Abstract
Background
Perkinsosis, a disease caused by the protist Perkinsus, is responsible for mass mortalities of many molluscan species worldwide. The rapid, early and accurate detection of Perkinsus infection is necessary to react to outbreaks, and manage disease transmission. Current methods for diagnosis of Perkinsus spp. are time-consuming or require professional equipment and experienced personnel, rendering them unsuitable for field application. Recombinase polymerase amplification (RPA) assay is a highly sensitive and selective isothermal amplification technique that operates at temperatures of 37–42 °C, requires minimal sample preparation, and is capable of amplifying as low as 1–10 target DNA copies in less than 20 minutes.
Methods
We report a novel RPA assay that amplifies the internal transcriber spacer (ITS) region of P. beihaiensis, which, followed by rapid detection of amplicons using a lateral flow (LF) strip, enables easy visualization of results by the naked eye.
Results
The LF-RPA assay successfully amplified P. beihaiensis DNA using a set of primers of 20–25 bp in length. After incubation at 37 °C for 25 min, results were read within 5 min by the naked eye on a lateral flow strip. Our LF-RPA assay was comparably sensitive to qPCR assay, and capable of detecting as few as 26 copies of P. beihaiensis DNA. Cross-amplification occurred with other two Perkinsus species, P. olseni and P. chesapeaki, but not with other potential pathogen taxa in culture environments. We compared the performance of LF-RPA, conventional PCR and qPCR assays on 60 oyster samples. While LF-RPA assay results were 86.2% as sensitive, 77.4% as specific, and generally in agreement with those of conventional PCR results, they were more (93.3%) sensitive, (86.7%) specific, and agreed better with qPCR assay results. Future research should focus on developing simple DNA extraction methods that do not require professional laboratories and complicated extraction procedures, to facilitate application of this LF-RPA assay in the field.
Conclusions
Our LF-RPA assay provides a rapid and efficient method for detecting species of Perkinsus. This novel assay has potential to be used in field applications.
Similar content being viewed by others
Background
Perkinsosis, a disease caused by the protist Perkinsus, has caused mass mortalities in species of mollusc worldwide [1, 2]. Most Perkinsus species are generalist parasites, have low host specificity, can switch between hosts and are geographically widespread [3, 4]. For example, P. olseni, an internationally reportable or notifiable molluscan pathogen [World Organization for Animal Health (OIE)], is known from 17 countries and 27 host species in five orders and six families [5].
The life-cycles of Perkinsus spp. involve direct transmission between the molluscan host without an intermediate host. All life stages (trophozoite, hypnospore and zoospore) are infective [6]. Without surveillance, widespread transmission of Perkinsus spp. can occur anywhere molluscs (especially oyster species, e.g. Crassostrea gigas, C. virginica and C. hongkongensis) are cultivated, particularly through movements of infected hosts during aquaculture production, and in trade. Given the economic loss associated with tissue degradation and potential lethality of Perkinsus spp. to host species, and the difficulties of applying chemotherapeutants in open-sea culture environments, the rapid, early and accurate detection of Perkinsus spp. is essential to control its transmission and manage outbreaks.
The current gold standard for detection and quantification of Perkinsus spp. in marine molluscs involves incubation of host tissues in Ray’s fluid thioglycollate medium (RFTM) [7]. Fresh molluscan tissue is added to a tube containing RFTM (for medium components and concentrations see [7]) and incubated in darkness at room temperature (20–25 °C) for 4–7 days; after incubation, the tissue is digested with 2-M NaOH, and washed several times with phosphate-buffered saline (pH 7.2); the resultant tissue is used to detect and quantify Perkinsus spp. by counting hypnospores using a haemocytometer beneath a microscope. This technique lacks specificity, is time-consuming and must be performed using fresh tissue [8,9,10]; it is also only sensitive to P. marinus at concentrations exceeding 1000 cells per gram of wet oyster tissue [10, 11]. Other diagnostic methods for detection of Perkinsus spp. include: immunoassays using monoclonal and polyclonal antibodies [12], histopathological assays [13], real-time polymerase chain reaction assay (qPCR) [14, 15], loop-mediated isothermal amplification (LAMP) assay [10] and in situ hybridization assay (ISH) [16]. Most of these assays are time consuming, and must be performed in laboratories, requiring costly infrastructure, reliable electrical supplies and skilled staff. Of these, LAMP, an isothermal amplification method that uses enzymes instead of thermal cycling for DNA synthesis, has been recently applied in detection of Perkinsus spp. infection in molluscs. This assay must be performed at temperatures exceeding room temperature (60–65 °C), requires complicated primers and completion of the detection process takes at least 49 minutes [10].
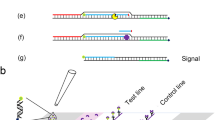
Recombinase polymerase amplification (RPA) is a highly sensitive and selective isothermal amplification technique that can be performed between 37 and 42 °C, requires minimal sample preparation and is capable of amplifying as few as 1–10 target DNA copies within 20 minutes [17, 18]. RPA can be detected by agarose gel electrophoresis (AGE), or in real-time using TwistAmp TM exo probes (TwistDx, Cambridge, UK) [19, 20]. Alternatively, oligochromatographic lateral flow (LF) strips can be used, the recognition of which is achieved by binding to a tag-specific antibody on the surface at the detection line and cross-linking it with a second tag-specific antibody on colloid gold nanoparticles, resulting in a coloured signal that can be semiquantitatively analysed and visualised by the naked eye [21]. As the LF strip assay takes 5–10 minutes to complete, requires no special equipment and can be visualised by the naked eye, combining it with RPA would create a sensitive, specific and quick system to identify the existence of Perkinsus in the field [22].
Given the advantages of RPA, this assay has been used to diagnose various pathogens, such as viruses [23,24,25], bacteria [19, 26, 27] and parasites [21, 22, 28]. However, most RPA assays have focused on detection of pathogens related to humans or domestic animals, and not aquatic animals [20, 29]. We are unaware of any published reports using this assay in which Perkinsus has been detected in molluscs. Accordingly, we developed a combined isothermal RPA and lateral flow strip detection assay (LF-RPA) for the detection of P. beihaiensis in the oyster C. hongkongensi. We evaluated the sensitivity of this LF-RPA assay by comparing it with that of qPCR and conventional PCR methods using McNemar’s Chi-square tests. Degrees of agreement among LF-RPA, qPCR and conventional PCR test results were measured using kappa (K) values.
Methods
Sample collection and DNA extraction
A total of 150 oysters (C. hongkongensis) were collected from farms in Zhanjiang, Guangdong Province, China, where P. beihaiensis was prevalent in local molluscs (e.g. C. hongkongensis, C. ariakensis, Soletellina acuta) [15, 30]. Oyster gills were excised using forceps and fixed in 95% (v/v) ethanol. Approximately 200 mg samples of gill tissue were cut and placed in 1.5-ml Eppendorf tubes. After evaporation of ethanol, samples were subjected to DNA extraction procedures using a HiPure Mollusc mini DNA Kit (Magen, Guangzhou, China) following the manufacturer protocols. DNA quality and concentration were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA); extracted DNA was stored at − 20 °C.
Conventional PCR detection
Diagnosis of Perkinsus spp. infection in oyster samples was performed using conventional polymerase chain reaction (PCR) amplification of the internal transcriber spacer (ITS) region, with Perkinsus genus-specific primer pairs PerkITS-85 and PerkITS-750 (Table 1) [31]. PCR was performed in a 25 μl reaction comprising 12.5 μl of PCR Mix (Dongsheng Biotech, Guangzhou, China), 1 μl of each primer (10 μM), 1 μl of extracted DNA and 9.5 μl of ultrapure water. PCR results were confirmed on a 1% agarose gel stained with SYBR green. A positive control using 1000 copies of P. beihaiensis-pMD 18T recombinant plasmid DNA and a negative control (nuclease-free water) were included in each run. Positive PCR products were sequenced to determine the specific Perkinsus species. When the target sequence was 99.5–100% identical to P. beihaiensis (GenBank accession no. JN054741), we considered it positive for infection of P. beihaiensis in the sample.
A total of 60 oyster samples (29 P. beihaiensis positive and 31 Perkinsus spp. negative) were used to evaluate the diagnostic performance of the RPA assay. These samples were also detected by a real time PCR assay (qPCR).
qPCR detection
The primers Q2-F and Q2-R (Table 1) were designed for the qPCR assay, which amplified a 291 bp ITS fragment of P. beihaiensis. qPCR sensitivity was determined using 10-fold serial dilutions of P. beihaiensis-pMD18T recombinant plasmid DNA (2.6 × 107 to 2.6 × 101 copies/μl). The qPCR was carried out in a 10 μl reaction, comprising 5 μl TB Green™ Premix (TaKaRa, Kusatsu, Japan), 0.5 μl of each primer (10 μM), 1 μl of extracted DNA and 3 μl of nuclease-free water. Reactions were conducted in triplicate on an Eco™ Real-Time PCR System (Illumina, San Diego, CA, USA) using the Eco System Software with the following temperature program: 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 60 °C for 40 s. In all reactions P. beihaiensis DNA was used as a positive control, and DNase-free water as the non-template control. A sample was considered positive if the qPCR threshold cycle (CT) value of the tissue rose above zero before the 35th cycle, and negative if the CT value was zero before the 35th cycle.
RPA detection
Primer and probe design
The highly repeated ITS region of P. beihaiensis (GenBank accession no. JN054741) was selected as a target sequence. Candidate RPA primers were first sought using Primer Premier v.5.0 software (Premier Biosoft, Palo Alto, CA, USA) following TwistAmp® reaction kit (TwistDx) guidelines (the best primers are 30–35 bases in length and have 30–70% GC content). No suitable RPA primers were screened, as gel-electrophoresis results lacked bands or produced unequally sized bands for the target fragment. Of 10 pairs of candidate RPA primers, one pair of efficient RPA primers (Pits6-F and Pits6-R, amplicon size 186 bp) was subsequently screened using primers of normal length (20–25 bases) (Table 1). For adaptation to the lateral flow detection system, the 5′ end of the reverse primer (Pits6-R) was labelled with a biotin (Table 1). Of 5 candidate RPA probes, one probe was screened out and its 5′ end labelled with a carboxyfluorescein (FAM) group, a C3 spacer (SpC3) on the 3′ end, and a tetrahydrofuran (THF) residue to replace an internal base (Table 1).
RPA assays
A TwistAmp Basic kit (TwistDx) was used to screen the best combinations of RPA primers following the manufacturer’s instructions. Each 50 μl reaction contained 29.5 μl of rehydration buffer, 2.4 μl of each RPA forward and reverse primer (10 μM), 12.2 μl of dH2O and 1 μl of DNA template; 2.5 μl of magnesium acetate (280 mM) was added to initiate the reaction. A positive control using 1000 copies of P. beihaiensis-pMD 18T recombinant plasmid DNA and a negative control (nuclease-free water) were included in each run. Reaction tubes were briefly centrifuged and vortexed to mix reagents, then incubated for 25 min in a heat block at 37 °C. After incubation, reaction products were purified using VAHTSTM DNA clean beads (Nanjing Vazyme Biotech Co., Nanjing, China) and visualised on a 2.5% agarose gel. Candidate RPA primers were regarded as suitable if purified RPA products were evident, a single band was apparent, the band size was the same as that of the primer-designed target fragment, and negative and non-template controls lacked bands.
Lateral-flow strip RPA assay (LF-RPA)
A TwistAmp nfo kit (TwistDx) was used in combination with HybriDetect 1 strips (Milenia Biotec, Giessen, Germany) to detect P. beihaiensis infection in oyster samples. The reaction procedure was similar to that for the TwistAmp Basic kit. To avoid contamination, all reaction procedures were performed in separate biological safety cabinets or pipetting hoods. Each 50 μl reaction contained 29.5 μl of rehydration buffer, 2.1 μl of RPA forward primer (10 μM), 2.1 μl of RPA biotinylated reverse primer (10 μM), 0.6 μl of FAM-labelled probe (10 μM), 12.2 μl of dH2O and 1 μl of DNA template; 2.5 μl of magnesium acetate (280 mM) was added to initiate the reaction. In all reactions 1000 copies of P. beihaiensis-pMD 18T recombinant plasmid DNA was used as a positive control, and DNase-free water as the non-template control. The reaction tube was incubated for 25 min in a heat block at 37 °C. After incubation, 10 μl of reaction product was diluted in 100 μl of running buffer to test the HybriDetect 1 lateral-flow strips. The strip was placed vertically into the diluted solution, then incubated at room temperature; the final result was read at 5 min. A result was regarded positive when the control and test lines were both visible and negative when only the control line was visible.
Evaluation of specificity and sensitivity
To determine primer specificity, DNA of seven common pathogens [P. beihaiensis, P. olseni, P. chesapeaki, Vibrio harveyi, Streptococcus agalactiae, ostreid herpesvirus 1(OsHV-1) and abalone herpes-like virus (AbHV)] of aquatic animals were used as templates. Pathogens were provided by the Fishery Organism Disease Control Division, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences. Each of P. beihaiensis, P. olseni and P. chesapeaki were obtained from molluscs in China’s coastal waters during a Perkinsus infection surveillance program performed by the authors (unpublished data); V. harveyi was provided by Dr Ruixuan Wang, S. agalactiae by Dr Youlu Su, OsHV-1 and AbHV by Dr Jingzhe Jiang.
The sensitivity of RPA, LF-RPA and qPCR assays was assessed using serial dilutions of P. beihaiensis-pMD18T recombinant plasmid DNA. Recombinant plasmids (including P. beihaiensis-, P. olseni- and P. chesapeaki-pMD18T) were constructed according to the method provided by Cui et al. [15]. qPCR standard curves were obtained in which CT values of each sample were plotted against the logarithm of the DNA starting concentration. The detection limit was determined as the lowest concentration (within the linear range) that produced an amplification signal on the Eco™ Real-Time PCR System.
Data analysis
Experimental data were analysed using SPSS Statistics v.19 (IBM Corporation, Armonk, NY, USA). Specificity, sensitivity and accuracy of the LF-RPA assay performance were evaluated. McNemar’s Chi-square test was used to compare sensitivities of LF-RPA, qPCR and conventional PCR assays. Degrees of agreement among LF-RPA, qPCR and conventional PCR test results were measured using kappa (K) values, with K < 0.4 considered to be a poor agreement and K ≥ 0.75 considered to be a good agreement. Statistical significance was set at P < 0.05.
Results
Specificity
The specificity of RPA and LF-RPA primers was evaluated using DNA of three recombinant plasmids (P. beihaiensis, P. olseni and P. chesapeaki) and four potential pathogens of aquatic animals (V. harveyi, S. agalactiae, OsHV-1 and AbHV). Both RPA and LF-RPA assays discriminated DNAs of three Perkinsus species from those of other pathogens (Fig. 1). RPA and LF-RPA assays designed for P. beihaiensis also detected P. olseni and P. chesapeaki.
Specificity of RPA and LF-RPA assays. Agarose gel electrophoresis of RPA purified products (a) and lateral-flow strip (b), showing successful amplification or visual detection of Perkinsus beihaiensis, P. olseni and P. chesapeaki, but not other potential pathogen DNAs. Abbreviations: AV, abalone herpes-like virus; C, control; LF, lateral flow; M, marker DL2000; NC, negative control (distilled water); OV, ostreid herpesvirus 1; PB, P. beihaiensis; PC, P. chesapeaki; PO, P. olseni; SA, Streptococcus agalactiae; RPA, recombinase polymerase amplification; T, test line; VH, Vibrio harveyi
Sensitivity
The RPA assay was least sensitive, with a lower detection limit of 260 copies of P. beihaiensis plasmid DNA (Fig. 2a). The LF-RPA assay had a detection limit as low as 26 copies (Fig. 2b), comparable to that of the universally recognised, highly sensitive qPCR assay (Fig. 3).
Sensitivity of RPA and LF-RPA assays for detecting Perkinsus beihaiensis. a RPA products, detection limit 260 copies of plasmid DNA on a stained agarose gel (2.5%). b LF-RPA assay, detection limit 26 copies of plasmid DNA on the lateral-flow strip. Abbreviations: C, control; LF, lateral flow; NC, negative control (distilled water); RPA, recombinase polymerase amplification; T, test line
Evaluation of the LF-RPA assay
Of the 60 oysters screened for P. beihaiensis infection by conventional PCR, 29 tested positive. Of these 29 oysters, 25 were LF-RPA-positive, equating to a test sensitivity of 86.2% (Table 2). Of 31 PCR-negative samples, 24 tested LF-RPA-negative, equating to a test specificity of 77.4%. In 49 of 60 test results (81.7%), LF-RPA assay results agreed with PCR results. A McNemar Chi-square test revealed LF-RPA assay results were not significantly different from those of the PCR assay (P = 0.549) (Table 2). Although results of LF-RPA and PCR assays were positively correlated, concordance was moderate (K = 0.634, i.e. K < 0.75) (Table 2).
Of 30 qPCR-positive samples, 28 were LF-RPA-positive, with test sensitivity of 93.3%. Of 30 qPCR-negative samples, 26 tested LF-RPA-negative, with test specificity of 86.7%. Assuming qPCR assay results are correct, the LF-RPA assay produced correct test results 90% of the time (in 54 of 60 samples). A McNemar Chi-square test demonstrated LF-RPA assay results were not significantly different from those of the qPCR assay (P = 0.687) (Table 2). LF-RPA assay results were positively correlated and highly concordant with qPCR assay results (K = 0.800, i.e. K > 0.75) (Table 2).
Discussion
We developed an RPA assay that amplifies the ITS region of P. beihaiensis, which, followed by rapid detection of amplicons on a lateral flow strip, enables rapid and easy visualization of test results by the naked eye. We demonstrated this assay to be more sensitive at detecting P. beihaiensis when combined with a lateral flow strip (LF-RPA) than when agarose gel electrophoresis (AGE-RPA), as detection limits in the former are roughly an order of magnitude lower than the latter. Improved detection arises from application of a fluorescence-labelled primer and probe in the LF-RPA assay; the reverse primer and probe were labelled by biotin and FAM, respectively, resulting in double-labelled RPA products being caught on lateral flow strips with gold-labelled anti-FAM antibodies. This fluorescence-labelled LF-RPA preparation process was simpler than immunoassays using monoclonal and polyclonal antibodies, but produced similar highly sensitive and specific results.
Although the LF-RPA assay provides results comparable to those of conventional PCR and qPCR, its advantages include: (i) a faster reaction time (less than 30 minutes for LF-RPA versus several hours for PCR and qPCR); (ii) convenient operation (simplified workload in LF-RPA using freeze-dried ready-to-use reagents); (iii) less energy requirement (37 °C for LF-RPA versus more than 55 °C for PCR and qPCR); and (iv) easier visualization of results (these can be read by the naked eye and interpreted by untrained personnel). Accordingly, the LF-RPA assay is a viable option for accurate and rapid diagnosis of perkinsosis, especially in environments where specialised equipment and trained personnel are not available.
Our P. beihaiensis LF-RPA assay detects plasmid DNA of P. beihaiensis, P. olseni and P. chesapeaki, possibly because of high sequence similarities in the ITS target region selected for primer and probe designs. Although longer primers would increase amplification specificity for P. beihaiensis, we could not screen these primers of 30–35 bp length [18, 24, 28]. It may be that these longer primers would lead to secondary structures and potential primer artefacts, promoting primer-primer interactions. Our shorter primers (20–25 bp) were efficient for amplification using RPA, and products were readily detected on lateral flow strips. Fortunately, although not specific to P. beihaiensis, these primers did not cross-react with the DNAs of other potential pathogens occurring in the culture environment. The use of shorter primers is advantageous in that primer secondary structures are avoided, and the primer-design process is simplified.
The LF-RPA assay can detect P. beihaiensis at levels as low as 26 copies, making it as sensitive as the qPCR assay [20, 25, 32]. This level of sensitivity theoretically renders the LF-RPA assay capable of detecting DNA from single P. beihaiensis zoospores, facilitating early diagnosis of Perkinsus infection. The LF-RPA assay appeared to be more sensitive than the conventional PCR assay. The lower sensitivity of the conventional PCR assay lead to more false negative results compared to the LF-RPA assay when detecting samples with low P. beihaiensis DNA copy numbers. In this study, 10% of LF-RPA assay results (6/60) disagreed with those from qPCR assays. This may be attributable to low concentrations of P. beihaiensis DNA in samples, as qPCR assays demonstrated most samples that produced disagreeing diagnoses had very low P. beihaiensis DNA copy numbers (4 qPCR-negative but LF-RPA-positive samples, 35 < Ct value < 40, data not shown). Perhaps increasing template volume, thus increasing the number of P. beihaiensis DNA copies, may improve problems associated with low copy numbers of P. beihaiensis DNA in samples [20, 24]. Further studies could also explore ways to optimize the LF-RPA assay to increase its detection sensitivity at low Perkinsus DNA concentrations. It should also be noted that false positive results also may be caused by contamination using LF-RPA assay, as amplification may occur when sterile gloves, tubes and pipets are cross-contaminated with target DNAs. However, we eliminated this risk by sterilizing all experimental materials, and performing all LF-RPA reactions in separate sterilized biological safety cabinets. Further optimization of the LF-RPA assay should endeavor to minimize the risk of cross-contamination, as ensuring sterilized environments in field conditions is difficult.
Conclusions
We developed a novel LF-RPA assay for detection of Perkinsus infection in molluscs that is relatively simple to use, provides accurate results and enables rapid diagnoses. While our intention had been to establish a P. beihaiensis-specific LF-RPA assay, the one we do develop detects multiple Perkinsus taxa. The short PCR primers we develop for this LF-RPA assay simplify the primer-design process. Our novel LF-RPA assay has a fast reaction time (less than 30 minutes) and can detect the DNA of multiple Perkinsus species as low as 26 copies. Future research should focus on developing simple DNA extraction methods that do not require professional laboratories and complicated extraction procedures, to facilitate application of this LF-RPA assay in the field.
Availability of data and materials
All data generated or analyzed during this study are included in the article.
Abbreviations
- AbHV:
-
abalone herpes-like virus
- AGE:
-
agarose gel electrophoresis
- CT:
-
threshold cycle
- FAM:
-
carboxyfluorescein
- FN:
-
false negative
- FP:
-
false positive
- ISH:
-
in situ hybridization assay
- ITS:
-
internal transcriber spacer
- LAMP:
-
loop-mediated isothermal amplification
- LF:
-
lateral flow
- OIE:
-
World Organization for Animal Health
- OsHV-1:
-
ostreid herpesvirus 1
- PCR:
-
polymerase chain reaction
- qPCR:
-
quantitative polymerase chain reaction
- RFTM:
-
Ray’s fluid thioglycollate medium
- RPA:
-
recombinase polymerase amplification
- THF:
-
tetrahydrofuran
- TN:
-
true negative
- TP:
-
true positive
References
Ray SM. Historical perspective on Perkinsus marinus disease of oysters in the Gulf of Mexico. J Shellfish Res. 1996;15:9–11.
Cho KS, Park KI. Review on the protozoan parasite Perkinsus olseni (Lester & Davis 1981) infection in Asian waters. In: Ishimatsu EA, Lie HA, editors. Coastal environmental and ecosystem issues of the east China Sea. Tokyo: Terra Scientific Publishing Company; 2010. p. 269–81.
Holt RD, Dobson AP, Begon M, Bowers RG, Schauber EM. Parasite establishment in host communities. Ecol Lett. 2003;6:837–42.
Taraschewski H. Hosts and parasites as aliens. J Helminthol. 2006;80:99–128.
Pagenkopp Lohan KM, Hill-Spanik KM, Torchin ME, Aguirre-Macedo L, Fleischer RC, Ruiz GM. Richness and distribution of tropical oyster parasites in two oceans. Parasitology. 2016;143:1119–32.
Villalba A, Reece KS, Ordas MC, Casas MC, Figueras A. Perkinsosis in molluscs: a review. Aquat Living Resour. 2004;17:411–32.
Dungan CF, Bushek D. Development and applications of Rayʼs fluid thioglycollate media for detection and manipulation of Perkinsus spp. pathogens of marine molluscs. J Invertebr Pathol. 2015;131:68–82.
Robledo JA, Gauthier JD, Coss CA, Wright AC, Vasta GR. Species-specificity and sensitivity of a PCR-based assay for Perkinsus marinus in the eastern oyster, Crassostrea virginica: a comparison with the fluid thioglycollate assay. J Parasitol. 1998;84:1237–44.
Ulrich PN, Ewart JW, Marsh AG. Prevalence of Perkinsus marinus (dermo), Haplosporidium nelsoni (MSX), and QPX in bivalves of Delawareʼs inland bays and quantitative, high-throughput diagnosis of dermo by qPCR. J Eukaryot Microbiol. 2007;54:520–6.
Feng CY, Wang CX, Lin XM, Zhang YN, Lvi JZ, Deng JH, et al. Development of a loop-mediated isothermal amplification method for detection of Perkinsus spp. in mollusks. Dis Aquat Org. 2013;104:141–8.
Bushek D, Ford SE, Allen SK Jr. Evaluation of methods using Rayʼs fluid thioglycollate medium for diagnosis of Perkinsus marinus infection in the eastern oyster, Crassostrea virginica. Annu Rev Fish Dis. 1994;4:201–17.
Bushek D, Dungan CF, Lewitus AJ. Serological affinities of the oyster pathogen Perkinsus marinus (Apicomplexa) with some dinoflagellates (Dinophyceae). J Eukaryot Microbiol. 2002;49:11–6.
Balseiro P, Montes J, Conchas RF, Novoa B, Figueras A. Comparison of diagnostic techniques to detect the clam pathogen Perkinsus olseni. Dis Aquat Organ. 2010;90:143–51.
Gauthier JD, Miller CR, Wilbur AE. Taqman® mcb real-time PCR approach to quantification of Perkinsus marinus and Perkinsus spp. in oysters. J Shellfish Res. 2016;25:619–24.
Cui YY, Ye LT, Wu L, Wang JY. Seasonal occurrence of Perkinsus spp. and tissue distribution of P olseni in clam (Soletellina acuta) from coastal waters of Wuchuan county, southern China. Aquaculture. 2018;492:300–5.
Moss JA, Burreson EM, Reece KS. Advanced Perkinsus marinus infections in Crassostrea ariakensis maintained under laboratory conditions. J Shellfish Res. 2006;25:65–73.
Piepenburg O, Williams CH, Stemple DL. DNA detection using recombination proteins. PLoS Biol. 2006;4:1115–21.
Lobato IM, O’Sullivan CK. Recombinase polymerase amplification: basics, applications and recent advances. TrAC-Trend Anal Chem. 2018;98:19–35.
Euler M, Wang Y, Otto P, Tomaso H, Escudero R, Anda P, et al. Recombinase polymerase amplification assay for rapid detection of Francisella tularensis. J Clin Microbiol. 2012;50:2234–8.
Jaroenram W, Owens L. Recombinase polymerase amplification combined with a lateral flow dipstick for discriminating between infectious Penaeus stylirostris densovirus and virus-related sequences in shrimp genome. J Virol Methods. 2014;208:144–51.
Rosser A, Rollinson D, Forrest M, Webster BL. Isothermal recombinase polymerase amplification (RPA) of Schistosoma haematobium DNA and oligochromatographic lateral flow detection. Parasit Vectors. 2015;8:446.
Sun K, Xing W, Yu X, Fu W, Wang Y, Zou M, et al. Recombinase polymerase amplification combined with a lateral flow dipstick for rapid and visual detection of Schistosoma japonicum. Parasit Vectors. 2016;9:476.
Boyle DS, Lehman DA, Lillis L, Peterson D, Singhal M, Armes N, et al. Rapid detection of HIV-1 proviral DNA for early infant diagnosis using recombinase polymerase amplification. MBio. 2013;4:e00135.
Teoh BT, Sam SS, Tan KK, Danlami MB, Shu MH, Johari J, et al. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J Clin Microbiol. 2015;53:830–7.
Tu PA, Shiu JS, Lee SH, Pang VF, Wang DC, Wang PH. Development of a recombinase polymerase amplification lateral flow dipstick (RPA-LFD) for the field diagnosis of caprine arthritis-encephalitis virus (CAEV) infection. J Virol Methods. 2017;243:98–104.
Clancy E, Higgins O, Forrest MS, Boo TW, Cormican M, Barry T, et al. Development of a rapid recombinase polymerase amplification assay for the detection of Streptococcus pneumoniae in whole blood. BMC Infect Dis. 2015;15:481.
Kim JY, Lee JL. Development of a multiplex real-time recombinase polymerase amplification (RPA) assay for rapid quantitative detection of Campylobacter coli and jejuni from eggs and chicken products. Food Control. 2017;73:1247–55.
Cabada MM, Malaga JL, Castellanos-Gonzalez A, Bagwell KA, Naeger PA, Rogers HK, et al. Recombinase polymerase amplification compared to real-time polymerase chain reaction test for the detection of Fasciola hepatica in human stool. Am J Trop Med Hyg. 2017;96:341–6.
Gao F, Jiang JZ, Wang JY, Wei HY. Real-time isothermal detection of abalone herpes-like virus and red-spotted grouper nervous necrosis virus using recombinase polymerase amplification. J Virol Methods. 2018;251:92–8.
Moss JA, Xiao J, Dungan CF, Reece KS. Description of Perkinsus beihaiensis n. sp., a new Perkinsus sp parasite in oysters of southern China. J Eukaryot Microbiol. 2008;55:117–30.
Casas SM, Villalba A, Reece KS. Study of perkinsosis in the carpet shell clam Tapes decussatus in Galicia (NW Spain). I. Identification of the aetiological agent and in vitro modulation of zoosporulation by temperature and salinity. Dis Aquat Org. 2002;50:51–65.
Cordray MS, Richards-Kortum RR. A paper and plastic device for the combined isothermal amplification and lateral flow detection of Plasmodium DNA. Malar J. 2015;14:472.
Acknowledgements
We thank Steve O’Shea from Edanz Group (http://www.edanzediting.com/ac), for editing an earlier draft of this manuscript. We would like to thank Drs Ruixuan Wang, Youlu Su and Jingzhe Jiang of the Fishery Organism Disease Control Division, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, for provision of molluscan pathogens used in this study. Special thanks are extended to two anonymous reviewers, who gave us many constructive suggestions and critical comments on the manuscript.
Funding
This study was supported by the Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (No. 2017YB08, 2017YB22), the Pearl River S&T Nova Program of Guangzhou (No. 201710010166), the Natural Science Foundation of Guangdong Province (No. 2018A030313349) and the earmarked fund for Modern Agro-industry Technology Research System (No. CARS-49).
Author information
Authors and Affiliations
Contributions
LY conceived the idea. LW conducted the experimental work as part of his Masters research project. ZW and YC collected oysters and assisted with data analysis. LY, LW and JW wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wu, L., Ye, L., Wang, Z. et al. Utilization of recombinase polymerase amplification combined with a lateral flow strip for detection of Perkinsus beihaiensis in the oyster Crassostrea hongkongensis. Parasites Vectors 12, 360 (2019). https://doi.org/10.1186/s13071-019-3624-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3624-3