Abstract
Background
Approximately 30% of children worldwide are infected with gastrointestinal parasites. Depending on the species, parasites can disrupt intestinal bacterial microbiota affecting essential vitamin biosynthesis.
Methods
Stool samples were collected from 37 asymptomatic children from a previous cross-sectional Argentinian study. A multi-parallel real-time quantitative PCR was implemented for Ascaris lumbricoides, Ancylostoma duodenale, Necator americanus, Strongyloides stercoralis, Trichuris trichiura, Cryptosporidium spp., Entamoeba histolytica and Giardia duodenalis. In addition, whole-genome sequencing analysis was conducted for bacterial microbiota on all samples and analyzed using Livermore Metagenomic Analysis Toolkit and DIAMOND software. Separate analyses were carried out for uninfected, Giardia-only, Giardia + helminth co-infections, and helminth-only groups.
Results
For Giardia-only infected children compared to uninfected children, DNA sequencing data showed a decrease in microbiota biodiversity that correlated with increasing Giardia burden and was statistically significant using Shannonʼs alpha diversity (Giardia-only > 1 fg/µl 2.346; non-infected group 3.253, P = 0.0317). An increase in diversity was observed for helminth-only infections with a decrease in diversity for Giardia + helminth co-infections (P = 0.00178). In Giardia-only infections, microbiome taxonomy changed from Firmicutes towards increasing proportions of Prevotella, with the degree of change related to the intensity of infection compared to uninfected (P = 0.0317). The abundance of Prevotella bacteria was decreased in the helminths-only group but increased for Giardia + helminth co-infections (P = 0.0262). Metagenomic analysis determined cobalamin synthesis was decreased in the Giardia > 1 fg/µl group compared to both the Giardia < 1 fg/µl and the uninfected group (P = 0.0369). Giardia + helminth group also had a decrease in cobalamin CbiM genes from helminth-only infections (P = 0.000754).
Conclusion
The study results may provide evidence for an effect of parasitic infections enabling the permissive growth of anaerobic bacteria such as Prevotella, suggesting an altered capacity of vitamin B12 (cobalamin) biosynthesis and potential impact on growth and development in children

.
Similar content being viewed by others
Background
Gastrointestinal (GI) parasites are estimated to infect more than two billion people throughout the world [1]. Both soil-transmitted helminths (STH) (Ascaris lumbricoides, hookworms, Strongyloides stercoralis, Trichuris trichiura) and protozoans (Giardia duodenalis, Cryptosporidium spp., Entamoeba histolytica) are prevalent in resource-limited areas [2, 3]. Symptoms include chronic diarrhea, severe anemia, and can lead to intestinal obstruction. Economically disadvantaged children have recurrent infections and malnutrition that may lead to growth and cognitive delays [4]. These children have more difficulties in school and, subsequently, in the job market [4]. The cycle continues when they remain in poverty and have their children [4]. The link between intestinal helminths and malnutrition leading to growth stunting and anemia has been found by others [5,6,7,8,9,10,11,12], and a Global Burden of Disease Study points to evidence that hookworm is a leading cause of anemia in resource-poor settings [13]. Valuable information from the Global Enteric Multicenter Study (GEMS) and studies related to returning travelers also reveals an unexpected global health impact caused by some protozoans, possibly including giardiasis [14, 15]. The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development (MAL-ED) study found an association with subclinical, non-diarrheal giardiasis and decreases in growth of children [16]. There are few studies attributing gut microbiome changes to giardiasis [17,18,19] and no published studies showing the impact on the human intestinal microbiome using multi-parallel real-time quantitative (qPCR) to detect the presence of Giardia and quantitating the burden of infection [20]. To date, most studies examining intestinal parasitism have not been able to study intestinal worms and protozoans simultaneously and to successfully dissect the relative contribution of each of the significant intestinal helminth or protozoan pathogens to specific diseases. The current state of diagnosing gastrointestinal parasites in resource-limited areas uses the subjective method of microscopy. Depending on the parasite, single stool microscopy exam sensitivity ranges from 50–80% [21]. As a result, large numbers of infected children are not being diagnosed correctly and treated.
Gastrointestinal parasites may modulate intestinal inflammation, malabsorption, and microbiome changes [22,23,24,25,26]. The microbiome is associated with digestion, nutrition and health, but alterations in biodiversity can increase disease states and induce intestinal inflammation [27]. Animal studies further show changes in microbial diversity due to G. duodenalis [18, 28]. There is a lack of literature studying the relationship between GI parasitesʼ impact on the human intestinal microbiome [19]. These few studies have presented discordant results of the impact parasites have on human intestinal microbiota biodiversity [29, 30].
The relationship between intestinal microbiota biodiversity may depend upon which specific parasite is present in the gut [30]. Since the burden of parasite infection is directly correlated to morbidity and disease, the intensity of infection may also impact the intestinal microbiota [30]. The qPCR quantitates the burden of helminths and protozoans, determining the correlation of burden to changes in intestinal microbiota biodiversity. Alterations in intestinal microbiota alter bacterial metabolites, such as vitamin B12 (cobalamin), reducing their availability for human use.
Interactions between parasites and intestinal microbiota may have a direct impact on child nutrition. Giardia duodenalis is known to cause malabsorption, steatorrhea, and diarrhea [31], with preliminary studies finding improvements in vitamin B12 serum levels after treatment for giardiasis [32, 33]. Vitamin B12 is a crucial microbiota-derived co-enzyme for humans who cannot produce it [34,35,36]. As vitamin B12 production is unique to specific intestinal bacteria, alterations in intestinal microbiota could diminish vitamin B12 availability for human use [37]. Advances in next-generation DNA sequencing allow for precise taxonomic comparisons between intestinal microbiotas and can simultaneously be used to scan the intestinal microbiota meta-genome for the presence of functional genes necessary for the specific functions, like cobalamin synthesis. In this pilot study, parasite qPCR and next-generation DNA sequencing was used to explore whether quantitative burden of specific parasites (Giardia duodenalis and soil-transmitted helminths) influence the composition of intestinal microbial communities. Using vitamin B12 as a representative bacteria-generated nutrient, we analyzed bacterial metagenomes as a surrogate for changes in intestinal bacteria functions associated with intestinal parasitic infections. This was a preliminary study using a population with high prevalence of intestinal parasites. It is meant as an introductory for the future direction of our research.
Methods
Study population
This descriptive study aimed to determine the effect of G. duodenalis and other intestinal parasites on bacterial microbiota and subsequent cobalamin metagenomics. Samples were randomly selected from a previously published study using qPCR in peri-urban Argentina [38]. No previous antiparasitics or antibiotics were administered 3 months prior to the sample collection. Samples consisted of four groups: (i) a control group with no parasites detected by qPCR (uninfected); (ii) a Giardia-only infected group; (iii) a Giardia and helminth co-infection group; and (iv) a helminth-only infected group. Helminths included in this study were Ascaris lumbricoides, Ancylostoma duodenale, Necator americanus and Strongyloides stercoralis (Table 1).
Multi-parallel real-time quantitative PCR
All stools were collected and immediately stored on ice and sealed in air-tight containers, frozen within 3 h, and DNA extracted using MP FastDNA Spin Kits for Soil (MP Biomedicals, Irvine, CA) [38]. This collection method was found to yield adequate microbiome data for study analysis [29]. Samples were processed in Salta, Argentina, using a modified bead-beating process described previously [38, 39]. DNA was processed for qPCR in Houston, Texas, as previously described [38, 39]. The intensity of parasite DNA was calculated using reference plasmids to create a standard curve, as previously reported [38]. The qPCR results from the previous Argentinian study were used for these analyses [38].
Whole-genome DNA sequencing
DNA sequencing and library construction were performed at New England Biolabs (NEB). All the samples underwent removal of methylated DNA with NEBNext® Microbiome enrichment kits (New England Biolabs, Ipswich, MA). No DNA size selection was made. DNA Library prep was done as per the manufacturerʼs protocol using 1 µg of sample DNA and eight cycles of PCR enrichment (NEBNext Ultra DNA Library Prep Kit for Illumina, Version 5.1, 9/17). Sequencing was performed using an Illumina NextSeq (Illumina, San Diego, CA) with paired or single ends, and 151 bp reads.
Bioinformatics
Fastq reads were filtered for quality at a minimum Phred score of 20 (99% accuracy) and a minimum length of 50 using Cutadapt version 1.8.3 [40]. Paired-end reads were interleaved using merge_fastq_reads_with_N_separator. Pl Perl script included with Livermore Metagenomics Analysis Toolkit (LMAT) software version 1.2.6 [41]. Fastq files were converted to fasta files using seqtk software version 1.0 (https://github.com/lh3/seqtk). Fasta files were processed by LMAT for taxonomic classification using LMAT kFull database. LMAT output text files were filtered for LMAT defined confidence score of 1 and minimum reads of 500 using tolineage.py script. Subsequently, output files were combined using merge_metaphlan_tables.py script using Metaphlan [42].
Alpha diversity was calculated using the Phyloseq R package [43]. Abundant different operational taxonomic units (OTU) among the four groups were identified using the LEfSe algorithm [44]. Most abundant bacteria for Lefse were run with a logarithmic LDA score threshold of 4.5 and other parameters set to default. Metagenomics analysis was performed using Diamond v0.8.4 using blastx mode with 90% minimum identity and e-value of 10−5 against nr database fasta file [45]. Results from Diamond analysis were exported to Megan version 6 using daa-meganizer program [46]. GenInfo identifier to Interpro identifier mapping within Megan program was used to annotate the vitamin B12 synthesis gene [47]. STAMP software was used for statistical analysis pertaining to taxonomic and metagenomic differences [48]. Microbial attributes were derived from (LMAT) taxonomic output using the Megan program.
Statistics
qPCR results were recorded for each patient as positive or negative, including the concentration of DNA (fg/µl) for each parasite. All statistics were performed using Prism v. 7.0b (GraphPad, La Jolla, CA). Mann-Whitney and one-way ANOVA tests were used to compare two and multiple groups, respectively. Spearmanʼs rank test was used to correlate the Giardia DNA concentration to Shannonʼs alpha diversity, including the “uninfected” and “Giardia-only” group. Shannonʼs alpha diversity is a commonly reported diversity metric that weights the numbers of species by their relative evenness [49]. Proportion of targeted sequences was used in calculating OTU and metagenomics. Vitamin B12 data was calculated comparing specific genes to all other vitamin B12 synthesis genes. All statistical models used P-values less than 0.05 as significant.
Results
Parasite DNA intensity
Parasite DNA intensity with the intestinal parasite detection by stool qPCR as described by Cimino et al. [38] (Table 1).
Diversity of intestinal microbiota
Measuring bacterial diversity in relation from children with no Giardia DNA to increasing Giardia DNA intensity of burden (fg/µl) showed an inverse correlation of bacterial diversity to Giardia DNA (fg/µl) (Spearmanʼs r(18) = − 0.4781, P = 0.0447) (Fig. 1a). From the scatter plot (Fig. 1a) a noticeable change in slope occurred at levels > 1 fg/µl. Since there are no guidelines for heavy Giardia burden, greater than 1 fg/µl was selected, and all further analyses validated this threshold.
a There was an inverse correlation of Giardia DNA to decreased Shannonʼs alpha diversity (Spearmanʼs r = − 0.4781, P = 0.0447); > 1 fg/µl was selected as heavy Giardia DNA burden since a noticeable change in slope occurred after 1 fg/µl. b The mean of the Giardia > 1 fg/µl group (dotted line) has a significant decrease in bacterial diversity compared with the uninfected group (P = 0.0317) and all other groups (P = 0.000166). *P < 0.05
Giardia > 1 fg/µl group had a significantly lower biodiversity compared to the uninfected children using Shannonʼs alpha diversity (Giardia-only > 1 fg/µl: 2.346; uninfected group: 3.253, U(10) = 2, Z = − 2.1934, P = 0.0317 (Fig. 1b). Giardia > 1 fg/µl had lower diversity than the helminth-only group U(17) = 0, Z = − 3.1096, P = 0.0003 (Fig. 1b). Giardia > 1 fg/µl also had lower diversity compared to all other groups F(3, 25) = 9.982, P = 0.000166 (Fig. 1b). Alpha diversity was also lower for the Giardia + helminth co-infected group compared to the helminth-only group U(19) = 7, Z = 2.958, P = 0.00178 (Fig. 1b).
Change in bacterial abundance
In Giardia-only infections, microbiome analysis data indicates decreased biodiversity in the infected parasite group compared to the non-infected group, a bias toward increased Prevotella, with the degree of change related to the intensity of infection (Fig. 2a). Giardia-only children had significantly higher proportions of the genus Prevotella bacteria directly correlating to above 1 fg/µl Giardia DNA versus uninfected group U(10) = 2, Z = 2.1934, P = 0.0317 (Fig. 2a). The helminth-only group with decreased Prevotella proportions relative to the Giardia-only group U(25) = 41, Z = 2.0125, P = 0.0457, but similar to the control group (Fig. 2b). Interestingly, the Giardia + helminth co-infected group had increased Prevotella proportions compared to the helminth-only group U(19) = 8, Z = − 2.8735, P = 0.00262 (Fig. 2b). Prevotella copri was the major species in each group, Giardia (37%), helminths-only (17%), uninfected (22%), Giardia + helminths (36%) (data not shown).
aGiardia > 1 fg/µl group had greater proportions of Prevotella DNA than the uninfected group (P = 0.0317) in their intestinal microbiota. b The helminths-only group had decreased proportions of Prevotella DNA compared with the Giardia-only group (P = 0.0457) in their intestinal microbiota. The Giardia + helminth co-infected group had increased proportion of Prevotella DNA compared to the helminth-only group (P = 0.00262) in their intestinal microbiota. *P < 0.05
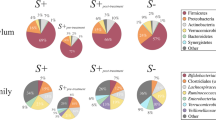
All four groups had different bacteria genera as their most abundant microbiota. Giardia- infected children, including Giardia + helminth co-infected, had higher Bacteroidales, including Prevotella species (Fig. 3). Specific cobalamin producing bacteria, such as Lactobacillus and Bifidobacterium, were found in higher abundance for the non-Giardia infected children (Fig. 3).
Metagenomics of cobalamin biosynthesis
Vitamin B12 InterPro identifier IPR002751 biosynthesis CbiM gene was used in the analysis. Children with Giardia > 1 fg/µl infections had fewer quantities of cobalamin DNA sequences than the Giardia < 1 fg/µl and uninfected group combined F(2, 15) = 4.145, P = 0.0369 (Fig. 4a). Children with Giardia + helminth co-infections had a less proportion of vitamin B12 pathway DNA sequences, compared to helminth-group U(19) = 5, Z = 3.1271, P = 0.000754 (Figs. 4b, 5).
a Children infected with > 1 fg/µl Giardia DNA concentrations had decreased cobalamin biosynthesis genes compared to the combined Giardia < 1 fg/µl and uninfected groups (P = 0.0369) with b the Giardia + helminth group having less cobalamin biosynthesis genes than the helminth-only infections (P = 0.000754). *P < 0.05
Discussion
The association of G. duodenalis with microbiome diversity was observed in this study using whole-genome sequencing. Giardia duodenalis plays a prominent role, perhaps as its primary site of infection and replication is in the small intestine.
Parasites alter the intestinal microbiota of children
This study determined that children infected with Giardia DNA > 1 fg/µl are associated with decreased microbial diversity and increases of Prevotella. DNA levels of Giardia > 1 fg/µl implies that there are more parasites to alter the intestinal microbiome, and thus have a higher impact on intestinal bacterial species.
It is unclear whether the parasites are impacting the intestinal microbiota or external factors such as age, diet, or sex differences altering the intestinal microbiota and making the subjects more susceptible to enabling a G. duodenalis infection. Certain bacteria can permit G. duodenalis colonization, as evidenced from a mouse model study where the mouse intestinal microbiota (enteroaggregative Escherichia coli), independent of G. duodenalis infection, can promote inflammation, but together synergistically increased signals of intestinal injury [18].
Changes in vitamin B12 due to parasite infections
Vitamin B12 synthesis primarily occurs in anaerobes, including Bifidobacterium and Lactobacillus species [37, 50,51,52,53]. These microorganisms may promote intestinal homeostasis and may protect against inflammatory diseases [54,55,56,57]. Vitamin B12 is absorbed in the small intestines [58,59,60] while the majority of microbiota reside in the colon [58], although, the small intestine is not sterile and does contain a robust microbiota that influences the absorption of vitamins [61,62,63]. Specific bacteria produce vitamin B12 [64], and the children infected with G. duodenalis with DNA levels above 1 fg/µl may be unable to synthesize the required amounts of vitamin B12 for nutritional benefit. The group with less than 1 fg/µl Giardia DNA infections had equal vitamin B12 bacterial genes to the uninfected group, both being higher than the Giardia > 1 fg/µl group, likely showing that lower intensity of Giardia infections has similar effects as the uninfected group on vitamin B12 synthesis.
Evidence of the impact of G. duodenalis on the diversity and available micronutrients was also observed in the Giardia + helminth co-infection group, compared to the alterations of the microbiome seen in the helminth-only group. While the helminth-only group did not have changes in diversity or decreased cobalamin synthesis genes, a possible explanation is that most of these helminths reside in the colon and do not alter the microenvironments as does G. duodenalis.
Study limitations
A limitation of this cross-sectional study is the small sample size. However, the results are consistent, and this potential limitation can be viewed in terms of the sheer number, and fidelity of, using enriched microbe DNA for whole gene sequencing, producing millions of reads for analysis. This study also did not take into account Giardia assemblages that can have differing amount of pathogenicity. Another limitation is that serum vitamin B12 levels were not measures in children and thus the decrease of vitamin B12 genes in Giardia infected children could not be translated into a loss of vitamin B12.
Conclusions
In this study, there is a possible link as to why G. duodenalis and other parasites may cause growth and developmental delays in infected children. Giardia-infected children with > 1 fg/µl DNA concentrations were associated with less microbiome diversity, and a higher abundance of Prevotella associated with the diminished presence of cobalamin synthesis genes. The influence of G. duodenalis appears to be evident regardless of the presence of STH, and was associated with altered microbiome composition or function as measured by cobalamin synthesis. This descriptive study is a preliminary evidence for future studies looking at the cobalamin biosynthesis pathway. We are currently extending these results to additional populations where G. duodenalis and other intestinal parasite infections are endemic, determining Giardia assemblages, and confirming the biochemical analysis of the vitamin B12 pathway products in affected children.
Availability of data and materials
The data supporting the conclusions of this article are available in the NCBI BioProject repository, accession PRJNA612291 https://www.ncbi.nlm.nih.gov/bioproject/PRJNA612291/.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- GEMS:
-
Global enteric multicenter study
- GI:
-
Gastrointestinal
- LMAT:
-
Livermore metagenomics analysis toolkit
- OTU:
-
Operational taxonomic unit
- qPCR:
-
Multi-parallel real-time quantitative polymerase chain reaction
- STH:
-
Soil-transmitted helminths
References
Brooker S, Clements AC, Bundy DA. Global epidemiology, ecology and control of soil-transmitted helminth infections. Adv Parasitol. 2006;62:221–61.
Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D, et al. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet. 2006;367:1521–32.
Baldursson S, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks—an update 2004–2010. Water Res. 2011;45:6603–14.
Hotez PJ. American Society for Microbiology. Forgotten people, forgotten diseases: the neglected tropical diseases and their impact on global health and development. 2nd ed. Washington, DC: ASM Press; 2013.
Joseph SA, Casapia M, Montresor A, Rahme E, Ward BJ, Marquis GS, et al. The effect of deworming on growth in 1-year-old children living in a soil-transmitted helminth-endemic area of Peru: a randomized controlled trial. PLoS Negl Trop Dis. 2015;9:e0004020.
Zhou H, Watanabe C, Ohtsuka R. Impacts of dietary intake and helminth infection on diversity in growth among schoolchildren in rural south China: a 4-year longitudinal study. Am J Hum Biol. 2007;19:96–106.
Thomas M, Woodfield G, Moses C, Amos G. Soil-transmitted helminth infection, skin infection, anaemia, and growth retardation in schoolchildren of Taveuni Island. Fiji. N Z Med J. 2005;118:U1492.
Savioli L, Neira M, Albonico M, Beach MJ, Chwaya HM, Crompton DW, et al. Treatment for intestinal helminth infection. Review needed to take account of all relevant evidence, not only effects on growth and cognitive performance. BMJ. 2000;321:1226–7.
Dickson R, Awasthi S, Williamson P, Demellweek C, Garner P. Effects of treatment for intestinal helminth infection on growth and cognitive performance in children: systematic review of randomised trials. BMJ. 2000;320:1697–701.
Hadju V, Stephenson LS, Mohammed HO, Bowman DD, Parker RS. Improvements of growth, appetite, and physical activity in helminth-infected schoolboys 6 months after single dose of albendazole. Asia Pac J Clin Nutr. 1998;7:170–6.
Hadju V, Stephenson LS, Abadi K, Mohammed HO, Bowman DD, Parker RS. Improvements in appetite and growth in helminth-infected schoolboys 3 and 7 weeks after a single dose of pyrantel pamoate. Parasitology. 1996;113:497–504.
Urban JF Jr, Romanowski RD, Steele NC. Influence of helminth parasite exposure and strategic application of anthelmintics on the development of immunity and growth of swine. J Anim Sci. 1989;67:1668–77.
Kassebaum NJ, Jasrasaria R, Naghavi M, Wulf SK, Johns N, Lozano R, et al. A systematic analysis of global anemia burden from 1990 to 2010. Blood. 2014;123:615–24.
Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22.
Ross AG, Cripps AW. Enteropathogens and chronic illness in returning travelers. N Engl J Med. 2013;369:784.
Rogawski ET, Liu J, Platts-Mills JA, Kabir F, Lertsethtakarn P, Siguas M, et al. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob Health. 2018;6:e1319–28.
Toro-Londono MA, Bedoya-Urrego K, Garcia-Montoya GM, Galvan-Diaz AL, Alzate JF. Intestinal parasitic infection alters bacterial gut microbiota in children. PeerJ. 2019;7:e6200.
Bartelt LA, Bolick DT, Mayneris-Perxachs J, Kolling GL, Medlock GL, Zaenker EI, et al. Cross-modulation of pathogen-specific pathways enhances malnutrition during enteric co-infection with Giardia lamblia and enteroaggregative Escherichia coli. PLoS Pathog. 2017;13:e1006471.
Iebba V, Santangelo F, Totino V, Pantanella F, Monsia A, Di Cristanziano V, et al. Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Côte dʼIvoire. J Infect Dev Ctries. 2016;10:1035–41.
Donowitz JR, Alam M, Kabir M, Ma JZ, Nazib F, Platts-Mills JA, et al. A prospective longitudinal cohort to investigate the effects of early life giardiasis on growth and all cause diarrhea. Clin Infect Dis. 2016;63:792–7.
Mosli M, Gregor J, Chande N, Lannigan R. Nonutility of routine testing of stool for ova and parasites in a tertiary care Canadian centre. Can J Microbiol. 2012;58:653–9.
Easton AV, Quinones M, Vujkovic-Cvijin I, Oliveira RG, Kepha S, Odiere MR, et al. The impact of anthelmintic treatment on human gut microbiota based on cross-sectional and pre- and postdeworming comparisons in western Kenya. mBio. 2019;10:2.
Hagel I, Cabrera M, Puccio F, Santaella C, Buvat E, Infante B, et al. Co-infection with Ascaris lumbricoides modulates protective immune responses against Giardia duodenalis in school Venezuelan rural children. Acta Trop. 2011;117:189–95.
Weatherhead J, Cortes AA, Sandoval C, Vaca M, Chico M, Loor S, et al. Comparison of cytokine responses in Ecuadorian children infected with Giardia, Ascaris, or both parasites. Am J Trop Med Hyg. 2017;96:1394–9.
Trelis M, Taroncher-Ferrer S, Gozalbo M, Ortiz V, Soriano JM, Osuna A, et al. Giardia intestinalis and fructose malabsorption: a frequent association. Nutrients. 2019;11:E2973.
Rogawski ET, Bartelt LA, Platts-Mills JA, Seidman JC, Samie A, Havt A, et al. Determinants and impact of Giardia infection in the first 2 years of life in the MAL-ED birth cohort. J Pediatric Infect Dis Soc. 2017;6:153–60.
Jaeggi T, Kortman GA, Moretti D, Chassard C, Holding P, Dostal A, et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut. 2015;64:731–42.
Barash NR, Maloney JG, Singer SM, Dawson SC. Giardia alters commensal microbial diversity throughout the murine gut. Infect Immun. 2017;85:e00948.
Cooper P, Walker AW, Reyes J, Chico M, Salter SJ, Vaca M, et al. Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS ONE. 2013;8:e76573.
Lee SC, Tang MS, Lim YA, Choy SH, Kurtz ZD, Cox LM, et al. Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl Trop Dis. 2014;8:e2880.
Cain GD, Moore P Jr, Patterson M. Malabsorption associated with Giardia lamblia infestation. South Med J. 1968;61:532–4.
Olivares JL, Fernandez R, Fleta J, Ruiz MY, Clavel A. Vitamin B12 and folic acid in children with intestinal parasitic infection. J Am Coll Nutr. 2002;21:109–13.
Ventura LL, Oliveira DR, Viana JC, Santos JF, Caliari MV, Gomes MA. Impact of protein malnutrition on histological parameters of experimentally infected animals with Giardia lamblia. Exp Parasitol. 2013;133:391–5.
Martens JH, Barg H, Warren MJ, Jahn D. Microbial production of vitamin B12. Appl Microbiol Biotechnol. 2002;58:275–85.
Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–81.
Lawrence JG, Roth JR. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics. 1996;142:11–24.
LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–8.
Cimino RO, Jeun R, Juarez M, Cajal PS, Vargas P, Echazu A, et al. Identification of human intestinal parasites affecting an asymptomatic peri-urban Argentinian population using multi-parallel quantitative real-time polymerase chain reaction. Parasit Vectors. 2015;8:380.
Mejia R, Vicuna Y, Broncano N, Sandoval C, Vaca M, Chico M, et al. A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Am J Trop Med Hyg. 2013;88:1041–7.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–20.
Ames SK, Hysom DA, Gardner SN, Lloyd GS, Gokhale MB, Allen JE. Scalable metagenomic taxonomy classification using a reference genome database. Bioinformatics. 2013;29:2253–60.
Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–4.
McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8(4):e61217.
Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60.
Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60.
Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–86.
Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43(1):213–21.
Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–21.
Reese AT, Dunn RR. Drivers of microbiome biodiversity: a review of general rules, feces, and ignorance. MBio. 2018;9:e01294.
Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, et al. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009;3:745.
Turroni F, Foroni E, Pizzetti P, Giubellini V, Ribbera A, Merusi P, et al. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl Environ Microbiol. 2009;75:1534–45.
Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 2012;20:467–76.
Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE. 2012;7:e36957.
Salminen S, Nybom S, Meriluoto J, Collado MC, Vesterlund S, El-Nezami H. Interaction of probiotics and pathogens—benefits to human health? Curr Opin Biotechnol. 2010;21:157–67.
Lomax AR, Calder PC. Probiotics, immune function, infection and inflammation: a review of the evidence from studies conducted in humans. Curr Pharm Des. 2009;15:1428–518.
Ghouri YA, Richards DM, Rahimi EF, Krill JT, Jelinek KA, DuPont AW. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol. 2014;7:473–87.
Sjoberg F, Barkman C, Nookaew I, Ostman S, Adlerberth I, Saalman R, et al. Low-complexity microbiota in the duodenum of children with newly diagnosed ulcerative colitis. PLoS ONE. 2017;12:e0186178.
Gille D, Schmid A. Vitamin B12 in meat and dairy products. Nutr Rev. 2015;73:106–15.
Glass GB, Boyd LJ, Stephanson L. Intestinal absorption of vitamin B12 in man. Science. 1954;120:74–5.
Glass GB, Boyd LJ, Stephanson L. Intestinal absorption of vitamin B12 in humans as studies by isotope technic. Proc Soc Exp Biol Med. 1954;86:522–6.
Onishi JC, Campbell S, Moreau M, Patel F, Brooks AI, Zhou YX, et al. Bacterial communities in the small intestine respond differently to those in the caecum and colon in mice fed low- and high-fat diets. Microbiology. 2017;163:1189–97.
Ghoshal UC, Shukla R, Ghoshal U. Small intestinal bacterial overgrowth and irritable bowel syndrome: a bridge between functional organic dichotomy. Gut Liver. 2017;11:196–208.
Ghoshal UC, Ghoshal U. Small intestinal bacterial overgrowth and other intestinal disorders. Gastroenterol Clin North Am. 2017;46:103–20.
Magnusdottir S, Ravcheev D, de Crecy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148.
Acknowledgments
We wish to thank Don Comb, Rich Roberts, Tom Evans, Andy Gardner and Tilde Carlow (from NEB) for support of this project and the NEB DNA Sequencing Core (Laurie Mazzola, Danielle Fuchs, Joanna Bybee).
Funding
Internal funds from New England Biolabs for DNA sequencing. Research funding support for RM was provided by the US Department of Health and Human Services, Health Resources and Services Administration for Baylor College of Medicine Center of Excellence in Health Equity, Training, and Research (Grant No: D34HP31024). Also, the Baylor College of Medicine Junior Faculty Seed Award was used for research supplies. Microsoft Azure Sponsorship provided funding for cloud computing. Funding also provided by the Texas Children’s Hospital Center for Vaccine Development and the National School of Tropical Medicine, Baylor College of Medicine.
Author information
Authors and Affiliations
Contributions
RM and AD were responsible for bioinformatics and statistical analysis. RM, RJ and PEB performed DNA extraction, qPCR, demethylation, and library preparation. BS, EL ED and CL were involved in the DNA sequencing sample reactions and preliminary analysis. PV, MJ, PSC, JN, AK and ROC collected patient samples in Argentina. RM, AD and BS wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The internal review boards approved this study of Baylor College of Medicine and Universidad Nacional de Salta Argentina. Informed written consent was obtained from each participant or a parent/guardian. Anti-parasitic treatment, based on microscopy findings, was provided per standard of care in the region. The majority of subjects were of preschool age and did not receive standard mass drug administration of anthelmintics. The bioethics committee approved the study design and protocols of Universidad Nacional de Salta, Argentina (UNSA).
Consent for publication
Not applicable.
Competing interests
RM receives research funding from Romark Laboratories, LC. This association did not impact this study. AD, RJ, PEB, PV, MJ, PSC, JN, AK, EL, CL, ED, ROC, BS declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mejia, R., Damania, A., Jeun, R. et al. Impact of intestinal parasites on microbiota and cobalamin gene sequences: a pilot study. Parasites Vectors 13, 200 (2020). https://doi.org/10.1186/s13071-020-04073-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04073-7