Abstract
Background
Avian coccidiosis is a widespread, economically significant disease of poultry, caused by several Eimeria species. These parasites have complex and diverse life-cycles that require invasion of their host cells. This is mediated by various proteins secreted from apical secretory organelles. Apical membrane antigen 1 (AMA1), which is released from micronemes and is conserved across all apicomplexans, plays a central role in the host cell invasion. In a previous study, some putative EtAMA1-interacting proteins of E. tenella were screened. In this study, we characterized one putative EtAMA1-interacting protein, E. tenella Eimeria -specific protein (EtEsp).
Methods
Bimolecular fluorescence complementation (BiFC) and glutathione S-transferase (GST) fusion protein pull-down (GST pull-down) were used to confirm the interaction between EtAMA1 and EtEsp in vivo and in vitro. The expression of EtEsp was analyzed in different developmental stages of E. tenella with quantitative PCR and western blotting. The secretion of EtEsp protein was tested with staurosporine when sporozoites were incubated in complete medium at 41 °C. The localization of EtEsp was analyzed with an immunofluorescence assay (IFA). An in vitro invasion inhibition assay was conducted to assess the ability of antibodies against EtEsp to inhibit cell invasion by E. tenella sporozoites.
Results
The interaction between EtAMA1 and EtEsp was confirmed with BiFC and by GST pull-down. Our results show that EtEsp is differentially expressed during distinct phases of the parasite life-cycle. IFA showed that the EtEsp protein is mainly distributed on the parasite surface, and that the expression of this protein increases during the development of the parasite in the host cells. Using staurosporine, we showed that EtEsp is a secreted protein, but not from micronemes. In inhibition tests, a polyclonal anti-rEtEsp antibody attenuated the capacity of E. tenella to invade host cells.
Conclusion
In this study, we show that EtEsp interacts with EtAMA1 and that the protein is secreted protein, but not from micronemes. The protein participates in sporozoite invasion of host cells and is maybe involved in the growth of the parasite. These data have implications for the use of EtAMA1 or EtAMA1-interacting proteins as targets in intervention strategies against avian coccidiosis.

Similar content being viewed by others
Background
Avian coccidiosis is a widespread, economically significant disease of poultry that results in annual global economic losses of approximately $2.4 billion, including both production losses and disease prevention and treatment costs [1]. It is an enteric disease caused by several species of the protozoan genus Eimeria, predominantly E. acervulina, E. brunetti, E. maxima, E. mitis, E. necatrix, E. praecox and E. tenella [2]. Of these, E. tenella is one of the species causing hemorrhagic pathologies and high mortality. Eimeria spp. belong to the phylum Apicomplexa, which includes important pathogens of humans and domestic animals, such as the causative agents of malaria (Plasmodium spp.), toxoplasmosis (Toxoplasma gondii), babesiosis (Babesia spp.), and coccidiosis (Eimeria spp.). Most apicomplexans are obligate intracellular parasites and are characterized by their apical complexes of specialized secretory organelles (micronemes, rhoptries and dense granules) [3]. They use actin-based motility coupled to regulated protein secretion from their apical organelles to actively invade host cells [4]. These parasites have complex and diverse life-cycles that involve the invasion of many different cell types, including erythrocytes, lymphocytes, macrophages, and digestive-tract cells. Despite the diversity of their target host cells, they maintain a highly conserved mechanism for this active invasion process [5].
The host-cell invasion mechanism involves four steps, i.e. attachment, apical reorientation, moving junction formation, and the formation of a protective parasitophorous vacuole. Each invasion step is mediated by various proteins, which are secreted from apical secretory organelles [6]. Apical membrane antigen 1 (AMA1), a type I transmembrane protein, is one of a number of proteins released from micronemes that are conserved across all apicomplexans. It is known to play several important roles during host-cell penetration [7]. For instance, previous reports have shown that antibodies against AMA1 or small specific AMA1-binding peptides inhibit the invasion of host cells by Toxoplasma spp., E. tenella, Babesia spp., Neospora spp. and Plasmodium spp. [3, 8,9,10,11]. AMA1 is also a long-standing effective candidate vaccine for some apicomplexans, including N. caninum, T. gondii and Plasmodium spp. [11,12,13]. In Toxoplasma spp. and Plasmodium spp., AMA1 is reportedly involved in apical reorientation [14], host-cell attachment [7, 15], invasion and establishment of the moving-junction [16], and the provision of a signal that initiates intracellular replication [17].
In contrast to the functions of AMA1 in other apicomplexan parasites, there are only a few reports of this conserved protein in Eimeria spp. In a previous in vitro study, AMA1 antibodies or specific EtAMA1-binding peptides inhibited the invasion of host cells by E. tenella sporozoites [10, 18]. EtAMA1 also partially protected host cells against homologous challenge with E. tenella when used as a recombinant protein vaccine and against heterologous challenge with E. maxima when the AMA1 protein from E. maxima was expressed as a live vectored vaccine [19]. Although AMA1 plays an important role in host-cell invasion by E. tenella sporozoites, its precise functions are unknown.
Proteins perform a vast number of cellular functions when they interact with one or multiple binding partners. Protein-protein interactions are essential in the mediation of almost all cellular processes, including replication, transcription, translation and signal transduction [20]. The biochemical analysis of protein complexes and the identification of their components have been fundamental to our understanding of their biological functions in cells [21].
To understand the precise functions of EtAMA1 during host-cell invasion, we screened EtAMA1-interacting proteins with a yeast two-hybrid system and identified 14 putative EtAMA1-interacting proteins in a previous study [22]. E tenella Eimeria-specific protein (EtEsp) (GenBank: JZ905773) is one of these putative interacting proteins. In this study, we cloned and characterized EtEsp. We systematically analyzed its interaction with EtAMA1 using bimolecular fluorescence complementation (BiFC) in vivo and a glutathione S-transferase (GST) pull-down assay in vitro. Our results show that the EtEsp is secreted protein, but not from micronemes, interacts with EtAMA1, and is involved in the invasion of host cells by E. tenella sporozoites.
Methods
Parasite collection
Eimeria tenella was obtained from the Key Laboratory of Animal Parasitology of the Ministry of Agriculture, Shanghai Veterinary Research Institute, the Chinese Academy of Agricultural Sciences, Shanghai, China. The parasites were maintained and propagated by passage through coccidia-free, 2-week-old chickens, as previously described [23]. Coccidia-free 14-day-old chickens were inoculated with 1 × 104 sporulated oocysts of E. tenella. Unsporulated oocysts (UO) were collected from infected chicken ceca at 7 days post-infection. Sporulated oocysts (SO) were derived from UO that had undergone sporulation in 2% potassium dichromate at a temperature of 28–30 °C for 72–120 h, under forced aeration with a suitable pump. When more than 90% of the oocysts had sporulated, the oocysts were collected and purified. The sporozoites (Spz) were purified from cleaned SO with in vitro excystation [24]. Second-generation merozoites (sMrz) were isolated from infected chicken ceca at 115 h post-inoculation, as described previously [25]. All parasites were collected and frozen in liquid nitrogen.
Chickens and rabbits were fed and used according to a protocol approved by the Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
The chicken embryo fibroblast cell line, DF-1, a derivative of the East Lansing Line (ELL-0) [10], was used for BiFC and in vitro infection experiments.
Molecular cloning and sequence analysis of E. tenella-specific protein
Total RNA was extracted from E. tenella sporozoites with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). GeneRacer™ primers (GR5P and GR5N) were provided for the random amplification of PCR ends (RACE) in the GeneRacer™ Kit (Invitrogen) and gene-specific primers (GS5P and GS5N) were designed based on the expressed sequence tag (EST) sequence (GenBank: JZ905773) which is 790 bp in length and contains a poly(A) at the 3’-end (Additional file 1: Table S1). The 5’-end of this gene was determined according to the manufacturer’s protocol. The PCR-amplified fragment was then ligated into the pGEM-T Easy Vector (Promega, Madison, WI, USA) and used to transform competent E. coli TOP10 cells. After PCR identification, the plasmid DNA was sequenced. After the resulting sequence was assembled and aligned with the original EST sequence, the full-length cDNA sequence of the gene was determined and submitted to the National Center for Biotechnology Information (NCBI) GenBank under the accession number MN161778). The full-length EtEsp cDNA sequence was used in a BLAST search of the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) and the E. tenella genome database (http://www.genedb.org/Homepage/Etenella). The deduced amino acid sequence was obtained with the ORF Finder tool at NCBI. The molecular mass and theoretical isoelectric point were calculated with ProtParam tools (http://web.expasy.org/protparam/). The signal peptide sequence was identified with the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/), and transmembrane regions were predicted with the TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Protein motifs were scanned with Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan).
Recombinant protein expression and polyclonal anti-rEtEsp serum
The EtEsp open reading frame (ORF) cDNA was amplified with PCR using primers EtEsp-UP and EtEsp-LP (Additional file 1: Table S1), which contained BamHI and XhoI restriction sites, respectively. The PCR fragment was then ligated into the prokaryotic expression vector pET28a(+) digested with the same restriction endonucleases, to construct the recombinant expression plasmid pET-EtEsp. The recombinant protein His-EtEsp (rEtEsp) was expressed in Escherichia coli BL21 cells at 37 °C with 1 mM isopropyl-thio-α-d-galactoside. The cell pellet was lysed with sonication and digested with 10 µg/ml lysozyme (Sigma-Aldrich, St. Louis, MO, USA). The lysate was then analyzed with 12% SDS-PAGE to confirm that the recombinant protein was present as a soluble protein or inclusion bodies. rEtEsp was purified with His·Bind® Resin (Merck, Darmstadt, Germany) and its concentration measured with a BCA Protein Assay Kit (Beyotime, Haimen, China).
Two 2-month-old male rabbits were inoculated with 200 μg of purified rEtEsp emulsified in Freund’s complete adjuvant (Sigma-Aldrich). After 14 days, a booster of 200 μg of purified rEtEsp in Freund’s incomplete adjuvant (Sigma-Aldrich) was administered, followed by a second and third booster on days 28 and 42. One week after the final booster, the rabbit serum was collected and stored at − 20 °C until use.
Analysis of EtEsp transcript levels with real-time quantitative PCR (qPCR)
The expression profiles of EtEsp mRNA were examined in four developmental stages of E. tenella (UO, SO, Spz and sMrz) with qPCR. cDNA samples were synthesized from DNaseI-treated total RNAs of the E. tenella developmental stages using SuperScript™ II Reverse Transcriptase (Invitrogen) and random pd(N)6 primer. The housekeeping 18S rRNA gene was used as the internal control. The primers used to amplify the EtEsp cDNA (EtEsp-SP and EtEsp-AP) and the 18S rRNA gene (18S-SP and 18S-AP) were designed with Primer3 v. 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) (Additional file 1: Table S1). qPCR was performed with the StepOnePlus™ Real-Time PCR System using the SYBR® Premix Ex Taq™ II kit (Takara, Tokoyo, Japan). All experiments were performed twice, with separate biological replicates. In each experiment, the reactions were performed in triplicate. A dilution series of cDNA templates of the sporozoites was used to establish standard curves, and all standard curves had correlation coefficients of R2 > 0.99. The comparative 2−ΔΔCq method was used to analyze the relative levels of gene expression.
SDS-PAGE and western blotting
Protein samples were prepared from the four E. tenella developmental stages (UO, SO, Spz and sMrz), and from DF-1 cells transfected with the recombinant plasmids, for western blotting. The protein concentrations were determined with a BCA Protein Assay Kit (Beyotime). The purified rEtEsp and protein lysates were separated with SDS-PAGE and transferred electrophoretically to polyvinylidene difluoride membranes. Rabbit antiserum (1:100) against sporozoite proteins, previously produced in our laboratory [26], a rabbit anti-rEtEsp antibody (1:100), a mouse monoclonal anti-α-tubulin antibody (1:1000) (Sigma-Aldrich), and a monoclonal anti-His antibody (1:1000) were used as the primary antibodies to detect rEtEsp or native EtEsp. Naïve rabbit serum (1:100) was used as the negative control. IRDye-800CW-labelled goat anti-rabbit IgG antibody (1:25,000) and IRDye-680RD-labeled donkey anti-mouse IgG antibody (1:25,000; LI-Cor, Lincoln, NE, USA) were used as the secondary antibodies. The IRDyes were detected with the Odyssey Infrared Imaging System (LI-Cor).
BiFC assay
The ORF fragments of EtEsp and the EtAMA1 ectodomain, with no stop codon, were amplified from the first-strand cDNA with two pairs of primers (BfEtEsp-UP/BfEtEsp-LP and BfEtAMA1-UP/BfEtAMA1-LP, respectively), which contained EcoRI and BglII restriction sites (EtEsp) or EcoRI and XhoII restriction sites (EtAMA1). The fragments were then digested with the appropriate restriction enzymes and ligated into the pBiFC-VN155 and pBiFC-VC155 vectors digested with the same enzymes, respectively, to construct the recombinant plasmids pBiFC-VN155-EtEsp and pBiFC-VC155-EtAMA1, respectively. Before the BiFC assay, the uptake of the expression vectors by the cells was confirmed. DF-1 cells were transfected with the recombinant plasmid pBiFC-VN155-EtEsp or pBiFC-VC155-EtAMA1 using Lipofectamine™ 2000 Transfection Reagent (Invitrogen), according to the manufacturer’s instructions. At 48 h after transfection, the cells were harvested, and the proteins were extracted with RIPA Lysis Buffer (Beyotime). Western blots were probed with rabbit anti-rEtEsp antibody and rabbit anti-rEtAMA1 antibody, which were previously produced in our laboratory [10]. After confirmation that the cells had expressed the two constructs, DF-1 cells were cotransfected with pBiFC-VN155-EtEsp and pBiFC-VC155-EtAMA1. DF-1 cells were also cotransfected with pBiFC-bJunVN55 (I152L) and pBiFC-bFosVC155 or pBiFC-VC155-EtAMA1 and pBiFC-VN155 empty vector, or pBiFC-VN155-EtEsp and pBiFC-VC155 empty vector as the positive or negative control, respectively. The DF-1 cells were observed with fluorescence microscopy 24 h after transfection with the different constructs.
GST pull-down
To confirm the interaction between EtAMA1 and EtEsp317 in vitro, a GST pull-down assay was performed with the Pierce™ GST Protein Interaction Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. The recombinant plasmid pGEX-6P-EtAMA1 was previously constructed in our laboratory [10]. The expression of the recombinant protein GST-EtAMA1 was induced and the protein purified with GST resin for use as the bait protein. The ORF of EtEsp was inserted into the pET-28a vector to express the recombinant protein His-EtEsp (rEtEsp) as the prey protein. GST-EtAMA1 was incubated with equilibrated glutathione-agarose to immobilize the bait protein. rEtEsp was then added to the glutathione-agarose and incubated with the bait protein. The bait and prey proteins were eluted from the glutathione-agarose. E. coli BL21 cells were transformed with recombinant plasmid pET-EtMIC2, constructed previously in our laboratory [27], to express the recombinant protein His-EtMIC2 as the negative control. Another, rEtEsp was loaded in an empty glutathione-agarose column as the negative. All the proteins were then resolved with 12% SDS-PAGE and detected with western blotting using the appropriate antibodies, as described above.
Assay of EtEsp secretion
Freshly excysted sporozoites (4 × 106) were incubated in 100 μl of complete medium (CM; Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin/streptomycin, 2 mM l-glutamine) for 2 h at 41 °C under 5% CO2 for the secretion experiments. They were then incubated with 5, 10 or 20 μM staurosporine (Sigma-Aldrich; dissolved in dimethylsulfoxide (DMSO)) or an appropriate volume of carrier DMSO, as described previously [28]. The secretion of EtMIC2 and EtGRA(TgGRA7 homologous protein) was used as the control. The sporozoites were then pelleted by centrifugation for 10 min at 6000× g. The supernatants and sporozoites were recovered and analyzed with western blotting using a rabbit anti-rEtEsp antibody and rabbit anti-rEtMIC2 antibody generated previously in our laboratory [27] and mouse anti-TgGAR7 antibody which had been generated previously in another laboratory at Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Immunofluorescence staining of parasites
Purified differentially developed parasites (Spz, sporocysts [Sporo], and sMrz) were transferred to glass slides and air-dried, as previously described [10, 29]. Freshly purified sporozoites were used to infect DF-1 cells after incubation in CM for 2 h at 41°C. At different time points after infection, the DF-1 cells were collected, washed, transferred to glass slides, and air-dried. The slides were then fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS) and placed in 1% Triton X-100 in PBS for 15 min to increase their permeability. Non-permeabilized sporozoites and sporocysts were used as a control. The slides were blocked with PBS containing 2% (w/v) bovine serum albumin for overnight at 4 °C. A rabbit anti-rEtEsp antibody (1:100) was added and the cells were incubated for 1 h at 37 °C. A 1:500 dilution of fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG antibody (Sigma-Aldrich) was then added and the cells incubated for 1 h at 37 °C. The cell nuclei were stained by incubation in 10 μg/ml 4′,6-diamidino-2-phenylindole (Beyotime) at room temperature for 10 min. After each step, the slides were washed three times for 10 min each with PBS containing 0.05% Tween 20. The slides were finally mounted with 50 μl of Fluoromount Aqueous Mounting Medium (Sigma-Aldrich) before observation with a fluorescence microscope (Olympus, Tokyo, Japan). At the same time, we performed the co-localization of EtEsp and EtAMA1 in sporozoites. Purified sporozoites were treated with mouse anti-rEtAMA1 antibody (1:100) and rabbit anti-rEtEsp antibody (1:100), then goat anti-rabbit IgG fluorescein isothiocyanate (FITC)-conjugated antibody (1:500) and goat anti-mouse IgG cyanine (Cy3)-conjugated antibody (1:500; Sigma-Aldrich) were used as secondary antibodies.
Invasion inhibition assay in vitro
The invasion inhibition assay was based on previous reports of the invasion of DF-1 cells by E. tenella sporozoites. Antibodies were purified with Protein A + G Agarose (Beyotime). DF-1 cells (2 × 105 cells per well) were cultured in 24-well plates (Corning, UN, USA) in CM for 24 h at 37 °C under 5% CO2. The freshly purified sporozoites were counted and labeled with carboxyfluorescein diacetate succinimidyl ester (Beyotime). The labeled sporozoites were incubated at 37 °C with 50, 100, 200, 300 or 400 μg/ml purified IgG directed against rEtEsp for 2 h. The same quantity of IgG from naïve rabbit serum (Sigma-Aldrich) was used as the negative control, and an equivalent volume of PBS as the normal control. After they were washed twice with sterile PBS, DF-1 cells (105/well) were infected with the labeled sporozoites (105/well) in 24-well plates and cultured for 16 h at 41 °C under 5% CO2. The cells were then collected and analyzed with flow cytometry on a Cytomics™ FC 500 (Beckman Coulter, Indianapolis, IN, USA). The controls were uninfected DF-1 cells. The infected cells, uninfected cells, and free sporozoites were gated using the CXP software (Beckman Coulter) to count the infected (labeled sporozoites) and uninfected (fluorescence-free) cells. All assays were performed in triplicate. The percentage of infected cells in the presence or absence of an anti-rEtEsp polyclonal antibody were used to calculate the inhibition rates, as previously described [10].
Results
Cloning and sequence analysis of full-length EtEsp cDNA
The 1108 bp full-length cDNA of EtEsp was obtained with RACE. The sequence analysis showed that the full-length cDNA included a 5’-untranslated region (UTR) of 70 bp, a 3’-UTR of 542 bp with a poly(A) tail, and an ORF of 501 bp, which encoded 166 amino acids with a calculated molecular weight of 18.1 kDa and a theoretical isoelectric point of 4.2 (Fig. 1). Analysis of the amino acid sequence showed a signal peptide of 19 amino acids at the N-terminus and no transmembrane region. Searches in the Motif Database and the Conserved Domain Database revealed the presence of 1 N-glycosylation site, 5 casein kinase II phosphorylation sites, 6 N-myristoylation sites, 1 tyrosine kinase phosphorylation site, 1 intein DOD-type homing endonuclease domain, and no conserved domains (Fig. 1). A BLAST search of the E. tenella genome database showed that the ORF sequence shared 100% sequence identity with ETH_00016590, which encodes an Eimeria-specific protein, on supercontig Eth_scaff124: 9216–10141.
Nucleotide sequence of the full-length cDNA of EtEsp and the deduced amino acid sequence. Start and stop codons are underlined. One putative intein DOD-type homing endonuclease domain is shown with wavy underlining. A putative N-glycosylation site has a double line. Five putative casein kinase II phosphorylation sites are shown in yellow. Six putative N-myristoylation sites are shown in gray
The amino acid sequence shared 100% homology with the E. tenella Eimeria-specific protein (XP_013228647.1) and 92% (152/170) identity with the E. necatrix Eimeria-specific protein (XP_013435139.1) in NCBI. Therefore, this gene was designated EtEsp and submitted to the GenBank database under the accession number MN161778. It also shared 68% (106/157) amino acid identity with E. brunetti conserved hypothetical protein (CDJ53027.1), 63% (108/172) identity with E. praecox conserved hypothetical protein (CDI81636.1), 74% (97/131) identity with E. maxima conserved hypothetical protein (XP_013336310.1), and 73% (91/124) identity with E. acervulina conserved hypothetical protein (XP_013248166.1). Also, this protein is not found in other apicomplexan parasites. These results show that the protein is conserved in Eimeria spp.
Expression and characterization of recombinant EtEsp
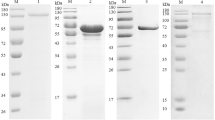
rEtEsp was expressed as a His6-tagged fusion protein. SDS-PAGE showed that rEtEsp was mainly present in the soluble fraction of the bacterial lysate. After the purification of rEtEsp with Ni-NTA chromatography, a protein of approximately 21 kDa was observed with SDS-PAGE. Because 3 kDa of the fusion protein was derived from the vector, the predicted molecular mass of EtEsp was about 18.1 kDa. Western blotting showed that purified rEtEsp was recognized by rabbit serum directed against sporozoites and by a monoclonal anti-His6 antibody. Naïve rabbit serum failed to recognize any protein corresponding to the expected size of rEtEsp (Fig. 2). These results indicate that rEtEsp was recognized specifically by rabbit serum directed against a soluble sporozoite protein and by a monoclonal anti-His antibody.
EtEsp mRNA and protein expression at different developmental stages of E. tenella
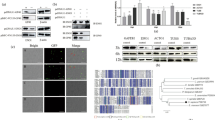
qPCR was used to analyze the UO, SO, Spz, and sMrz stages of E. tenella for the presence of EtEsp mRNA. The levels of EtEsp mRNA were much higher in the sMrz stage than in the other three stages, and EtEsp mRNA was almost undetectable in UO (Fig. 3a).
EtEsp expression at different developmental stages of E. tenella. a qPCR of EtEsp at different developmental stages of E. tenella. Bars with different letters indicate significant differences (P < 0.05) and the error bars indicate standard deviations. b Western blot showing EtEsp at different developmental stages, probed with rabbit anti-rEtEsp serum or mouse monoclonal anti-α-tubulin antibody. c The densitometric intensity of western blot images was analyzed using ImageJ software. Bars with different letters indicate significant differences (P < 0.05) and the error bars indicate standard deviations. Abbreviations: UO, unsporulated oocysts; SO, sporulated oocysts; Spz, sporozoites; sMrz, second-generation merozoites
The expression of EtEsp in the 4 developmental stages was also determined with immunoblotting using rabbit antiserum against rEtEsp. A monoclonal anti-α-tubulin antibody was used as the control. Western blotting showed that the anti-rEtEsp antibody reacted with a band of approximately 18 kDa in the parasite lysates prepared from the four different developmental stages of E. tenella. The expression levels of EtEsp were higher in sporozoites than in other three stages (Fig. 3b, c).
Confirmation of the interaction between EtAMA1 and EtEsp
To characterize the interaction between EtAMA1 and EtEsp in vivo, a BiFC assay was performed. For the BiFC assay, fragments of the EtEsp ORF and the EtAMA1 ectodomain sequence were cloned into the plasmids pBiFC-VN155 and pBiFC-VC155, respectively, to generate the constructs pBiFC-VN155-EtEsp and pBiFC-VC155-EtAMA1, respectively. The total proteins were extracted from DF-1 cells transfected separately with one or the other construct. Western blotting showed that the two constructs were expressed individually in the DF-1 cells at 48 h after transfection (Fig. 4a). Strong green fluorescence was observed in DF-1 cells 48 h after they were co-transfected with both constructs. Green fluorescence was also observed in the positive control. However, there was no visible fluorescence in the DF-1 cells co-transfected with pBiFC-VC155-EtAMA1 and pBiFC-VN155 empty vector or pBiFC-VN155-EtEsp and pBiFC-VC155 empty vector. These results indicate that EtEsp interacts with EtAMA1 in cells (Fig. 4b).
Interaction between EtAMA1 and EtEsp in DF-1 cells assessed with BiFC. a DF-1 cells were transfected with VC155-EtAMA1 and VN155-EtEsp and the cellular lysates were analyzed with immunoblotting using antisera against EtAMA1 and EtEsp. Lane 1: anti-r EtAMA1 antibody; Lane 2, anti-r EtEsp antibody. b BiFC was performed. b1 DF-1 cells were co-transfected with VC155-EtAMA1 and VN155-EtEsp. b2 DF-1 cells were co-transfected with positive controls bFos and bJun. b3 DF-1 cells were co-transfected with pBiFC-VC155-EtAMA1 and pBiFC-VN155 empty vector. b4 DF-1 cells were co-transfected with pBiFC-VN155-EtEsp and pBiFC-VC155 empty vector
GST pull-down
To confirm the interaction between EtAMA1 and EtEsp in vitro, a GST pull-down assay was performed. GST-EtAMA1 and His-EtEsp were expressed individually in E. coli and purified. GST-EtAMA1 was bound to an equilibrated glutathione-agarose column, and then His-EtESP was added to the column. The proteins bound to the glutathione-agarose, and any non-specifically bound proteins were removed by elution with buffer. The proteins retained on the column were then eluted and detected with immunoblotting using anti-rEtAMA1 and anti-rEtEsp antibodies (Fig. 5). The results clearly indicated a direct interaction between the EtAMA1 and EtEsp proteins.
In vitro pull-down assay between EtAMA1 and EtEsp. Lane M: protein marker; Lane 1: rEtMIC2 protein incubated with rEtEsp, detected with anti-rEtAMA1 and anti-rEtMIC2 as a negative control; Lane 2: rEtAMA1 protein incubated with rEtEsp, detected with anti-rEtAMA1 and anti-rEtEsp; Lane 3: rEtEsp loaded in an empty glutathione-agarose column, detected with anti-rEtEsp as a negative control
EtEsp is not secreted from the microneme
To examine the secretion of EtEsp, sporozoites were incubated in CM at 41 °C. The supernatant containing the excretory-secretory antigens (ESA) from the incubated sporozoites and sporozoites pellets were analyzed with western blotting. Immunoblots of the ESA samples and sporozoites were probed with an anti-rEtEsp antibody and showed that EtEsp was secreted when the sporozoites were incubated at 41 °C under 5% CO2 in CM. Rabbit serum raised against the micronemal protein EtMIC2 was used as the experimental control [27]. To demonstrate whether EtEsp secretion is dependent on the micronemal pathway, we added staurosporine to the CM because staurosporine is a protein kinase inhibitor known to specifically inhibit microneme secretion [28]. In the parasites treated with 5, 10 or 20 μM staurosporine, the secretion of EtEsp and EtGRA into the supernatant was not affected, but the secretion of EtMIC2 in supernatant was significant reduced compared with their secretion in the presence of the DMSO solvent only (Fig. 6a, b). These results show that EtEsp is a secreted protein, but not a micronemal protein.
Western blotting analysis of secretion assays (supernatants and sporozoites pellet). a The supernatants (Sup). b The sporozoites pellet (Spz). Lane M: protein marker; Lanes 1–3: 5, 10 and 20 μM staurosporine dissolved in DMSO; Lanes 4–6: volumes of DMSO solvent corresponding to 5, 10 or 20 μM staurosporine
Immunolocalization of EtEsp at different developmental stages of E. tenella
To investigate the localization and distribution of the EtEsp protein in different development stages of E. tenella, including sporozoites, second-generation merozoites, immature schizonts, and mature schizonts, the protein was localized with immunofluorescence in vitro using an antibody against rEtEsp. The EtEsp protein was mainly distributed on the surfaces of the permeabilized parasite sporozoites, sporocysts, and second-generation merozoites (Fig. 7a1, b1, k). The protein was also mainly located on the surface of non-permeabilized sporozoites and sporocysts (Fig. 7a2, b2). After incubation in CM for 2 h, the fluorescence increased and the protein was mainly localized to the anterior and surface of the parasites (Fig. 7c). EtEsp protein was also mainly located on the surface of the parasites 2 h after their invasion of DF-1 cells (Fig. 7d). At 12 h after the sporozoites were added to DF-1 cells, EtEsp also localized to the cytoplasm of the sporozoites, except for the refractile body in the posterior section of the parasites, and the intensity of EtEsp staining had increased (Fig. 7e). At 24–72 h post-infection, the EtEsp protein was uniformly distributed in trophozoites, immature schizonts, and mature schizonts, and the protein’s expression had increased (Fig. 7f–j).
Immunofluorescent localization of EtEsp at different developmental stages of E. tenella. Parasites were immuno-stained with anti-rEtEsp antibody. a Sporozoites (Spz) were incubated in PBS (a1 permeabilized sporozoites, a2 non-permeabilized sporozoites). b Sporocysts (Sporo) were incubated in PBS (b1 permeabilized sporocysts, b2 non-permeabilized sporocysts). c Sporozoites (Spz) were incubated in complete medium (CM) for 2 h at 41 °C. d, e, intracellular sporozoites (iSpz) at 2 h and 12 h post-infection, respectively. f Trophozoites (iTropho) at 24 h post-infection. g, h Immature schizonts (iSc) at 48 and 60 h post-infection, respectively. i Mature schizonts (mSc) at 68 h post-infection. j First-generation merozoites (fMrz) at 72 h post-infection. k Second-generation merozoites (sMrz) in PBS
Co-localization of EtAMA1 and EtEsp
IFAs were performed to determine the location of EtAMA1 and EtEsp. Purified sporozoites were treated with mouse anti-rEtAMA1 antibody and rabbit anti-rEtEsp antibody. The results showed EtEsp was mainly located on the surface of sporozoites, EtAMA1 was distributed throughout the cytoplasm and the membrane of sporozoites except for refractile bodies (Fig. 8).
Anti-rEtEsp antibodies inhibit DF-1 cell invasion
To evaluate the effect of the EtEsp protein on the invasion of DF-1 cells by E. tenella sporozoites, an invasion inhibition assay of sporozoites was performed in vitro. When the sporozoites were incubated with purified anti-rEtEsp antibody before infection, their capacity to invade the DF-1 cells was significantly reduced. After pre-treatment with 50, 100, 200, 300 or 400 μg/ml anti-rEtEsp IgG antibody, their invasion of cells was highly significantly reduced compared with that of sporozoites treated with naïve rabbit IgG (negative control) (P < 0.01). Under these experimental conditions, an inhibition plateau of 62.9% was reached at an antibody concentration of 300 μg/ml. In a comparative analysis, the same dose of the naïve rabbit serum IgG antibody did not significantly affect invasion (Fig. 9).
Inhibition of sporozoite invasion in vitro. All assays were performed in triplicate. Abbreviations: anti-rEtEsp, rabbit antiserum generated against recombinant EtESP protein; NI, IgG from naïve rabbit serum. **P < 0.01 for differences between treatment with antibody against rEtEsp or with naïve rabbit serum with the same IgG concentration
Discussion
In this study, we cloned and characterized the E. tenella Eimeria-specific protein, a putative EtAMA1-interacting protein, using a yeast two-hybrid system in our laboratory [22]. Although the yeast two-hybrid system is a widely used and powerful method for identifying the partners of proteins in regulatory complexes and in the analysis of protein-protein interactions [30], the system has several limitations, including the possibility of isolating very large numbers of clones with no biological relevance [31]. Therefore, the interaction between EtAMA1 and EtEsp required validation with an alternative technique, such as a BiFC assay or GST pull-down assay. The BiFC assay is a versatile technique for investigating protein-protein interactions in living systems, and is based on the reconstitution of a fluorescent protein in vivo [32]. GST pull-down is amenable to more specific investigations of protein-protein interactions in vitro, but relies on purified proteins that may not fully mimic the protein’s native conformation or post-translational modification, which mediate its interactions [33]. Although these assays have some advantages in identifying protein-protein interactions, each also has its drawbacks. Therefore, in many research fields, these methods are often combined to identify the interactions between two proteins [33,34,35]. In this study, the interaction between EtAMA1 and EtEsp was confirmed with a GST pull-down assay in vitro and a BiFC assay in vivo. These results indicated that EtEsp interacts with EtAMA1.
Proteins perform a vast number of cellular functions through their interactions with one or multiple binding partners. Moreover, many protein-protein interactions are regulated by post-transcriptional modification (e.g. phosphorylation) of the protein of interest, and these modifications are induced by exposure to certain circumstances [36]. In the present study, an amino acid sequence analysis predicted that EtEsp contains one N-glycosylation site, five casein kinase II phosphorylation sites, six N-myristoylation sites, one tyrosine kinase phosphorylation site and one intein DOD-type homing endonuclease domain. Inteins, also called protein introns, are parasitic genetic elements that excise themselves at the protein level by self-splicing, allowing the formation of functional, non-disrupted proteins [37]. These data suggest that its functions may be regulated by post-translational modification. We supposed that the interaction of EtAMA1 with EtEsp may be regulated by post-translational modification.
To understand the expression of EtEsp in the different developmental stages of the parasite, we examined its expression patterns with qPCR and western blotting. Our results indicated that EtEsp mRNA levels were higher in second-generation merozoites and sporozoites than in sporulated oocysts or unsporulated oocysts. But western blotting showed that the expression of EtEsp was higher in sporozoites than other developmental stages of E. tenella. Immunofluorescent localization showed that the expression of the protein increased with the development of the parasites in DF-1 cells. Previous proteomic and transcriptomic data confirm that EtAMA paralogues are tightly stage-regulated [38, 39]. EtAMA1 is a sporozoite-specific protein involved in the invasion process of sporozoites [10, 19, 38]. While another EtAMA1 paralogues, EtAMA2 is a merozoites-specific protein not involved in the parasite invasion. All these finding indicate that E. tenella parasites harbour stage-specific AMA proteins that could be relevant during specific phases of the parasite cycle [19]. In this study, EtEsp is differentially expressed during the distinct phases of the parasite life-cycle and may be very important in the invasion and development of the parasite life-cycle.
Immunofluorescent localization showed that EtEsp was located on the surface of the sporozoite and concentrated around the anterior of the parasite during its incubation in CM. However, the protein has no transmembrane region or glycophosphatidyl inositol (GPI)-anchor sequence, but has a signal peptide, and six N-myristoylation sites. The presence of a signal peptide is necessary for the translocation of proteins from their ribosomal sites of translation into the lumen of the endoplasmic reticulum, from where they are trafficked in the endomembrane system to their final locations within the cell or beyond [40]. We speculated that EtEsp also undergoes post-translational modification according to the amino acid sequence analysis, including phosphorylation, myristoylation, and glycosylation. Among these modifications, myristoylation is the key factor in the membrane localization of signal-transducing proteins [41].
Most surface antigens are involved in the invasion, pathogenesis and immune evasion of parasites. For example, in Plasmodium, merozoite surface proteins are critical for parasite invasion, and represent attractive targets for antibody-based therapies against clinical malaria [42]. We also found that the expression of EtEsp increased and that the protein mainly localized on the anterior and surface of the parasites after incubation in CM for 2 h. This suggests that the protein is involved in the sporozoite invasion of host cells. To investigate the function of EtEsp in the invasion process, we performed an invasion test in vitro and found that polyclonal rabbit anti-rEtEsp serum efficiently reduced the sporozoite invasion of cultured DF-1 cells. EtAMA1 also localized to the anterior of the sporozoites after their invasion of DF-1 cells [10]. Previous reports have shown that monospecific mouse anti-rEtAMA1 serum or polyclonal rabbit antiserum against rEtAMA1 also blocked the invasion of host cells in vitro [10, 19]. In the present study, EtEsp is involved in invasion as demonstrated by using antibodies raised against EtEsp in vitro. We tested whether EtEsp is an interacting protein with EtAMA1 by using BiFC, GST-pull down and yeast two-hybrid system. Therefore, we speculated that EtEsp mediates sporozoites invasion in host cells by interacting with EtAMA1. The exact function of EtEsp requires further study.
Previous studies have shown that in T. gondii and Plasmodium, AMA1 interacts directly with rhoptry neck protein 2 (RON2), which is secreted from the parasite rhoptries and specifically localizes at the moving junction. The RON2-AMA1 interaction is a critical step in the moving-junction-dependent invasion of host cells by apicomplexan parasites [43, 44]. Although the interaction between AMA1 and RON2 has not been reported in Eimeria spp., E. tenella is an apicomplexan and AMA1 is conserved in this phylum. Therefore, we inferred that EtAMA1 may also interact with EtRON2 and specifically localize to the moving junction during the invasion of host cells by E. tenella. In this study, we have shown that EtEsp is an EtAMA1-interacting protein, but whether it localizes to the moving junction during invasion requires further study.
Conclusions
In this study, we have shown that EtEsp interacts with EtAMA1 using a BIFC assay in vivo and a GST pull-down assay in vitro. Using staurosporine, we showed that EtEsp is a secreted protein of sporozoites but not from micronemes. An invasion inhibition assay revealed that an antibody against rEtEsp also blocked parasite invasion of its host cells by more than 62%. These data have implications for the use of EtAMA1 or EtAMA1-interacting proteins as targets in therapeutic intervention strategies against avian coccidiosis.
Availability of data and materials
The datasets used and/or analysed during the present study are presented in the article and its additional file or are available from the corresponding author upon reasonable request.
Abbreviations
- EtAMA1:
-
Eimeria tenella apical membrane antigen 1
- EtEsp:
-
E. tenella Eimeria-specific protein
- BiFC:
-
Bimolecular fluorescence complementation
- GST pull-down:
-
Glutathione S-transferase pull-down
- RACE:
-
Random amplification of PCR ends
- UO:
-
Unsporulated oocysts
- SO:
-
Sporulated oocysts
- Spz:
-
Sporozoites
- sMrz:
-
Second-generation merozoites
- NCBI:
-
National Center for Biotechnology Information
- ORF:
-
Open reading frame
- CM:
-
Complete medium
References
Quiroz-Castaneda RE, Dantan-Gonzalez E. Control of avian coccidiosis: future and present natural alternatives. Biomed Res Int. 2015;2015:430610.
Blake DP, Tomley FM. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014;30:12–9.
Tyler JS, Treeck M, Boothroyd JC. Focus on the ringleader: the role of AMA1 in apicomplexan invasion and replication. Trends Parasitol. 2011;27:410–20.
Sibley LD. Invasion strategies of intracellular parasites. Science. 2004;304:248–53.
Sharma P, Chitnis CE. Key molecular events during host cell invasion by apicomplexan pathogens. Curr Opin Microbiol. 2013;16:432–7.
Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J Cell Biol. 1978;77:72–82.
Bargieri DY, Andenmatten N, Lagal V, Thiberge S, Whitelaw JA, Tardieux I, et al. Apical membrane antigen 1 mediates apicomplexan parasite attachment but is dispensable for host cell invasion. Nat Commun. 2013;4:255.
Collins CR, Withers-Martine C, Hackett F, Blackman MJ. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS Pathog. 2009;5:e1000273.
Lamargue M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 2011;7:e1001276.
Jiang L, Lin J, Han H, Dong H, Zhao Q, Zhu S, et al. Identification and characterization of Eimeria tenella apical membrane antigen-1 (AMA1). PLoS ONE. 2012;7:e41115.
Zhang H, Nishikawa Y, Yamagishi J, Zhou J, Ikehara Y, Kojima N, et al. Neospora caninum: Application of apical membrane antigen 1 encapsulated in the oligomannose-coated liposomes for reduction of offspring mortality from infection in BALB/c mice. Exp Parasitol. 2010;125:130–6.
Lagal V, Dinis M, Cannella D, Bargieri D, Gonzalez V, Andenmatten N, et al. AMA1-deficient Toxoplasma gondii parasites transiently colonize mice and trigger an innate immune response that leads to long-lasting protective immunity. Infect Immun. 2015;83:2475–86.
Srinivasan P, Baldeviano GC, Miura K, Diouf A, Ventocilla JA, Leiva KP, et al. A malaria vaccine protects Aotus monkeys against virulent Plasmodium falciparum infection. NPJ Vaccines. 2017;2:14.
Mitchell GH, Thomas AW, Margos G, Dluzewski AR, Bannister LH. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect Immun. 2004;72:154–8.
Mital J, Meissner M, Soldati D, Ward GE. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol Biol Cell. 2005;16:4341–9.
Lamarque M, Besteiro S, Papoin J, Roques M, Vulliez-Le Normand B, Morlon-Guyot J, et al. The RON2-AMA1 interaction is a critical step in moving junction-dependent invasion by apicomplexan parasites. PLoS Pathog. 2011;7:e1001276.
Santos JM, Ferguson DJ, Blackman MJ, Soldati-Favre D. Intramembrane cleavage of AMA1 triggers Toxoplasma to switch from an invasive to a replicative mode. Science. 2010;331:473–7.
Ma D, Huang Y, Ma C, Zhang L, Wang J, Wang D, et al. Eimeria tenella: specific EtAMA1-binding peptides inhibit sporozoite entry into host cells. Poult Sci. 2019;98:4480–91.
Pastor-Fernadez I, Kim S, Billington K, Bumstead J, Marustead-Hernandez V, Kuster T, et al. Development of cross-protective Eimeria-vectored vaccines based on apical membrane antigens. Int J Parasitol. 2018;48:505–18.
Pratt EP, Owens JL, Hockerman GH, Hu CD. Bimolecular fluorescence complementation (BiFC) analysis of protein-protein interactions and assessment of subcellular localization in live cells. Methods Mol Biol. 2016;1474:153–70.
Foltman M, Sanchez-Diaz A. Studying protein-protein interactions in budding yeast using co-immunoprecipitation. Methods Mol Biol. 2016;1369:239–56.
Han H, Xue P, Dong H, Zhu S, Zhao Q, Huang B. Screening and characterization of apical membrane antigen 1 interacting proteins in Eimeria tenella. Exp Parasitol. 2016;170:116–24.
Tomley F. Techniques for isolation and characterization of apical organelles from Eimeria tenella sporozoites. Methods. 1997;13:171–6.
Han HY, Lin JJ, Zhao QP, Dong H, Jiang LL, Xu MQ, et al. Identification of differentially expressed genes in early stages of Eimeria tenella by suppression subtractive hybridization and cDNA microarray. J Parasitol. 2010;96:95–102.
Zhou BH, Wang HW, Wang XY, Zhang LF, Zhang KY, Xue FQ. Eimeria tenella: effects of diclazuril treatment on microneme genes expression in second-generation merozoites and pathological changes of caeca in parasitized chickens. Exp Parasitol. 2010;125:264–70.
Han H, Kong C, Dong H, Zhu S, Zhao Q, Zhai Q, et al. Molecular characterization and functional analysis of subunit 7 of eukaryotic initiation factor 3 from Eimeria tenella. Exp Parasitol. 2015;154:118–26.
Yan M, Cui X, Zhao Q, Zhu S, Huang B, Wang L, et al. Molecular characterization and protective efficacy of the microneme 2 protein from Eimeria tenella. Parasite. 2018;25:60.
Carruthers VB, Giddings OK, Sibley LD. Secretion of micronemal proteins is associated with toxoplasma invasion of host cells. Cell Microbiol. 1999;1:225–35.
Peroval M, Pery P, Labbe M. The heat shock protein 90 of Eimeria tenella is essential for invasion of host cell and schizont growth. Int J Parasitol. 2006;36:1205–15.
Miller J, Stagljar I. Using the yeast two-hybrid system to identify interacting proteins. Methods Mol Biol. 2004;261:247–62.
Ferro E, Baldni E, Trabalzini L. Use of the yeast two-hybrid technology to isolate molecular interactions of Ras GTPases. Methods Mol Biol. 2014;1120:97–120.
Pham CD. Detection of protein-protein interaction using bimolecular fluorescence complementation assay. Methods Mol Biol. 2015;1278:483–95.
Bain AL, Harris JL, Khanna KK. Identification of ATM-interacting proteins by co-immunoprecipitation and glutathione-S-transferase (GST) pull-down assays. Methods Mol Biol. 2017;1599:163–81.
Lv L, Huang B, Zhao Q, Zhao Z, Dong H, Zhu S, et al. Identification of an interaction between calcium-dependent protein kinase 4 (EtCDPK4) and serine protease inhibitor (EtSerpin) in Eimeria tenella. Parasit Vectors. 2018;11:259.
Ma F, Ni L, Liu L, Li X, Zhang H, Zhang A, et al. ZmABA2, an interacting protein of ZmMPK5, is involved in abscisic acid biosynthesis and functions. Plant Biotechnol J. 2016;14:771–82.
Takahashi Y. Co-immunoprecipitation from transfected cells. Methods Mol Biol. 2015;1278:381–9.
Naor A, Altman-Price N, Soucy SM, Green AG, Mitiagin Y, Turgeman-Grott I, et al. Impact of a homing intein on recombination frequency and organismal fitness. Proc Natl Acad Sci USA. 2016;113:E4654–61.
Lal K, Bromley E, Oakes R, Prieto JH, Sanderson SJ, Kurian D, et al. Proteomic comparison of four Eimeria tenella life-cycle stages: unsporulated oocyst, sporulated oocyst, sporozoite and second-generation merozoite. Proteomics. 2009;9:4566–76.
Reid AJ, Blake DP, Ansari HR, Billington K, Browne HP, Bryant J, et al. Genomic analysis of the causative agents of coccidiosis in domestic chickens. Genome Res. 2014;24:1676–85.
Marugan-Hernandez V, Long E, Blake D, Crouch C, Tomley F. Eimeria tenella protein trafficking: differential regulation of secretion versus surface tethering during the life cycle. Sci Rep. 2017;7:4557.
McCabe JB, Berthiaume LG. N-terminal protein acylation confers localization to cholesterol, sphingolipid-enriched membranes but not to lipid rafts/caveolae. Mol Biol Cell. 2001;12:3601–17.
Babacar M, Fall MM, Varela ML, Loucoubar C, Joos C, Fall B, et al. Analysis of antibody responses to selected Plasmodium falciparum merozoite surface antigens in mild and cerebral malaria and associations with clinical outcomes. Clin Exp Immunol. 2019;196:86–96.
Tonkin ML, Roques M, Lamarque MH, Pugnière M, Douguet D, Crawford J, et al. Host cell invasion by apicomplexan parasites: insights from the co-structure of AMA1 with a RON2 peptide. Science. 2011;333:463–7.
Shen B, Sibley LD. The moving junction, a key portal to host cell invasion by apicomplexan parasites. Curr Opin Microbiol. 2012;15:449–55.
Acknowledgements
We would like to thank all organizations which funded this work and all the teachers who cooperated in technical assistance. We would also like to thank Dr Wang Quan who provided the mouse anti-TgGRA7 antibody of T.gondii .
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31572266 and 31672551), the National Key Research and Development Program of China (2018YFD0500302), the National Sharing Service Platform for Parasite Resources (No. TDRC-22) and Shanghai Minhang District talent development special funds.
Author information
Authors and Affiliations
Contributions
HYH and HD conceived and designed the study. CL, QPZ, SHZ and QJW performed the experiments. CL, BH and HZZ analyzed the data. CL, QPZ, HXW, SLY, YY and SSL collected parasites. CL, QPZ and HYH wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The protocol was approved and authorized by the Animal Care and Use Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Primer sequences used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, C., Zhao, Q., Zhu, S. et al. Eimeria tenella Eimeria-specific protein that interacts with apical membrane antigen 1 (EtAMA1) is involved in host cell invasion. Parasites Vectors 13, 373 (2020). https://doi.org/10.1186/s13071-020-04229-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04229-5