella species, including B. capreoli and two novel Bartonella species. Hematophagous arthropods, such as deer keds and ticks, were frequently detected on sika deer that tested positive for Bartonella bacteria. These observations suggest that these ectoparasites are involved in the transmission of Bartonella bacteria between sika deer. To date, there have been no studies investigating the role of hematophagous arthropods in the transmission of Bartonella bacteria between sika deer. The aim of the study reported here was to determine whether deer keds and/or ticks serve as vectors of Bartonella bacteria in sika deer. To achieve this aim, we used a variety of bacteriological, molecular biological and immunohistochemical techniques.
Methods
Sample collection
Between 2009 and 2012, 17 Honshu deer were captured in Nara (N = 8) and Shizuoka (N = 9) Prefectures, Japan. Blood samples were collected from the deer and immediately transferred into EDTA-containing collection tubes. The blood samples collected from Nara Prefecture were utilized for the isolation of Bartonella bacteria in our previous study [16]; the blood samples from Shizuoka Prefecture were used in the present study. The frozen blood samples were sent to the Laboratory of Veterinary Public Health, Department of Veterinary Medicine, College of Bioresource Sciences, Nihon University and stored at − 70 °C until examined.
A total of 38 wingless deer keds were collected from the deer captured in Nara (N = 10) and Shizuoka Prefectures (N = 28). All of the keds were morphologically identified as Lipoptena fortisetosa. Five deer keds from Shizuoka Prefecture were used for immunofluorescence analysis to determine the distribution of Bartonella bacteria within the bodies of the insects. A total of 36 ticks were collected from the deer in Nara (N = 33) and Shizuoka Prefectures (N = 3). Based on morphological analysis under stereomicroscopic observations these ticks were identified as Haemaphysalis flava (N = 16), H. megaspinosa (N = 15), H. longicornis (N = 4) and Ixodes monospinosus (N = 1). The live arthropod samples were immediately sent to the same laboratory under room temperature or refrigeration conditions (approx. 4 °C).
Isolation of Bartonella bacteria from deer blood samples and arthropods
Isolation of Bartonella bacteria from deer blood samples was performed according to the procedure reported by Sato et al. [16]. To isolate Bartonella bacteria from the keds and ticks, first the surface of each arthropod was sterilized for 10 min with 500 μl of 70% ethanol containing 0.1% povidone-iodine, following which the arthropod was washed twice (1 min each wash) with 0.01 M phosphate buffered saline (PBS) containing 0.5% fetal bovine serum (FBS; Life Technologies, Carlsbad, CA, USA). Each arthropod was then homogenized in 400 μl of sodium sucrose glutamic acid buffer (10 mM sodium phosphate, 220 mM sucrose and 0.50 mM l-glutamic acid) for 1 min using a Micro Smash MS-100R homogenizing system (Tomy Seiko Co., Ltd., Tokyo, Japan) set at 3000 rpm, 4 °C. An aliquot (200 μl) of the homogenate was mixed with 200 μl of medium 199 supplemented with 1 mM sodium pyruvate solution and 20% volume of FBS, and 200 μl of this mixture was inoculated onto a heart infusion agar (HIA) plate (Difco, Becton Dickinson, Spark, MD, USA) containing 5% rabbit blood.
The inoculated plates were incubated at 35 °C in a moist atmosphere under 5% CO2 for up to 4 weeks. Bacterial colonies on HIA were tentatively identified as Bartonella based on colony morphology (small, gray or cream-yellow, round colonies), and five colonies per sample were subcultured on a fresh HIA plate using the same conditions as for the primary culture.
Bartonella DNA detection by real-time PCR
A 200-µl aliquot of the homogenate from each arthropod was used for the extraction of DNA using InstaGene Matrix (Bio-Lad, Hercules, CA, USA). Real-time PCR targeting the 16S−23S internal transcribed spacer (ITS) region of Bartonella [17, 18] was used as a molecular screening for Bartonella DNA. To avoid false-negative reactions, TaqMan Exogenous Internal Positive Control (Exo IPC) Reagents (Applied Biosystems, Foster City, CA, USA) were also added to each well according to the manufacturer’s instructions. The reaction mixture (25 μl) consisted of 2.5 μl of the DNA extracts, 1.25 μl of each primer (10 μM), 12.5 μl of 2× TaqMan Fast PCR Master Mixture, 1.25 μl of FAM-labeled probe (5 μM), 2.5 μl of 10 × Exo IPC Mix including the specific primers and VIC-labeled probe, 0.5 μl of Exo IPC DNA (Applied Biosystems) and 3.25 μl of nuclease-free water. Three non-template controls (nuclease-free water) were prepared as a negative control for each PCR. Real-time PCR was performed under the following conditions: 50 °C for 2 min to activate uracil-N glycosylase; 95 °C for 10 min to activate the DNA polymerase; then 95 °C/15 s and 60 °C/1 min for 45 cycles. The targeted DNA was amplified using the 7500 Fast/Real-Time PCR system (Applied Biosystems), and fluorescence was detected through the FAM channel for ITS amplification and the VIC channel for IPC amplification.
Estimation of the total number of Bartonella bacteria in keds by quantitative real-time PCR
We used quantitative real-time PCR (qRT-PCR) targeting the ITS of Bartonella species to estimate the total number of Bartonella bacteria in each culture-positive ked. A standard curve for estimating ITS copy number in each sample was constructed using serial dilutions of plasmid DNAs; the ITS amplicons from ked strains amplified by the 16SF and 23S1 primers [19] were inserted into a pGEM-T easy plasmid vector (Promega, Madison, WI, USA) and serially diluted from 1 × 101 to 1 × 107 copies/μl with nuclease-free water. As the Bartonella genome carries two ITS copies [20], the total number of Bartonella per ked was calculated by dividing the detected number of ITS copies by two copies.
Immunofluorescence analysis to determine the distribution of Bartonella bacteria in deer keds
Immunofluorescent analysis was performed to investigate the distribution of Bartonella bacteria in deer keds. After fixation in 4% paraformaldehyde/PBS, the keds were sectioned toward the sagittal direction using a cryofracture technique and adhesive film [21]. Bartonella bacteria inside the ked body were visualized by indirect immunofluorescence staining. First, mouse anti-sera was made by immunizing mice with the four deer Bartonella strains (Honshu-8.1, Honshu-9.1, Honshu-9.3 and Yezo-25.1) [16] in our laboratory. Then, the anti-sera (1:100) diluted with 1% bovine serum albumin/PBS were applied to the sections and incubated for 2 h at room temperature; binding of the primary antibodies was detected using Alexa Fluor 488 conjugated goat anti-mouse IgG antibodies (Invitrogen, Carlsbad, CA, USA) for 1 h at room temperature. The actin fiber of the cytoskeleton of the deer ked was counterstained with Alexa Fluor 568 phalloidin (Invitrogen). Normal mouse serum was used as a negative control for each sample. The distribution of the Bartonella bacteria and actin fibers were visualized using a fluorescence microscope (model IX71) with the appropriate fluorescence filters (Olympus, Tokyo, Japan). Digital images of each section were captured for processing and analysis using the imaging software ‘cellSens’ (Olympus).
Genogrouping of Bartonella strains based on the gltA and rpoB sequences
Genomic DNA was extracted from whole bacterial cells using InstaGene Matrix (Bio-Rad) and subjected to genus-specific PCR targeting the citrate synthase gene (gltA) [22] and RNA polymerase beta-subunit-encoding gene (rpoB) [23]. The PCR amplicons were purified using the Spin Column PCR product purification kit (Bio Basic Inc., Markham, ON, Canada) and sequenced directly using Applied Biosystems’ BigDye Terminator Cycle Sequencing Ready Reaction kit and Genetic Analyzer 3130. The obtained gltA and rpoB sequences were compared with those of the representative deer Bartonella strains from 11 genogroups [16] and genomic sequences of prokaryotes registered in the International Nucleotide Sequence Database (INSD) by BLAST search. Novel genogroup numbers were assigned to strains with new sequence variants of gltA and/or rpoB.
Classification of Bartonella strain lineages based on phylogenetic analysis
The lineages of the genogroups were classified as described previously [16]. Thus, a representative strain of each novel genogroup was submitted to additional PCR and DNA sequencing analyses of the cell-division protein gene (ftsZ) [24] and the riboflavin synthase gene (ribC) [25]. A phylogenetic tree was constructed from the concatenated sequences of gltA, rpoB, ftsZ, and ribC using the maximum-likelihood method with the General Time Reversible model in MEGA 6 [26].
Results
Prevalence of Bartonella bacteria in sika deer and hematophagous arthropods
Bartonella bacteria were isolated from eight of the nine deer (88.9%) captured in Shizuoka Prefecture and subsequently isolated from 17 of 33 (51.5%) deer keds, whereas only one (H. megaspinosa) of 36 ticks (2.8%) was positive for Bartonella bacteria. Bartonella DNA was detected in 29 of 33 keds (87.9%), but only in three of 36 ticks (8.3%). The Bartonella-positive ticks were identified as H. megaspinosa (N = 1) and H. longicornis (N = 2). IPC amplification was confirmed in all of the reactions.
Bartonella cell counts by qRT-PCR and the distribution of Bartonella bacteria within deer keds
In the 17 keds from which Bartonella bacteria were isolated (see above), a qRT-PCR analysis indicated that the total number of Bartonella bacteria per ked ranged from 1.4 × 104 to 7.0 × 106 cells, with a mean of 9.2 × 105 per ked (Table 1).
Examination of immunostained ked tissue sections showed that specific fluorescence indicating Bartonella aggregates was only present in the midguts of three keds (Fig. 1a-1, b-1, c-1). No fluorescence were observed in other organs of these keds with normal mouse sera (Fig. 1a-2, b-2, c-2).
Distribution of Bartonella bacteria inside wingless deer keds (Lipoptena fortisetosa). a, b, c Sagittal sections of three different deer keds, respectively. a-1, b-1, c-1 Immunostained sagittal sections of the keds using anti-sika deer strains mouse sera and Alexa Fluor 488 conjugated goat anti-mouse IgG antibodies. a-2, b-2, c-2 Sagittal sections of the keds treated with normal mouse serum as a negative control. Actin fibers in the deer keds were counterstained with Alexa Fluor 568 phalloidin in order to aid determination of Bartonella distribution. Scale bar: 100 µm.
Genogrouping of Bartonella strains
A total of 120 Bartonella isolates (5 isolates per one sample) were recovered from 15 keds, one tick, and eight deer. More three isolates were obtained from two keds (ID# D91-Ked1 and D91-Ked5) because a few colonies of Bartonella bacteria were grown on the agar plates. As a result, a total of 123 isolates were obtained in the present study and were classified into 15 genogroups (I to XV); the genogroups XII (10 strains), XIII (three strains), XIV (two strains), and XV (10 strains) were newly found in the present study. The new sequence variants of the gltA, rpoB, ftsZ, and ribC in the representative strains from novel genogroups have been registered at INSD and accession numbers assigned to each of the variant sequences (Table 2).
Lineage classification and sequence homology analysis of novel Bartonella genogroups
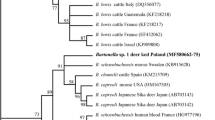
A phylogenetic analysis of representative strains of the 15 genogroups based on the concatenated sequences of the four housekeeping genes (Fig. 2) revealed that the genogroups were divided into five lineages (A, B, C, D and E) (Fig. 2). None of the ked strains belonged to lineage A. Ked strain D13-Ked2.1 (XII) was grouped in lineage B along with the seven deer strains consisting of Honshu-9.1 (IV) to Honshu-11.1 (X). Ked strain D93-Ked3.1 (XV) belonged to lineage C along with deer strain Honshu-16.1 (XI). Ked strains D8-Ked2.1 (XIII) and D8-Ked2.2 (XIV) formed monophyletic clades as lineages D and E, respectively.
Phylogenetic tree of Bartonella strains from deer ked and Japanese sika deer and other ruminants. The tree was produced using the maximum-likelihood method based on the General Time Reversible model. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [G, parameter = 0.4518]). Four representative strains (D8-Ked2.1, D8-Ked2.2, D13-Ked2.1 and D93-Ked3.1) from deer keds, 11 representative strains (Honshu-8.1, -9.1, -9.3, -11.1, -12.1, -16.1, -18.2, -18.5, -53.5, -58.5 and Yezo-25.1) from Japanese sika deer, the ruminant-associated Bartonella species (B. bovis 91-4T, B. capreoli IBS193T, B. capreoli B28980, B. chomelii A828T, B. schoenbuchensis R1T and candidatus B. melophagi K-2C) and several other known Bartonella species were included in the tree. The tree was rooted using Brucella melitensis strain 16MT as an outgroup. Bootstrap values with > 70% confidence are indicated at the tree nodes. Branch lengths are measured as the number of substitutions per site.
The gltA and rpoB sequences of representative strains from the novel genogroups were compared with those of the bacterial genomic sequences registered in INSD using BLAST search (Table 3). The gltA and rpoB sequences of ked strain D13-Ked2.1 (XII) were identical to those of deer strains Honshu-9.1 and Honshu-18.5. The rpoB sequence of ked strain D13-Ked2.1 (XII) showed 98.8% similarity with that of deer strain Honshu-12.1. The gltA sequence of ked strain D8-Ked2.1 (XIII) showed 97.6% similarity with strain MUD detected from Lipoptena sp. in the USA. The rpoB sequence of ked strain D8-Ked2.1 (XIII) showed 99.6% similarity with five uncultured Bartonella strains from L. cervi in Poland. The gltA sequence of ked strain D8-Ked2.2 (XIV) showed 96.7% similarity with deer strains Honshu-9.1 and Honshu-18.5 from sika deer, whereas the rpoB sequence showed 97.2% similarity with an uncultured Bartonella strain detected from a white-tailed deer (Odocoileus virginianus) in the USA. The gltA and rpoB sequences of ked strain D93-Ked3.1 (XV) showed sequence similarity of 100% and 99.4% for gltA and rpoB, respectively, to deer strain Honshu-16.1.
Relationship of Bartonella lineages among hematophagous arthropods and sika deer
Fourteen deer keds and one tick harbored Bartonella strains of lineage B, whereas two keds (ID# D93-Ked3 and D96-Ked2) harbored lineage C strains (Table 4). Another ked, D8-Ked2, harbored two Bartonella strains classified in lineages D and E. Five of eight deer harbored Bartonella strains classified in lineage B, and the others harbored Bartonella strains classified in lineage A.
Deer 11, 12, 89 and 93 harbored Bartonella strains of the same lineage (B) as the hematophagous arthropods D11-Tick12, D12-Ked1, D89-Ked2, D93-Ked1, D93-Ked2 and D93-Ked4. In contrast, deer 8, 13, 91 and 96 harbored Bartonella strains of lineage A or B, but no strains of the same lineages were detected from the keds that infested the deer.
Discussion
In the present study, wingless keds collected from Japanese sika deer were morphologically identified as Lipoptena fortisetosa and found to harbor Bartonella bacteria at a high rate (87.9%). Halos et al. [11] first demonstrated that Bartonella bacteria were present at a high rate (93.8%) in wingless L. cervi collected from French roe deer. Since then, it has been reported that Bartonella bacteria are prevalent also in the keds collected from red deer and roe deer in Hungary [14] and Poland [27] and from moose in Norway [28]. Likewise, Bartonella DNAs were detected from both wingless L. cervi (83.3%) [13] and wingless L. mazamae (28.9%) collected from white-tailed deer in the USA [29], and from 100% of wingless L. mazamae collected from gray brocket deer in Brazil [30]. These epidemiological data show that Lipoptena keds harbor high levels of Bartonella bacteria and are likely to play an important role in transmitting the bacteria between various deer species.
We isolated Bartonella bacteria from 51.5% of the wingless L. fortisetosa collected. More than 1000 Bartonella colonies were recovered from a ked (ID# D93-Ked4) using the culture method, and a very high number of Bartonella cell counts (7.0 × 106) were estimated by Bartonella-specific qRT-PCR. These data suggest that L. fortisetosa offer an optimum environment for Bartonella bacteria. Results from a survey of deer in Germany demonstrated that a large number of Bartonella colonies (> 1000 per L. cervi) could be recovered from L. cervi by the culture method, with the immunohistochemical and transmission electron microscopic analyses also showing that bacterial aggregates were only present in the ked midguts, suggesting that Bartonella bacteria proliferated in the midgut of L. cervi [12]. In the present study, our immunofluorescent analysis also found that Bartonella aggregates were only detected in the midgut of L. fortisetosa. This result is consistent with those of the previous study [12] and suggests that Bartonella bacteria may also propagate in the midgut of L. fortisetosa.
Although Bartonella DNAs were detected from Ixodes ricinus ticks in France [31] and Poland [32], it is unknown whether the Bartonella DNAs were host-borne or not. Moreover, it is suggested that the mere presence of Bartonella DNA in ticks is not enough to prove vector competence of Bartonella bacteria [33]. Tijsse-Klasen et al. [34] reported that Bartonella DNA was not found in 1719 questing I. ricinus nymph and adult ticks collected in the field in the Netherlands. In the present study, the isolation rate from ticks was considerably lower than that from keds, as Bartonella bacteria were isolated only from one engorged Haemaphysalis tick collected on a bacteremic sika deer. The lineage of the Bartonella isolated from tick D11-Tick12 was the same (lineage B) as that of the deer from which the tick was collected. This suggests that the Bartonella bacteria might have been present in the deer blood ingested by the tick. Therefore, it is possible that Ixodes and Haemaphysalis ticks do not play a significant role in the transmission of Bartonella in deer.
The phylogenetic analysis shows that novel genogroups XII and XV were included in lineages B and C, respectively. A BLAST search also indicated that ked strain D13-Ked2.1 (genogroup XII) was closest to deer strains Honshu-9.1, Honshu-18.5 and Honshu-12.1 in lineage B, whereas ked strain D93-Ked3.1 (genogroup XV) was closest to deer strain Honshu-16.1 in lineage C. In our previous study [16], the Bartonella strains in lineage A were identified as B. capreoli; however, the strains in lineages B and C did not cluster with known Bartonella species, suggesting that these strains are novel Bartonella species. From these results, genogroup XII in lineage B and genogroup XV in lineage C are also suggested to be novel Bartonella species. The ked strains D8-Ked2.1 (genogroup XIII) and D8-Ked2.2 (genogroup XIV) formed the new independent lineages D and E, respectively. Furthermore, the gltA and rpoB sequences of ked strain D8-Ked2.1 (genogroup XIII) were closest to those of uncultured Bartonella species detected from Lipoptena keds in the USA and Poland. Although the gltA sequence of ked strain D8-Ked2.2 (genogroup XIV) was similar to deer strains Honshu-9.1 and Honshu-18.5, the rpoB sequence of the same genogroup was similar to uncultured Bartonella species detected from white-tailed deer in the USA [35]. These findings support the suggestion that genogroup XIII in lineage D and genogroup XIV in lineage E may also be novel Bartonella species.
In our previous study, the deer Bartonella strains from Hokkaido, Wakayama and Nara Prefectures were classified in lineages A to C [16]. In the present study, the Bartonella strains from the deer in Shizuoka Prefecture were classified in lineages A and B, while most of the strains from L. fortisetosa were classified in lineages B and C. These data indicate that L. fortisetosa harbors two novel Bartonella species, namely lineages B and C, and may transmit the bacteria to Japanese sika deer. The other novel Bartonella species, namely lineages D and E, were found in only one ked (D8-Ked2), but not in sika deer or other deer species in other countries. Further analyses are required to determine whether these new lineages are deer ked-specific Bartonella species.
Deer keds are generally thought to stay on the same deer body after dropping their wings. However, Samuel and Trainer [9] found evidence that wingless L. mazamae on white-tailed deer can mechanically transfer between individuals during direct contact between deer. In the present study, the Bartonella lineages of deer keds differed from those of four sampled deer (deer 8, 13, 91, and 96), raising the possibility that wingless L. fortisetosa may also be able to transfer between individuals within the deer population. Given that Bartonella bacteria propagate in the ked midgut, Lipoptena keds may serve as a biological vector for transmitting Bartonella bacteria to deer.
It has been suggested that B. schoenbuchensis can be transmitted not only vertically from adult L. cervi to offspring [14], but also transstadially to the next growth stage of L. cervi [10]. Unfortunately, we did not obtain any pupae and winged adult keds and so could not examine these possibilities in our study. Further studies are needed to resolve the questions of transovarial and transstadial transmission of Bartonella bacteria in L. fortisetosa.
Interestingly, B. capreoli (lineage A) was not found in any deer keds examined in our study, although this Bartonella species was isolated from three sika deer. To date, B. capreoli has not been detected from L. cervi and/or L. mazamae in Europe and the USA although this species has frequently been isolated from several deer species. The absence of this Bartonella species in Lipoptena species suggests that other hematophagous arthropods may be involved in the transmission of B. capreoli between deer populations.
Conclusions
Our data indicate that L. fortisetosa likely serves as a vector of at least two Bartonella species in Japanese sika deer, similar to L. cervi and L .mazamae transmitting Bartonella bacteria to deer in Europe and the USA. In contrast, Ixodes and Haemaphysalis ticks may not play a significant role in the transmission of Bartonella in the sika deer.
Availability of data and materials
The nucleotide sequences of the ked strains were submitted to INSD under accession numbers LC485114–LC485129.
References
Kosoy M, Hayman DT, Chan KS. Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol. 2012;12:894–904.
Caceres AG. Geographic distribution of Lutzomyia verrucarum (Townsend, 1913) (Diptera, Psychodidae, Phlebotominae), vector of human bartonellosis in Peru. Rev Inst Med Trop Sao Paulo. 1993;35:485–90.
Fournier PE, Minnick MF, Lepidi H, Salvo E, Raoult D. Experimental model of human body louse infection using green fluorescent protein-expressing Bartonella quintana. Infect Immun. 2001;69:1876–9.
Seki N, Kasai S, Saito N, Komagata O, Mihara M, Sasaki T, et al. Quantitative analysis of proliferation and excretion of Bartonella quintana in body lice, Pediculus humanus L.. Am J Trop Med Hyg. 2007;77:562–6.
Chomel BB, Kasten RW, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, et al. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–6.
Tsai YL, Chang CC, Chuang ST, Chomel BB. Bartonella species and their ectoparasites: selective host adaptation or strain selection between the vector and the mammalian host? Comp Immunol Microbiol Infect Dis. 2011;34:299–314.
Kortet R, Harkonen L, Hokkanen P, Harkonen S, Kaitala A, Kaunisto S, et al. Experiments on the ectoparasitic deer ked that often attacks humans; preferences for body parts, colour and temperature. Bull Entomol Res. 2010;100:279–85.
Rantanen T, Reunala T, Vuojolahti P, Hackman W. Persistent pruritic papules from deer ked bites. Acta Derm Venereol. 1982;62:307–11.
Samuel WM, Trainer DO. Lipoptena mazamae Rondani, 1878 (Diptera: Hippoboscidae) on white-tailed deer in southern Texas. J Med Entomol. 1972;9:104–6.
Korhonen EM, Perez Vera C, Pulliainen AT, Sironen T, Aaltonen K, Kortet R, et al. Molecular detection of Bartonella spp. in deer ked pupae, adult keds and moose blood in Finland. Epidemiol Infect. 2015;143:578–85.
Halos L, Jamal T, Maillard R, Girard B, Guillot J, Chomel B, et al. Role of Hippoboscidae flies as potential vectors of Bartonella spp. infecting wild and domestic ruminants. Appl Environ Microbiol. 2004;70:6302–5.
Dehio C, Sauder U, Hiestand R. Isolation of Bartonella schoenbuchensis from Lipoptena cervi, a blood-sucking arthropod causing deer ked dermatitis. J Clin Microbiol. 2004;42:5320–3.
Matsumoto K, Berrada ZL, Klinger E, Goethert HK, Telford SR 3rd. Molecular detection of Bartonella schoenbuchensis from ectoparasites of deer in Massachusetts. Vector Borne Zoonotic Dis. 2008;8:549–54.
de Bruin A, van Leeuwen AD, Jahfari S, Takken W, Foldvari M, Dremmel L, et al. Vertical transmission of Bartonella schoenbuchensis in Lipoptena cervi. Parasites Vectors. 2015;8:176.
Nagata J, Masuda R, Tamate HB, Hamasaki S, Ochiai K, Asada M, et al. Two genetically distinct lineages of the sika deer, Cervus nippon, in Japanese islands: comparison of mitochondrial D-loop region sequences. Mol Phylogenet Evol. 1999;13:511–9.
Sato S, Kabeya H, Yamazaki M, Takeno S, Suzuki K, Kobayashi S, et al. Prevalence and genetic diversity of Bartonella species in sika deer (Cervus nippon) in Japan. Comp Immunol Microbiol Infect Dis. 2012;35:575–81.
Raoult D, Roblot F, Rolain JM, Besnier JM, Loulergue J, Bastides F, et al. First isolation of Bartonella alsatica from a valve of a patient with endocarditis. J Clin Microbiol. 2006;44:278–9.
Angelakis E, Roux V, Raoult D, Rolain JM. Real-time PCR strategy and detection of bacterial agents of lymphadenitis. Eur J Clin Microbiol Infect Dis. 2009;28:1363–8.
Houpikian P, Raoult D. 16S/23S rRNA intergenic spacer regions for phylogenetic analysis, identification, and subtyping of Bartonella species. J Clin Microbiol. 2001;39:2768–78.
Lee ZM, Bussema C III, Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37(Database issue):D489–93.
Kawamoto T. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histol Cytol. 2003;66:123–43.
Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–803.
Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. Use of rpoB gene analysis for detection and identification of Bartonella species. J Clin Microbiol. 2001;39:430–7.
Zeaiter Z, Liang Z, Raoult D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J Clin Microbiol. 2002;40:3641–7.
Inoue K, Kabeya H, Kosoy MY, Bai Y, Smirnov G, McColl D, et al. Evolutional and geographical relationships of Bartonella grahamii isolates from wild rodents by multi-locus sequencing analysis. Microb Ecol. 2009;57:534–41.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Szewczyk T, Werszko J, Steiner-Bogdaszewska Z, Jezewski W, Laskowski Z, Karbowiak G. Molecular detection of Bartonella spp. in deer ked (Lipoptena cervi) in Poland. Parasites Vectors. 2017;10:487.
Duodu S, Madslien K, Hjelm E, Molin Y, Paziewska-Harris A, Harris PD, et al. Bartonella infections in deer keds (Lipoptena cervi) and moose (Alces alces) in Norway. Appl Environ Microbiol. 2013;79:322–7.
Reeves WK, Nelder MP, Cobb KD, Dasch GA. Bartonella spp. in deer keds, Lipoptena mazamae (Diptera: Hippoboscidae), from Georgia and South Carolina, USA. J Wildl Dis. 2006;42:391–6.
Souza U, Dall’Agnol B, Michel T, Webster A, Klafke G, Martins JR, et al. Detection of Bartonella sp. in deer louse flies (Lipoptena mazamae) on gray brocket deer (Mazama gouazoubira) in the Neotropics. J Zoo Wildl Med. 2017;48:532–5.
Halos L, Jamal T, Maillard R, Beugnet F, Le Menach A, Boulouis HJ, et al. Evidence of Bartonella sp. in questing adult and nymphal Ixodes ricinus ticks from France and co-infection with Borrelia burgdorferi sensu lato and Babesia sp. Vet Res. 2005;36:79–87.
Skotarczak B, Adamska M. Detection of Bartonella DNA in roe deer (Capreolus capreolus) and in ticks removed from deer. Eur J Wildlife Res. 2005;51:287–90.
Telford SR 3rd, Wormser GP. Bartonella spp. transmission by ticks not established. Emerg Infect Dis. 2010;16:379–84.
Tijsse-Klasen E, Fonville M, Gassner F, Nijhof AM, Hovius EK, Jongejan F, et al. Absence of zoonotic Bartonella species in questing ticks: first detection of Bartonella clarridgeiae and Rickettsia felis in cat fleas in the Netherlands. Parasites Vectors. 2011;4:61.
Tate CM, Mead DG, Luttrell MP, Howerth EW, Dugan VG, Munderloh UG, et al. Experimental infection of white-tailed deer with Anaplasma phagocytophilum, etiologic agent of human granulocytic anaplasmosis. J Clin Microbiol. 2005;43:3595–601.
Acknowledgements
We would like to express our gratitude all of our laboratory staff involved in this study.
Funding
This work was supported in part by the Strategic Research Base Development Program, International Research on Epidemiology of Zoonoses and Training for Young Researchers by the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Contributions
SS had a central role in performing the study, analyzing the data and writing the final manuscript. SM planned the study design and revised the manuscript. HK helped with analyses of the data and revised the manuscript. SI isolated the Bartonella bacteria and performed PCR and DNA sequencing together with SS. YS helped with immunofluorescence staining of the deer keds. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sato, S., Kabeya, H., Ishiguro, S. et al. Lipoptena fortisetosa as a vector of Bartonella bacteria in Japanese sika deer (Cervus nippon). Parasites Vectors 14, 73 (2021). https://doi.org/10.1186/s13071-021-04585-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-021-04585-w