Abstract
Background
Bartonella is a genus of Gram-negative facultative intracellular Alphaproteobacteria of public health importance. Although they are known to mainly infect mammalian hosts with some blood-feeding arthropods having been confirmed as vectors, there is some evidence of Bartonella association with non-mammalian hosts including birds.
Methods
Here we used high-throughput sequencing of 16S rRNA and Sanger sequencing of the citrate synthase (gltA) genes to test for the presence of Bartonellaceae in the blood of three migratory cavity nesting bird species, purple martins (Progne subis), tree swallows (Tachycineta bicolor) and eastern bluebirds (Sialia sialis) and their most prevalent and abundant nest ectoparasites, Dermanyssus prognephilus (mite), Ceratophyllus idius (flea) and Protocalliphora sialia (bird blow fly larva). We constructed maximum likelihood phylogenetic trees to verify the placement of the resulting sequences in the Bartonellaceae.
Results
We found evidence of Bartonella in all three bird species and all three arthropod species tested. We report multiple instances of identical Bartonella sequences in both birds and parasites, leading to the likely hypothesis that these ectoparasites are potential vectors of Bartonella. Our phylogenetic analysis suggests that ‘avian Bartonella’ may form its own sub-clade within the genus Bartonella.
Conclusions
To the best of our knowledge, we provide the first confirmation of overlapping Bartonella strains among bird hosts and various species of nest-associated ectoparasites from the same system, suggesting a possible Bartonella host–vector relationship between these arthropods and a non-mammalian host. Our study adds to the growing appreciation of the Bartonellaceae as a phylogenetically diverse group with a wide range of hosts.

Similar content being viewed by others
Background
Bartonella is a genus of Gram-negative facultative intracellular Alphaproteobacteria whose niche is traditionally described as within the red-blood cells of mammalian hosts, from which it is transferred between hosts by blood-feeding arthropods [1, 2]. The genus comprised a single species (Bartonella quintana) until as recently as 1993, but since then increased diversity has been recognized with 35 species with confirmed nomenclature and a further 10 putative species now in the literature [3]. In parallel with the increasing recognition of diversity within Bartonella, there has also been an expanding list of known host species. Indeed, our knowledge of Bartonella is no longer strictly limited to mammal-arthropod systems, as it has now been isolated by two studies in non-mammalian vertebrate hosts. One study found it in loggerhead sea turtles (Caretta caretta) [4] and another study found B. henselae and B. koehlerae in blood samples from North American wild birds [northern mockingbirds (Mimus polyglottos), red-winged blackbirds (Agelaius phoeniceus), red-bellied woodpeckers (Melanerpes carolinus) and common loons (Gavia immer)] [5]. If Bartonella is circulating in wild birds, it may also be assumed to be in some subset of hematophagous avian ectoparasites. Accordingly, the presence of B. grahamii has been confirmed in the tick species Ixodes turdus which was collected from migratory birds in Korea [6] and in the hematophagous mites of the bird-associated genus Dermanyssus in the Czech Republic [7]. Furthermore, a recent study found a predominance of ‘Bartonella-like’ bacteria in the microbiome of the poultry ectoparasite, Dermanyssus gallinae [8]. Although this study did not achieve a high taxonomic resolution on these bacteria (leading the authors to conservatively label it as ‘Bartonella-like’ bacteria), this could be of high commercial and animal-health interest as D. gallinae has a worldwide distribution with over 80% of poultry farms reporting infestations [9].
Although evidence for Bartonella in avian host–parasite systems is clearly growing, there has not so far been any study to link presence of Bartonella between avian-associated arthropods (e.g. hematophagous ectoparasites) and birds. Cavity-nesting birds and especially those using man-made nest boxes [10] are likely candidates for providing a fitting model as they are associated with a high load of hematophagous nest ectoparasites [11, 12]. Three species which fit this profile are purple martins (Progne subis), eastern bluebirds (Sialia sialis) and tree swallows (Tachycineta bicolor). They are all wide-ranging migrant cavity-nesting birds, which are sympatric throughout much of their breeding ranges and regularly (or exclusively in the case of the eastern subspecies of the purple martin) make their nests in man-made nest boxes [13,14,15]. While the taxonomic composition of their nest parasites appears to vary throughout their ranges [13,14,15,16], at our field site in western New York State, three hematophagous ectoparasite species (a mite, Dermanyssus prognephilus; a flea, Ceratophyllus idius; and a bird blow fly larva, Protocalliphora sialia) are found at high prevalence (and generally high abundance) in the nests of all three bird species as well as on the birds (HMW, unpublished data).
Here we use high-throughput sequencing of the bacterial 16S rRNA paired with Sanger sequencing of the citrate synthase (gltA) gene to detect Bartonella DNA in these three species of nest-associated ectoparasites and in the blood of nestlings of three of their avian host species. We discuss the phylogenetic placement of these sequences in terms of known Bartonella diversity and the potential for in-nest vector relationships under the hypothesis that overlapping sequences will be identified in both birds and their ectoparasites.
Methods
Sample collection
Fieldwork was carried out at Iroquois National Wildlife Refuge and Tonawanda Wildlife Management Area in Genesee County, New York State in the summers of 2017 and 2018. Purple martin, eastern bluebird and tree swallow nests were monitored as part of a wider project documenting the effect of nest-associated ectoparasites on nestling fitness. Blood samples were drawn from nestlings of 49 purple martins from 33 different nests; from 6 eastern bluebirds from 4 nests and from 6 tree swallows from 4 nests. The unequal sample size reflects the priorities of the aforementioned wider project. Nestlings were sampled at approximately 2/3 of their way through the nestling period (purple martin nestlings were aged between 16–20 days-old at the time of blood draw, with a mean fledging age of 28 days, whereas bluebird and tree swallow nestlings were sampled between 11–12 days-old and typically fledge at 18 and 20 days-old, respectively) to allow a sufficient period for a potential Bartonella infection to proliferate in the blood without risking prompting premature fledging by handling birds too late in their nestling stage [17]. We chose to sample nestlings rather than adults to reduce the number of possible of origins of a potential Bartonella infection. In each case blood was drawn from the brachial vein using a 27.5 gauge needle to collect a maximum of 140 µl of blood in heparinized microhematocrit capillary tubes. Blood samples were kept on ice in the field until being stored at − 70 °C on the same day until samples were processed.
Nest materials were collected after the birds fledged and were placed in Berlese funnels for 2–3 h to allow nest-dwelling invertebrates to collect in ethanol. Three hematophagous parasites were common in the sampled nests of all three bird species, the mite Dermanyssus prognephilus, the flea Ceratophyllus idius and the bird blow fly larva Protocalliphora sialia. Nest materials from purple martins were also sampled at regular intervals during the nesting period as part of a wider study. The same three species of nest-associated ectoparasites were commonly present at every sampling period. Parasites were identified via morphology following Moss [18], Traub et al. [19] and Sabrosky et al. [16], respectively, using a Zeiss microscope. Specimens were stored in 95% ethanol at − 70 °C and washed in PBS prior to DNA extraction. All samples (blood and ectoparasites) were collected between the last week in June and the first week in August in both years.
DNA extraction
DNA was extracted from blood samples (n = 61) using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA), directly following the handbook Spin-Column protocol for nucleated animal blood. In brief, 20 μl of proteinase K was added to 10 μl of blood, 190 μl of PBS and 200 μl of buffer AL and vortexed prior to incubation at 56 °C for 10 min. 200 μl of ethanol was added to the sample prior to centrifugation at 8000× rpm for 1 min in a spin column. Two further centrifugation steps were carried out, with 500 μl of buffer AW1 and then 500 μl of buffer AW2 added at each step. We used two elutions using 100 μl of buffer AE and 1 min of centrifugation at 8000× rpm before the DNA eluant was transferred to a stock tube.
Ectoparasite samples (D. prognephilus: n = 69; C. idius: n = 76; P. sialia: n = 12) were washed in PBS, then homogenized using sterile razor blades and incubated at 56 °C overnight in 180 μl ATL and 20 μl proteinase K. DNA was then extracted as per the Qiagen DNeasy Blood and Tissue Kit handbook protocol for Spin-Column animal tissues extractions. In summary, samples were vortexed and incubated for 1 h at 56 °C. We then added 200 μl of buffer AL and 200 μl of ethanol, transferred the solution to a spin column and centrifuged at 8000× rpm for 1 min. Two further centrifugation steps were carried out using buffer AW1 and buffer AW2 as was done for the blood samples. Finally, we used two elutions of 75 μl buffer AE rather than the standard 200 μl so as to increase the DNA concentration. Each sample contained multiple ectoparasite individuals of the same species from the same nest (8 for D. prognephilus, 4 for C. idius and 2 for P. sialia) to ensure sufficient DNA concentrations for analysis.
16S rRNA microbiome sequencing
We sequenced the hypervariable V3–V4 region of bacterial 16S rRNA from all samples as part of a separate microbiome study. PCR, library preparation and sequencing were carried out by the University at Buffalo Genomics and Bioinformatics Core facility following the 16S metagenomics sequencing library preparation guide for the Illumina MiSeq system. Initial DNA concentrations were measured using a Qubit Fluorometer and were adjusted to 5 ng/μl. In the first stage PCR, 2.5 μl of DNA was mixed with 12.5 μl 2× KAPA HiFi HotStart Readymix and 5 μl each of 1 μM forward and reverse primers (forward: 5′-CCT ACG GGN GGC WGC AG-3′, reverse: 5′-GAC TAC HVG GGT ATC TAA TCC-3′). This primer set was chosen as it has been found to have the best overall coverage for this bacterial region [20]. PCR was performed in a thermal cycler at initial denaturation at 95 °C for 3 min, followed by 25 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s followed by a final elongation step for 5 min at 72 °C. Second stage PCR allowed for attachment of Illumina sequencing adapters. 5 μl of first stage PCR product was added to each well on a new plate with 5 μl of each index primer, 25 μl 2× KAPA HiFi HotStart Readymix and 10 μl of PCR-grade water. PCR was performed in a thermal cycler at initial denaturation at 95 °C for 3 min, followed by 8 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s followed by a final elongation step for 5 min at 72 °C. PCR clean-up was performed after each reaction using 20 μl of AMPure XP beads in each well, final concentrations were measured using a Quant IT DsDNA Assay Kit and adjusted to 4 nM using EB buffer. Libraries were denatured using NaOH, diluted with hybridization buffer and heat denatured. Samples were loaded with 5% PhiX internal controls and were sequenced using the Illumina Miseq with 300-cycle paired-end sequencing with a negative control (no DNA added) on each plate.
Raw sequences were imported to QIIME2 (https://qiime2.org) as paired-end sequences after primer sequence removal and demultiplexed. We used DADA2 [21] to remove chimeras and to denoise our sequences by trimming the first 5 bases from both the forward and reverse sequences and truncating the reverse sequences at 223 bases based on sequence quality. We trained a naïve Bayesian classifier using the Greengenes 99% OTU database from version 13.8 [22] and extracted reads from the reference database which match the sequencing primer pair. We then used the classifier to cluster our reads to 1% Amplicon Sequence Variants (ASVs) and assign taxonomy.
16S rRNA Bartonella verification
In this microbiome study, 40 different ASVs identified to the family Bartonellaceae in our classification system, none of which occurred in the 4 negative controls (one per plate). We cross-referenced these sequences with NCBI GenBank using the BLASTn tool to perform a search limited to the Bartonellaceae. All 40 ASVs were associated with Bartonella sequences with low e-values and generally high coverage (Additional file 1: Table S1).
From GenBank we obtained a total of 93 homologous 16S rRNA sequences including 33 Bartonella species and sister taxa including 10 species of Mesorhizobium, 10 species of Ochrobactrum, 12 species of Brucella and 1 species of Rhizobium to use as outgroup (Additional file 2: Table S2). We also included 10 sequences classified as ‘Bartonella-like’ derived from Dermanyssus gallinae [8] (a taxon related to our mite species, D. prognephilus) (Additional file 2: Table S2). We were not able to include comparison sequences from the only other study of avian Bartonella [5] or from the study of Bartonella in avian ticks [6] as they sequenced different genomic regions. Sequences were aligned in Seaview v.4.6.5 [23] using Clustal-O. The aligned sequences were trimmed to consistently include a 430 bp of the 16S rRNA V3–V4 region across all samples.
To maintain a conservative analysis, we removed some ASVs on from our analysis at this point based on the following criteria: (i) two ASVs (Mite_2 and Mite_4) were removed due to a relatively poor alignment; (ii) four ASVs (Mite_8, Mite_9, Mite_10, Mite_12) were removed as they had shorter sequences than the other ASVs; (iii) Nine ASVs (Mite_7, Mite_14, Flea_1, Flea_3, Flea_4, Flea_5, PUMA_1, PUMA_3, TRES_2) were removed as there were fewer than 20 reads in the full dataset; and (iv) seven ASVs (Mite_1, Mite_5, Mite_6, Mite_11, Mite_16, Flea_2, PUMA_2) were removed as their percentage identity with the Bartonellaceae from BLASTn was in the lower quartile of our samples (i.e. below 98%). After quality filtering, we were left with 22 ASVs which were submitted to NCBI GenBank: MN320509-MN320530.
We used MEGA7 [24] to find the best-fit nucleotide substitution model for our data according to BICc. As K2 + G + I was selected as the best-fit model (Additional file 3: Table S3), we created a maximum likelihood tree in Seaview using the K80 substitution model, optimized invariable sites (I) and optimized across site rate variation (G), with 100 bootstrap replicates and the best of nearest neighbor and subtree pruning with 5 random starts. The resulting tree was exported from Seaview to FigTree v.1.4.4 [http://tree.bio.ed.ac.uk/software/figtree/] for editing.
PCR confirmation of Bartonella gltA
While the 16S rRNA gene is an appropriate target for metagenomics studies, it does not always provide high resolution for Bartonella sequences [25]. To support our identification of Bartonella, we therefore performed PCRs for the Bartonella citrate synthase gene (gltA) for a representative subset of our samples, which contained 16S rRNA Bartonella sequences. Two samples from eastern bluebird blood, 2 samples from tree swallow blood, 9 samples from purple martin blood, 7 samples from C. idius and 13 samples of D. prognephilus were selected for analysis.
PCR amplifications were performed in a 25 μl reaction including 1 μl of DNA, 0.2 μl of 5 U/μl Taq, 0.5 μl each of 10 μM forward and reverse primers, 2.5 μl of 50 mM dNTPs, 2.5 μl of 10× buffer, 1.5 μl of MgSO4 and 13.8 μl of PCR grade water. We used BhCS781.p (5′-GGG GAC CAG CTC ATG GTG G-3′) and BhCS1137.n (5′-AAT GCA AAA AGA ACA GTA AAC A-3′) primers to generate a 379-bp amplicon of the Bartonella gltA gene [26]. PCR reactions were performed with the following cycling conditions: one 5-min cycle at 95 °C, followed by 30 cycles of 95 °C for 30 s, 50 °C for 30 s and 72 °C for 40 s, followed by 1 final cycle at 72 °C for 7 min. PCR clean-up was performed using AMPure XP beads. Positive amplicons were sequenced with an ABI 3730XL sequencer at TACGen (http://www.tacgen.com). PCR-positive bands were obtained in all D. prognephilus samples (n = 13), all but 2 of the C. idius samples (n = 8) and all but 2 of the avian blood samples (n = 13). Paired-end sequences were merged, and the quality of the resulting consensus sequences was assessed from chromatograms in Geneious v.8.0.5 (https://www.geneious.com). However, sequence quality was very poor for all blood and C. idius samples and three of the D. prognephilus samples and these samples conservatively recorded as negative for Bartonella and excluded from further analysis. All of the 10 mite samples with high quality consensus gltA sequences were identified to Bartonella sequences using a BLASTn search (Additional file 1: Table S1). These 10 ASVs were submitted to NCBI GenBank: MN371264–MN371273.
As individual samples generated multiple Bartonella ASVs in the 16S rRNA sequencing, we did not concatenate our sequences, but rather generated a second phylogeny based on the gltA data to compare with the 16S rRNA tree. To that end we downloaded 51 representative homologous sequences including 13 species of Bartonella, 2 species of Mesorhizobium along with 7 species of Brucella sequences from GenBank, along with 3 species of Rhizobium to use as the outgroup (Additional file 2: Table S2). Sequences were aligned in Seaview using the Clustal-O algorithm and trimmed to the 379 bp region of interest. MEGA7 indicated that K2 + G was the best substitution model for the data (Additional file 4: Table S4), so Seaview was used with the K80 model and optimized across site rate variation (G), with 100 bootstrap replicates and the best of nearest neighbor and subtree pruning with 5 random starts. The resulting tree was exported from Seaview to FigTree for editing.
Results
Bartonella sequences from 16S rRNA sequencing
Sequences assigned to Bartonella were found in all three nest-associated ectoparasite species and all three cavity-nesting bird species (Additional file 5: Table S5). Prevalence of Bartonella in the bird species ranged between 33% (2/6) for eastern bluebirds, 39% (19/49) and 83% (5/6) for tree swallows. Ectoparasite samples were comprised of multiple individuals (8 for D. prognephilus, 4 for C. idius and 2 for P. sialia) which complicates the calculation of prevalence. At the sample level, a quarter of the P. sialia samples (3/12) and 36% of the C. idius samples (27/76) were Bartonella-positive. Only one D. prognephilus sample out of 57 did not contain Bartonella, but as that sample had a low total number of reads compared with other samples, we consider prevalence to approach 100%. Considering the limits of individual prevalence, these ranged between 13–25% in P. sialia, 9–36% in C. idius and 13–100% in D. prognephilus. The mites had a very high relative abundance of Bartonella sequences in their microbiome compared with the other arthropod and avian species (Table 1). Every nest we sampled contained some evidence of Bartonella, whether that be in the ectoparasites, in the birds, or (typically) both (Additional file 5: Table S5).
Multiple ASVs of Bartonella were found in all species tested; many individual birds harbored multiple Bartonella ASVs and many nests contained ectoparasite species containing multiple Bartonella sequences (Table 2). Of the 22 unique sequences of Bartonella found in our dataset after quality filtering, 5 ASVs were found only in birds and 7 ASVs were only found in nest-associated ectoparasites. Of particular interest, however, are the 10 strains which were isolated in both birds and ectoparasites (Fig. 1). Three of these cross-taxa strains included examples of the same Bartonella sequence existing in the nestlings and parasites from the same nest, providing strong support for a likely vector–host relationship.
Venn diagram showing distribution of unique 16S rRNA Bartonella ASVs between the three ectoparasite hosts [mites (Dermanyssus prognephilus), blow fly (Protocalliphora sialia) and fleas Ceratophyllus idius] and the three bird species tested [purple martins (Progne subis), tree swallows (Tachycineta bicolor) and eastern bluebirds (Sialia sialis)]. Ten Bartonella ASVs were found in all host species tested
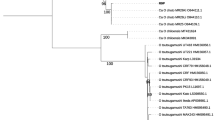
Comparison of the putative Bartonella 16S rRNA sequences from our dataset with other Bartonella sequences in the literature and homologous sequences from sister taxa showed that all of our sequences are situated in a well-supported clade with the Bartonella sequences from previous efforts (Fig. 2). Mesorhizobium formed its own clade and Brucella and Ochrobactrum grouped together in a well-supported clade. One sequence of Bartonella apis did not group with the other Bartonella sequences and although not a well-supported division, appeared closer to the Ochrobactrum/Brucella clade. ‘Bartonella-like’ sequences isolated from Dermanyssus gallinae [8], clearly grouped with our own sequences and appear to form their own clade. There was no clear separation between our putative Bartonella strains isolated from bird blood or ectoparasites, with many cases of the same sequence being isolated in both birds and arthropods (Figs. 1, 2).
Maximum likelihood phylogenetic tree showing the placement of our 16S rRNA Bartonella sequences within a wider taxonomy. The tree used the K80 substitution model, optimized invariable sites (I) and optimized across site rate variation (G), with 100 bootstrap replicates and the best of nearest neighbor and subtree pruning with 5 random starts. Numbers on branches show bootstrap support values. Only bootstrap supports above 0.75 are shown and monophyletic nodes are collapsed. The scale-bar indicates substitutions per site. “Dermanyssus gallinae” refers to Bartonella sequences obtained from the microbiome of Dermanyssus gallinae by Hubert et al. [8]. “Avian and ectoparasite” refers to ASVs from this study
Bartonella confirmation with gltA PCR and sequencing
Phylogenetic analysis showed that our mite Bartonella gltA sequences were within a single well-supported clade along with Bartonella sequences, supporting the findings from the 16S rRNA tree. Our sequences were situated in two sub-clades, with the sequences from mites parasitizing purple martins (Group B) being apparently closer to Bartonella tamiae and sequences from mites parasitizing tree swallows and eastern bluebirds (Group A) as a sister clade to the other Bartonella sequences. Mesorhizobium and Brucella both formed their own well-supported clades (Fig. 3).
Maximum likelihood phylogenetic tree showing the placement of our gltA Bartonella sequences within a wider taxonomy. The tree used the K80 substitution model, optimized invariable sites (I) and optimized across site rate variation (G), with 100 bootstrap replicates and the best of nearest neighbor and subtree pruning with 5 random starts. Numbers on branches show bootstrap support values. Only bootstrap supports above 0.75 are shown and monophyletic nodes are collapsed. The scale-bar indicates substitutions per site. ASVs in ‘Group A’ and ‘Group B’ were all isolated from Dermanyssus prognephilus in this study
Discussion
Results from this study have shown that there is a likely Bartonella host–vector relationship in an avian system. Our results support the findings of Mascarelli et al. [5] that Bartonella occurs in birds and nearly double the list of avian species known to carry Bartonella. We also add three species to the pool of avian-associated arthropods known to carry Bartonella spp. [6,7,8, 27]. Our phylogenetic analyses highlight potential expanded diversity within the Bartonellaceae associated with avian-system derived Bartonella ASVs. Our work implies that we need to broaden our view of the Bartonellaceae both in terms of the diversity of its reservoir and vector host range and in terms of phylogenetic diversity within the group.
Our finding of multiple identical Bartonella ASVs in birds and arthropods, sometimes within the same nest, strongly suggests a host–vector relationship. All three nest-associated ectoparasites sampled (Dermanyssus prognephilus, Ceratophyllus idius and Protocalliphora sialia) shared Bartonella ASVs with nestling birds and all three species were found in nests continuously throughout the nesting period (HMW, unpublished data), implying that any or all of these species could be vectoring Bartonella to the birds. Of the three, D. prognephilus may be the most likely vector candidate for two reasons. First, D. prognephilus is the most abundant and prevalent nest-associated ectoparasite of all three bird species at our field location, with prevalence approaching 100% for all three species and abundance estimates ranging from means of 300 mites per nest for eastern bluebirds, 600 for tree swallows and almost 2000 for purple martins (HMW, unpublished data). Given a positive correlation between vector abundance and transmission in other host–parasite systems [28,29,30], D. prognephilus may have a significant role in transmission in this system, simply due to the higher encounter rate between birds and mites compared to with the other ectoparasites. Secondly, our microbiome study of D. prognephilus suggests a much higher relative abundance of Bartonella than in either of the other two ectoparasites considered, or than observed in the birds themselves.
While vector-transmission appears to be a parsimonious explanation of our finding of identical ASVs in birds and ectoparasites, especially given most of the previous evidence that has been gathered for the Bartonellaceae, our study does not specifically address transmission modes for the bartonellae. Vector transmission of the Bartonellaceae by arthropods has been recorded both directly via blood-feeding [1, 31] and indirectly through contamination with the feces of infected arthropods, for example in humans scratching themselves due to infection with the body louse Pediculus humanus [32], or when humans acquire cat-scratch fever due to inoculation with infected fleas after cutaneous trauma from an infected cat [2]. In our system of cavity-nesting birds, either mechanism is plausible as birds are not only exposed to bites from the nest-associated ectoparasites, but also to their feces, which accumulate in the nest material.
It is also possible that the three nest-associated ectoparasites studied here are not the only potential Bartonella vectors in this system. Other hematophagous arthropods such as chewing lice and biting midges are known to inhabit or visit nests of all three bird species at low abundance and may equally transmit Bartonella horizontally to the nestlings. Alternatively, given that Bartonella has been recorded as being transmitted maternally in mammals [33], it is also plausible that the nestling birds acquired Bartonella vertically. In either of these scenarios, the bacteria could then have been acquired by D. prognephilus, C. idius and P. sialia during blood-feeding. Experimental studies documenting vector competence would be needed to distinguish between these scenarios and to definitively designate vectors and transmission mechanisms in this system. Furthermore, given the relatively short duration of the breeding period in the migratory birds in this study, our study does not capture potential seasonal variation in the prevalence and relative abundance of Bartonella in the microbiomes. As previous work has shown a variety of seasonal patterns in Bartonella infection status in different systems [34,35,36,37], further work could usefully determine whether the patterns in Bartonella prevalence and relative abundance observed fluctuate through the seasons.
Our phylogenetic analysis of two genomic regions, supports placement of our sequences within the genus Bartonella. However, the sequences do appear to form their own sub-clade within the genus, supported by the close phylogenetic relationship between the 16S rRNA sequences from this study and those found in the poultry mite, Dermanyssus gallinae [8]. It is possible that avian systems are harboring an, as yet, little documented additional source of diversity within the genus Bartonella and will lead to a further expansion in our view of the group in the coming years [1, 3].
A growing subset of Bartonella species has been associated with human disease [3]. Although we were not able to associate our ASVs with any known species of Bartonella, some authors have suggested that any species of Bartonella may be capable of human infection [38]. In this case, if Dermanyssus mites, in particular, are proven to be a vector of Bartonella, there may be consequences of this from a public-health perspective. The Bartonella sequences in our samples of D. prognephilus group with the sequences obtained in a recent study of the microbiome of the poultry mite D. gallinae [8]. Cases of human infestation with D. gallinae and associated dermatological lesions are increasing in the medical literature [39, 40] both in poultry workers and in urbanites where infection is associated with infestations from (mainly) pigeon nests on or inside houses and apartment buildings [41]. Furthermore, one study in the Czech Republic reported rashes, fever and tibialgia in a family of people exposed to mites of the Dermanyssus genus which were found to be carrying B. quintana [7]. Given the possibilities for human contact with these mite species and their demonstrated high relative abundance of Bartonella in their microbiomes, further work should examine whether Dermanyssus have the potential to vector Bartonella to humans and/or wildlife and whether avian-derived Bartonella species are pathogenic to humans.
Conclusions
We have uncovered additional non-mammalian reservoir host species of Bartonella and their most likely arthropod vectors. We also presented evidence of expanded diversity within the Bartonella genus. Future research may continue to expand our view of the diversity within the Bartonellaceae and may seek to confirm the vector relationships suggested here and the potential for avian strains of Bartonella to be pathogenic to humans.
Availability of data and materials
All sequencing data is submitted to and available from NCBI GenBank under the Accession Numbers MN320509–MN320530 and MN3711264–MN371273. All other data in the study are available from the corresponding author upon reasonable requests.
Abbreviations
- ASV:
-
amplicon sequence variant
References
Harms A, Dehio C. Intruders below the radar: molecular pathogenesis of Bartonella spp. Clin Microbiol Rev. 2012;25:42–78.
Chomel BB, Kasten RW. Bartonellosis, an increasingly recognized zoonosis. J Appl Microbiol. 2010;109:743–50.
Okaro U, Addisu A, Casanas B, Anderson B. Bartonella species, an emerging cause of blood-culture-negative endocarditis. Clin Microbiol Rev. 2017;30:709–46.
Valentine KH, Harms CA, Cadenas MB, Birkenheuer AJ, Marr HS, Braun-McNeill J, et al. Bartonella DNA in loggerhead sea turtles. Emerg Infect Dis. 2007;13:949–50.
Mascarelli PE, McQuillan M, Harms CA, Harms RV, Breitschwerdt EB. Bartonella henselae and B. koehlerae DNA in birds. Emerg Infect Dis. 2014;20:490–2.
Kang JG, Kim HC, Choi CY, Nam HY, Chae HY, Chong ST, et al. Molecular detection of Anaplasma, Bartonella, and Borrelia species in ticks collected from migratory birds from Hong-do Island, Republic of Korea. Vector Borne Zoonotic Dis. 2013;13:215–25.
Melter O, Arvand M, Votypka J, Hulinska D. Bartonella quintana transmission from mite to family with high socioeconomic status. Emerg Infect Dis. 2012;18:163–5.
Hubert J, Erban T, Kopecky J, Sopko B, Nesvorna M, Lichovnikova M, et al. Comparison of microbiomes between red poultry mite populations (Dermanyssus gallinae): predominance of Bartonella-like bacteria. Microb Ecol. 2017;74:947–60.
George DR, Finn RD, Graham KM, Mul MF, Maurer V, Moro CV, et al. Should the poultry red mite Dermanyssus gallinae be of wider concern for veterinary and medical science? Parasit Vectors. 2015;8:178.
Wesołowski T, Stanska M. High ectoparasite loads in hole-nesting birds—a nestbox bias? J Avian Biol. 2001;32:281–5.
Møller AP, Allander K, Dufva R. Fitness effects of parasites on passerine birds: a review. In: Blondel J, editor. Population biology of passerine birds. Heidelberg: Springer; 1990. p. 269–80.
Tripet F, Richner H. Host responses to ectoparasites: food compensation by parent blue tits. Oikos. 1997;78:557–61.
Tarof S, Brown C. Purple martin (Progne subis). In: Poole A, editor. The birds of North America. Ithaca: Cornell Laboratory of Ornithology; 2013. https://doi.org/10.2173/bna.287.
Winkler DW, Hallinger KK, Ardia DR, Robertson RJ, Stutchbury BJ, Cohen RR. Tree swallow (Tachycineta bicolor). In: Poole A, editor. The birds of North America. Ithaca: Cornell Laboratory of Ornithology; 2011. https://doi.org/10.2173/bna.11.
Gowaty PA, Plissner JH. Eastern bluebird (Sialia sialis). In: Poole A, editor. The birds of North America. Ithaca: Cornell Laboratory of Ornithology; 2015. https://doi.org/10.2173/bna.381.
Sabrosky CW, Bennett GF, Whitworth TL. Bird blow flies (Proto calliphora) in North America (Diptera: Caliphoridae), with notes on the Palearctic species. Washington D.C.: Smithsonian Institution Press; 1989.
Pietz PJ, Granfors DA, Grant TA. Hatching and fledging times from grassland passerine nests. In: Ribic CA, Thompson III FR, Pietz PJ, editors. Video surveillance of nesting birds. Berkeley: University of California; 2012. p. 47–60.
Moss WW. An illustrated key to the species of the acarine genus Dermanyssus (Mesostigmata: Laelapoidea: Dermanyssidae). J Med Entomol. 1968;5:67–84.
Traub R, Rothschild M, Haddow JF. The Ceratophyllidae: key to the genera and host relationships. Cambridge: Rothschild and Traub; 1983.
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72.
Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Kosoy M, Hayman DT, Chan KS. Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol. 2012;12:894–904.
Kosoy M, Bai Y, Lynch T, Kuzmin IV, Niezgoda M, Franka R, et al. Bartonella spp. in bats, Kenya. Emerg Infect Dis. 2010;16:1875–81.
Cerutti F, Modesto P, Rizzo F, Cravero A, Jurman I, Costa S, et al. The microbiota of hematophagous ectoparasites collected from migratory birds. PLoS ONE. 2018;13:e0202270.
Atkinson CT, Forrester DJ, Greiner EC. Epizootiology of Haemoproteus melegirdis (Protozoa: Haemosproina) in Florida: seasonal transmission and vector abundance. J Med Entomol. 1988;25:45–51.
Ebel GD, Rochlin I, Longacker J, Kramer LD. Culex restuans (Diptera: Culicidae) relative abundance and vector competence for West Nile virus. J Med Entomol. 2005;42:838–43.
Diuk-Wasser MA, Toure MB, Dolo G, Bagayoko M, Sogoba N, Traore SF, et al. Vector abundance and malaria transmission in rice-growing villages in Mali. Am J Trop Med Hyg. 2005;72:725–31.
Reis C, Cote M, Le Rhun D, Lecuelle B, Levin ML, Vayssier-Taussat M, et al. Vector competence of the tick Ixodes ricinus for transmission of Bartonella birtlesii. PLoS Negl Trop Dis. 2011;5:e1186.
Billeter SA, Levy MG, Chomel BB, Breitschwerdt EB. Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med Vet Entomol. 2008;22:1–15.
Kosoy MY, Regnery RL, Kosaya OI, Jones DC, Marston EL, Childs JE. Isolation of Bartonella spp. from embryos and neonates of naturally infected rodents. J Wildl Dis. 1998;34:305–9.
Sanguinetti-Morelli D, Angelakis E, Richet H, Davoust B, Rolain JM, Raoult D. Seasonality of cat-scratch disease, France, 1999–2009. Emerg Infect Dis. 2011;17:705–7.
Telfer S, Clough HE, Birtles LR, Bennett M, Carslake D, Helyar S, et al. Ecological differences and coexistence in a guild of microparasites: Bartonella in wild rodents. Ecology. 2007;88:1841–9.
Janecek E, Mietze A, Goethe R, Schnieder T, Strube C. Bartonella spp. infection rate and B. grahamii in ticks. Emerg Infect Dis. 2012;18:1689–90.
Bai Y, Kosoy MY, Ray C, Brinkerhoff RJ, Collinge SK. Temporal and spatial patterns of Bartonella infection in black-tailed prairie dogs (Cynomys ludovicianus). Microb Ecol. 2008;56:373–82.
Lin EY, Tsigrelis C, Baddour LM, Lepidi H, Rolain JM, Patel R, et al. Candidatus Bartonella mayotimonensis and endocarditis. Emerg Infect Dis. 2010;16:500–3.
Kavallari A, Kuster T, Papadopoulos E, Hondema LS, Oines O, Skov J, et al. Avian mite dermatitis: diagnostic challenges and unmet needs. Parasite Immunol. 2018;40:e12539.
George TL, Harrigan RJ, LaManna JA, DeSante DF, Saracco JF, Smith TB. Persistent impacts of West Nile virus on North American bird populations. Proc Natl Acad Sci USA. 2015;112:14290–4.
Raele DA, Galante D, Pugliese N, La Salandra G, Lomuto M, Cafiero MA. First report of Coxiella burnetii and Borrelia burgdorferi sensu lato in poultry red-mites, Dermanyssus gallinae (Mesostigmata, Acari) related to urban outbreaks of dermatitis, in Italy. New Microbes New Infect. 2018;23:103–9.
Acknowledgements
The authors are grateful to the field volunteers and students who helped with data collection and sample processing including Carl Zenger, Celeste Morien, Bob Schmidt, John McCarthy, Daniel Muldoon, Ramya Sridhar, Corinne Birchard, Joseph Toth, Logan Fahrenkopf, Alyssa Gooding and Samantha Wilcox. They are also grateful to Terry Whitworth and Ashley Dowling for assistance in identifying bird blow fly and mites, respectively.
Funding
This work was supported by Grant DEB-1556577 from the National Science Foundation, as well as Grants from the Purple Martin Conservation Association and the New York State and North American Bluebird Societies.
Author information
Authors and Affiliations
Contributions
HMW conducted analyses and interpretation and was the main author of the manuscript. KD provided guidance on interpretation of phylogenetic relationships and methodology. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All fieldwork was completed under IACUC BIO02016N at the State University of New York at Buffalo, USA. All relevant permits and permissions were obtained including US Fish and Wildlife Service monitoring and special use permit. #2016-002); US Fish and Wildlife Service scientific collecting permit (MB83070B-0); Federal bird banding permit (#08066); New York State Department of Environmental Conservation temporary revocable permit; New York State Department of Environmental Conservation license to collect of possess (scientific) (#1976) and New York State Department of Environmental Conservation license to collect or possess (banding) (#179).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
BLASTn analysis of ASVs identified as Bartonella spp. in this study.
Additional file 2: Table S2.
Sequences obtained from GenBank and used in phylogenetic trees.
Additional file 3: Table S3.
BICc ranking of nucleotide substitution models for the 16S rRNA tree.
Additional file 4: Table S4.
BICc ranking of nucleotide substitution models for the gltA tree.
Additional file 5: Table S5.
Showing prevalence of sequences identified to the Bartonella genus in 16S rRNA sequences via naïve Bayesian taxonomic classification against the Greengenes database.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Williams, H.M., Dittmar, K. Expanding our view of Bartonella and its hosts: Bartonella in nest ectoparasites and their migratory avian hosts. Parasites Vectors 13, 13 (2020). https://doi.org/10.1186/s13071-020-3896-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-3896-7