Abstract
Background
Anaplasma spp. are tick-borne Gram-negative obligate intracellular bacteria that infect humans and a wide range of animals. Anaplasma capra has emerged as a human pathogen; however, little is known about the occurrence and genetic identity of this agent in wildlife. The present study aimed to determine the infection rate and genetic profile of this pathogen in wild animals in the Republic of Korea.
Methods
A total of 253 blood samples [198 from Korean water deer (Hydropotes inermis argyropus), 53 from raccoon dogs (Nyctereutes procyonoides) and one sample each from a leopard cat (Prionailurus bengalensis) and a roe deer (Capreolus pygargus)] were collected at Chungbuk Wildlife Center during the period 2015–2018. Genomic DNA was extracted from the samples and screened for presence of Anaplasma species by PCR/sequence analysis of 429 bp of the 16S rRNA gene marker. Anaplasma capra-positive isolates were genetically profiled by amplification of a longer fragment of 16S rRNA (rrs) as well as partial sequences of citrate synthase (gltA), heat-shock protein (groEL), major surface protein 2 (msp2) and major surface protein 4 (msp4). Generated sequences of each gene marker were aligned with homologous sequences in the database and phylogenetically analyzed.
Results
Anaplasma capra was detected in blood samples derived from Korean water deer, whereas samples from other animal species were negative. The overall infection rate in tested samples was 13.8% (35/253) and in the water deer the rate was 17.8% (35/198), distributed along the study period from 2015 to 2018. Genetic profiling and a phylogenetic analysis based on analyzed gene markers revealed the occurrence of two distinct strains, clustered in a single clade with counterpart sequences of A. capra in the database.
Conclusions
Anaplasma capra infection were detected in Korean water deer in the Republic of Korea, providing insight into the role of wildlife as a potential reservoir for animal and human anaplasmosis. However, further work is needed in order to evaluate the role of Korean water deer as a host/reservoir host of A. capra.
Similar content being viewed by others
Background
The cosmopolitan genus Anaplasma includes six species of Gram-negative obligate intracellular bacteria that are transmitted by ticks to a wide range of animals, including humans [1,2,3,4,5], resulting in considerable economic losses in the livestock industry and serious public health concerns [6, 7]. Anaplasma phagocytophilum, A. ovis and recently reported A. capra are human pathogens [8,9,10,11,12], whereas other species in the genus have no known zoonotic potential. However, A. platys may have zoonotic potential after frequent reports of human infection [13, 14].
The provisional name Anaplasma capra was assigned after its initial characterization in goats (Capra aegagrus hircus) in China [12]. Later, it was isolated from sheep, goats and cattle in different geographical regions [15,16,17,18,19] as well as from various tick species (Haemaphysalis qinghaiensis, H. longicornis, Ixodes persulcatus) [12, 20,21,22,23]. Infection of A. capra was also reported in six wild animals in China including three takins (Budorcas taxicolor), two Reeves’s muntjacs (Muntiacus reevesi) and one forest musk deer (Moschus berezovskii) [24].
Anaplasma species usually parasitize bone marrow-derived elements, including neutrophils (A. phagocytophilum), erythrocytes (A. marginale, A. centrale and A. ovis), monocytes (A. bovis) and platelets (A. platys) [7, 9, 10, 12]. However, A. capra seems to infect endothelial cells, rendering its microscopic detection in blood smears unreliable [12, 15]. In humans, the disease caused by A. capra is generally characterized by undifferentiated fever, headache, malaise, dizziness, myalgia and chills, with potential progression to CNS involvement and cerebrospinal fluid pleocytosis [12].
Although different Anaplasma species have been detected in wildlife [23,24,25,26,27,28,29], little is known about the prevalence and genetic identity of A. capra in these animals in Korea. Using molecular tools, the present study aimed at investigating the occurrence and characterizing the genetic profile of this pathogen in wildlife in the Republic of Korea.
Methods
Collection of samples
Chungbuk Wildlife Center is located in Cheongju city, Chungcheongbuk-do province in the Republic of Korea (36°38′13.99ʺN, 127°29′22.99ʺE). The center receives terrestrial and avian wild animals for purposes of treatment from sickness/injuries and/or rehabilitation. Blood samples are collected for diagnosis and treatment of wildlife referred to the Chungbuk Wildlife Center. Blood samples are archived in EDTA-treated tubes and stored at – 80 °C. A total of 253 blood samples including 198 from Korean water deer (Hydropotes inermis argyropus), 53 from raccoon dogs (Nyctereutes procyonoides) and one sample each from a leopard cat (Prionailurus bengalensis) and a roe deer (Capreolus pygargus), collected from January 2015 to June 2018, were used.
DNA extraction and PCR amplification
Frozen blood samples were thawed at room temperature and genomic DNA was extracted from 200 µl of blood using a Magpurix® Blood Kit and Magpurix® 12s automated nucleic acid purification system (Zinexts Life Science Corp., Taipei, Taiwan), according to the manufacturer’s recommendations. DNA preparations were tested for the presence of Anaplasma species by PCR/sequence analysis of 429 bp of the 16S rRNA gene as described previously [30]. Anaplasma capra-positive isolates were genetically profiled by the amplification of a longer fragment of 16S rRNA (rrs) gene as well as partial sequences of citrate synthase (gltA), heat-shock protein (groEL), major surface protein 2 (msp2) and major surface protein 4 (msp4) genes as described previously (Table 1). Amplified fragments were electrophoresed on 1.2% gel loaded with EcoDye™ stain (BIOFACT, Daejeon, Korea) and visualized using UV light.
DNA sequence analysis
The PCR products (rrs and groEL) or secondary PCR product (for other gene markers) were purified and sequenced, either directly or after cloning in the pGEM-T vector (Promega, Madison, WI, USA), in both directions. Generated sequences were assembled using ChromasPro v.2.1.8 (https://technelysium.com.au/wp/chromaspro/).
Phylogenetic analysis
The obtained sequences from each genetic locus were aligned with each other and reference sequences, available in GenBank (https://www.ncbi.nlm.nih.gov/), using ClustalX (http://www.clustal.org/) to determine the identity of Anaplasma spp. Evolutionary relationships were inferred based on partial sequences of 16S rRNA, citrate synthase (gltA), heat-shock protein (groEL), major surface protein 2 (msp2) and major surface protein 4 (msp4) genes using the maximum likelihood (ML) method implemented in MEGA7 (http://www.megasoftware.net/). The ML phylogenetic analysis was conducted using the Kimura 2-parameter model and 1000 bootstrap replicates. The ML tree was rooted against the nucleotide sequences L36221 (Rickettsia typhi), KY124257 (Rickettsia parkeri), U96733 (Rickettsia rickettsii) and BDDN01000175 (Ehrlichia ruminantium) for 16S rRNA, gltA, groEL and msp4 gene markers, respectively.
Results
The overall infection rate of A. capra in tested animals was 13.8% (35/253); however, samples from raccoon dogs (n = 53), leopard cat (n = 1) and roe deer (n = 1) were negative. The infection rate in KWD was 17.7% (35/198), distributed as follows: 24.6% (14/57) in 2015; 13.2% (5/38) in 2016; 17.3% (14/81) in 2017; and 9.1% (2/22) in 2018 (Table 2).
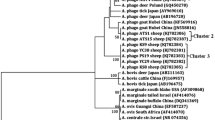
Molecular and phylogenetic analyses indicated to occurrence of two genetically distinct strains [named Cheongju (23 isolates) and Chungbuk (12 isolates)] of this pathogen. Sequences obtained from both strains were similar to those derived from A. capra from goats, sheep, cattle, ticks and humans; however, they had striking genetic differences, suggesting that they are novel strains. Sequences of the rrs gene fragment of both strains showed an identity of ~ 99.5% with counterparts in database and clustered in the clade of A. capra from different hosts (Fig. 1). Both strains had single nucleotide polymorphisms (SNPs), resulting in four genotypes at this gene locus. Phylogenetic analysis revealed that three sequences designated A. centrale (GenBank: AB211164, AF283007 and GU064903) and two sequences designated Anaplasma spp. (GenBank: AB454075 and AB509223) clustered within the A. capra clade, even though other A. centrale sequences from different hosts and geographical regions formed separate clusters in the ML tree.
Maximum-likelihood phylogenetic trees of Anaplasma species based on partial sequences of 16S rRNA gene. The tree was constructed using MEGA7 with the Kimura 2-parameter model. The newly generated sequences are indicated by diamonds. The numbers at nodes represent bootstrap values. The scale-bar represents the number of nucleotide substitutions per site
The gltA gene of the Cheongju strain shared a similarity of 99.5% (with two substitutions, A/G at position 456 and T/C at position 533) with gltA sequences KM206274, KJ700628 and MH029895 isolated from a human, goat and tick, respectively [12, 23]. Sequences of the Chungbuk strain showed a similarity of 98–99% with KX685885, KX685886 and MF071308 of A. capra from ticks and sheep [13, 19]. Both strains clustered with their homologous sequences in the A. capra clade (Fig. 2). groEL gene sequences derived from the Cheongju strain shared a similarity of 99% (one substitution) with their counterparts from humans (GenBank: KM206275), goats (GenBank: KJ700629), sheep (GenBank: KX417356) and ticks (GenBank: KR261633 and KR261635), whereas sequences from the Chungbuk strain shared a similarity of 91% with the reference sequences (Fig. 3). The msp2 sequences showed extensive intra- and inter-sequence variations, including multiple InDels and single nucleotide substitutions; however, all sequences remained clustered in the A. capra clade (Fig. 4). A hypervariable stretch was detected between positions 285 and 414 of the generated sequences (corresponding to positions 550 and 679 in the reference sequence KM206276 of A. capra from humans). The msp4 sequences were identical in the two strains and showed an identity of 100% with those from humans (GenBank: KM206277) and ticks (GenBank: KR261637 and KR261640) (Fig. 5).
Maximum-likelihood phylogenetic trees of Anaplasma species based on partial sequences of gltA gene. The tree was constructed using MEGA7 with the Kimura 2-parameter model. The newly generated sequences are indicated by diamonds. The numbers at nodes represent bootstrap values. The scale-bar represents the number of nucleotide substitutions per site
Maximum-likelihood phylogenetic trees of Anaplasma species based on partial sequences of groEL gene. The tree was constructed using MEGA7 with the Kimura 2-parameter model. The newly generated sequences are indicated by diamonds. The numbers at nodes represent bootstrap values. The scale-bar represents the number of nucleotide substitutions per site
Maximum-likelihood phylogenetic trees of Anaplasma species based on partial sequences of msp2 gene. The tree was constructed using MEGA7 with the Kimura 2-parameter model. The newly generated sequences are indicated by diamonds. The numbers at nodes represent bootstrap values. The scale-bar represents the number of nucleotide substitutions per site
Maximum-likelihood phylogenetic trees of Anaplasma species based on partial sequences of msp4 gene. The tree was constructed using MEGA7 with the Kimura 2-parameter model. The newly generated sequences are indicated by diamonds. The numbers at nodes represent bootstrap values. The scale-bar represents the number of nucleotide substitutions per site
Discussion
Wild animals act as reservoirs for a wide range of pathogens [31,32,33]. The emergence of infectious disease agents of wildlife origin is a prominent challenge to public health and the livestock industry [34,35,36]. Anaplasma capra has recently been isolated from human patients in China with non-specific clinical manifestations, with potential progression to CNS complications, suggesting that this species could pose a substantial threat to public health [12, 37]. We detected A. capra DNA in blood samples of 35 out of 198 KWD (17.7% infection rate) at the Chungbuk Wildlife Center, Korea. Epidemiological data for this pathogen in wildlife are lacking in Korea; however, our findings are similar to those obtained from wildlife (five takins, three Himalayan gorals, three Reeves’s muntjacs, one forest musk deer and one wild boar) in China [24]. In addition, a low percentage of infection rate was reported cattle, sheep and goats in China, Sweden and Korea [15,16,17,18,19], indicating that A. capra has a broad host range. Occurrence of infection during the study period from 2015 to 2018 indicates the persistence of the infection in KWD, suggesting that the species may act as a reservoir for this pathogen. However, it is difficult to explain the negative results from raccoon dogs in the present study. This may be attributed to persistent infection making the pathogen below detectable level in the blood of these animals. In support of this view, A. capra has been reported to infect endothelial cells [12, 15], making its detection in the blood possible in case of considerable bacteremia or released endothelial cells, resembling Rickettsia species [12]. Furthermore, the sample size and species and/or the age of animals may play a role in these findings. Further investigations are needed to clarify these points.
Our genetic profiling results indicated that the newly generated 16S rRNA gene sequences shared a homology of > 99.5% with sequences of A. capra strains from humans, sheep, goats, cattle and ticks [12, 15, 16, 18, 20, 22], suggesting that they likely are within the same species of bacteria [38, 39]. Clustering of sequences named A. centrale from deer (Cervus nippon nippon) (GenBank: AB211164) and cattle (GenBank: AF283007) in Japan and from ticks (Haemaphysalis longicornis) in Korea (GenBank: GU064903), as well as Anaplasma spp. from deer (GenBank: AB454075; direct submission) and Japanese serow (Capricornis crispus) (GenBank: AB509223) in the same clade of A. capra indicate a close phylogenetic relationship. The clustering pattern of these sequences in the A. capra clade does not support their assignment as sister taxa and suggests that these isolates are in fact A. capra [15, 25] and may need re-description, since these sequences were more related to Chungbuk strain in the A. capra clade.
The results obtained using different gene markers showed considerable sequence variation, suggesting that A. capra has a high degree of genetic diversity. Notably, extensive genetic variation was detected in msp2. Consistent with our results, sequence variation at the studied gene markers has been reported previously among isolates from ticks, sheep and goats [12, 15,16,17,18, 20,21,22,23,24,25]. Similarly, genetic variation is common in other Anaplasma species [40,41,42,43,44,45]. Although genetic diversity is reportedly related to infectivity, virulence, pathogenicity, niche preference, immune evasion, and/or host adaptability [46,47,48,49], this has not been established in A. capra and further studies are needed to evaluate these relationships.
Due to the extinction of natural predators, the KWD is thriving in Korea and has been designated as “harmful wildlife” by the Ministry of Environment in 1994 owing to harmful interactions with humans and their properties. This close interaction poses substantial threats to domestic animals and human health in Korea. This study was limited by analyzing samples from one geographical area and few animal species, which my lead to biases in the results. A large-scale study is underway to fully elucidate the host range of wildlife, vector ticks, pathogenicity and geographical distribution of this organism in Korea.
Conclusions
To our knowledge, the results presented herein provide the first evidence for the presence of A. capra in Korean water deer in Korea. As an emerging human pathogen, the detection of A. capra in deer provides insight into the role of wildlife as a potential reservoir for human anaplasmosis. Furthermore, the obtained results expand the known geographical and host range of the Anaplasma capra.
Availability of data and materials
Data supporting the conclusions of this article are provided within the article. The nucleotide sequences generated in this study are available in the GenBank database under the accession numbers LC432092–LC432126 (rrs), LC432127–LC432161 (gltA), LC432162–LC432196 (groEL), LC432232–LC432266 (msp2) and LC432197–LC432231 (msp4).
Abbreviations
- rrs :
-
16S rRNA
- gltA :
-
citrate synthase
- groEL :
-
heat-shock protein
- msp2:
-
major surface protein 2
- msp4:
-
major surface protein 4 gene
- KWD:
-
Korean water deer (Hydropotes inermis argyropus)
- ML:
-
maximum likelihood
References
Petrovec M, Lotric Furlan S, Zupanc TA, Strle F, Brouqui P, Roux V, et al. Human disease in Europe caused by a granulocytic Ehrlichia species. J Clin Microbiol. 1997;35:1556–9.
Lotric-Furlan S, Rojko T, Petrovec M, Avsic-Zupanc T, Strle F. Epidemiological, clinical and laboratory characteristics of patients with human granulocytic anaplasmosis in Slovenia. Wien Klin Wochenschr. 2006;118:708–13.
Parkins MD, Church DL, Jiang XY, Gregson DB. Human granulocytic anaplasmosis: first reported case in Canada. Can J Infect Dis Med Microbiol. 2009;20:e100–2.
McFee RB. Tick borne illness–anaplasmosis. Dis Mon. 2018;64:181–4.
Chvostáč M, Špitalská E, Václav R, Vaculová T, Minichová L, Derdáková M. Seasonal patterns in the prevalence and diversity of tick-borne Borrelia burgdorferi sensu lato, Anaplasma phagocytophilum and Rickettsia spp. in an urban temperate forest in southwestern Slovakia. Int J Environ Res Public Health. 2018;15:994.
Dumler JS, Choi KS, Garcia-Garcia JC, Barat NS, Scorpio DG, Garyu JW, et al. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg Infect Dis. 2005;11:1828–34.
Araya-Anchetta A, Busch JD, Scoles GA, Wagner DM. Thirty years of tick population genetics: a comprehensive review. Infect Genet Evol. 2015;29:164–79.
Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–95.
Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophilum. Int J Syst Evol Microbiol. 2001;51:2145–65.
Ismail N, McBride JW. Tick-borne emerging infections: ehrlichiosis and anaplasmosis. Clin Lab Med. 2017;37:317–40.
Kim SW, Kim CM, Kim DM, Yun NR. Manifestation of anaplasmosis as cerebral infarction: a case report. BMC Infect Dis. 2018. https://doi.org/10.1186/s12879-018-3321-4.
Li H, Zheng YC, Ma L, Jia N, Jiang BG, Jiang RR, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15:663–70.
Arraga-Alvarado CM, Qurollo BA, Parra OC, Berrueta MA, Hegarty BC, Breitschwerdt EB. Case report: molecular evidence of Anaplasma platys infection in two women from Venezuela. Am J Trop Med Hyg. 2014;91:1161–5.
Maggi RG, Mascarelli PE, Havenga LN, Naidoo V, Breitschwerdt EB. Co-infection with Anaplasma platys, Bartonella henselae and Candidatus mycoplasma haematoparvum in a veterinarian. Parasites Vectors. 2013;6:103.
Yang J, Liu Z, Niu Q, Liu J, Han R, Guan G, et al. A novel zoonotic Anaplasma species is prevalent in small ruminants: potential public health implications. Parasites Vectors. 2017;10:264.
Grandi G, Aspán A, Pihl J, Gustafsson K, Engström F, Jinnerot T, et al. Detection of tick-borne pathogens in lambs undergoing prophylactic treatment against ticks on two Swedish farms. Front Vet Sci. 2018. https://doi.org/10.3389/fvets.2018.00072.
Yang J, Han R, Niu Q, Liu Z, Guan G, Liu G. Occurrence of four Anaplasma species with veterinary and public health significance in sheep, northwestern China. Ticks Tick Borne Dis. 2018;9:82–5.
Peng Y, Wang K, Zhao S, Yan Y, Wang H, Jing J, et al. Detection and phylogenetic characterization of Anaplasma capra: an emerging pathogen in sheep and goats in China. Front Cell Infect Microbiol. 2018;8:283.
Seo MG, Ouh IO, Lee H, Geraldino PJL, Rhee MH, Kwon OD, et al. Differential identification of Anaplasma in cattle and potential of cattle to serve as reservoirs of Anaplasma capra, an emerging tick-borne zoonotic pathogen. Vet Microbiol. 2018;226:15–22.
Sun XF, Zhao L, Wen HL, Luo LM, Yu XJ. Anaplasma species in China. Lancet Infect Dis. 2015;15:1263–4.
Yang J, Liu Z, Niu Q, Liu J, Han R, Liu G, et al. Molecular survey and characterization of a novel Anaplasma species closely related to Anaplasma capra in ticks, northwestern China. Parasites Vectors. 2016;9:603.
Zhang H, Sun Y, Jiang H, Huo X. Prevalence of severe febrile and thrombocytopenic syndrome virus, Anaplasma spp. and Babesia microti in hard ticks (Acari: Ixodidae) from Jiaodong Peninsula, Shandong Province. Vector Borne Zoonotic Dis. 2017;17:134–40.
Han R, Yang JF, Mukhtar MU, Chen Z, Niu QL, Lin YQ, et al. Molecular detection of Anaplasma infections in ixodid ticks from the Qinghai-Tibet Plateau. Infect Dis Poverty. 2019;8:12.
Yang J, Liu Z, Niu Q, Mukhtar MU, Guan G, Liu G, et al. A novel genotype of “Anaplasma capra” in wildlife and its phylogenetic relationship with the human genotypes. Emerg Microbes Infect. 2018;7:210.
Qin XR, Han FJ, Luo LM, Zhao FM, Han HJ, Zhang ZT, et al. Anaplasma species detected in Haemaphysalis longicornis tick from China. Ticks Tick Borne Dis. 2018. https://doi.org/10.1016/j.ttbdis.2018.03.014.
Ebani VV, Rocchigiani G, Nardoni S, Bertelloni F, Vasta V, Papini RA, et al. Molecular detection of tick-borne pathogens in wild red foxes (Vulpes vulpes) from central Italy. Acta Trop. 2017;172:197–200.
Lee M, Seo MG, Lee SH, Ouh IO, Kim YH, Kim JK, et al. Molecular detection and phylogenetic analysis of tick-borne pathogens in wild Korean water deer and farmed elk in Gyeongbuk and Gangwon provinces of Korea. J Vet Med Sci. 2018;80:1473–8.
Kang JG, Chae JB, Cho YK, Jo YS, Shin NS, Lee H, et al. Molecular detection of Anaplasma, Bartonella, and Borrelia theileri in raccoon dogs (Nyctereutes procyonoides) in Korea. Am J Trop Med Hyg. 2018;98:1061–8.
Penzhorn BL, Netherlands EC, Cook CA, Smit NJ, Vorster I, Harrison-White RF, Oosthuizen MC. Occurrence of Hepatozoon canis (Adeleorina: Hepatozoidae) and Anaplasma spp. (Rickettsiales: Anaplasmataceae) in black-backed jackals (Canis mesomelas) in South Africa. Parasites Vectors. 2018;11:158.
Seong G, Han YJ, Oh SS, Chae JS, Yu DH, Park J, et al. Detection of tick-borne pathogens in the Korean water deer (Hydropotes inermis argyropus) from Jeonbuk Province, Korea. Korean J Parasitol. 2015;53:653–9.
Duscher GG, Leschnik M, Fuehrer HP, Joachim A. Wildlife reservoirs for vector-borne canine, feline and zoonotic infections in Austria. Int J Parasitol Parasites Wildl. 2014;4:88–96.
Plowright RK, Parrish CR, McCallum H, Hudson P, Ko A, Graham AL, et al. Pathways to zoonotic spillover. Nat Rev Microbiol. 2017;15:502–10.
Waltner-Toews D. Zoonoses, one health and complexity: wicked problems and constructive conflict. Philos Trans R Soc B. 2017;372(1725):20160171.
Daszek P, Cunningham AA, Hyatt A. Emerging infectious diseases of wildlife—threats to biodiversity and human health. Science. 2000;287:443–9.
Cunningham AA, Daszak P, Wood JL. One health, emerging infectious diseases and wildlife: two decades of progress? Philos Trans R Soc B. 2017;372:20160167.
Titcomb G, Allan B, Ainsworth T, Henson L, Hedlund T, Pringle R, et al. Interacting effects of wildlife loss and climate on ticks and tick-borne disease. Proc R Soc B. 2017;284:20170475.
Beyer AR, Carlyon JA. Of goats and men: rethinking anaplasmoses as zoonotic infections. Lancet Infect Dis. 2015;15:619–20.
Clarridge JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17:840–62.
Adékambi T, Drancourt M, Raoult D. The rpoB gene as a tool for clinical microbiologists. Trends Microbiol. 2009;17:37–45.
Wang Y, Chen C, Zhang L. Molecular characterization of Msp2/P44 of Anaplasma phagocytophilum isolated from infected patients and Haemaphysalis longicornis in Laizhou Bay, Shandong Province, China. PLoS ONE. 2013;8:e78189.
Kocan KM, de la Fuente J, Cabezas-Cruz A. The genus Anaplasma: new challenges after reclassification. Rev Sci Tech. 2015;34:577–86.
Graça T, Silva MG, Kostyukova AS, Palmer GH. Structural basis for recombinatorial permissiveness in the generation of Anaplasma marginale Msp2 antigenic variants. Infect Immun. 2016;84:2740–7.
Quiroz-Castañeda RE, Amaro-Estrada I, Rodríguez-Camarillo SD. Anaplasma marginale: diversity, virulence, and vaccine landscape through a genomics approach. Biomed Res Int. 2016;2016:9032085.
Battilani M, De Arcangeli S, Balboni A, Dondi F. Genetic diversity and molecular epidemiology of Anaplasma. Infect Genet Evol. 2017;49:195–211.
Yang J, Han R, Liu Z, Niu Q, Guan G, Liu G, et al. Insight into the genetic diversity of Anaplasma marginale in cattle from ten provinces of China. Parasites Vectors. 2017;10:565.
den Bakker HC, Moreno Switt AI, Govoni G, Cummings CA, Ranieri ML, et al. Genome sequencing reveals diversification of virulence factor content and possible host adaptation in distinct subpopulations of Salmonella enterica. BMC Genom. 2011;12:425.
Iraola G, Pérez R, Naya H, Paolicchi F, Pastor E, Valenzuela S, et al. Genomic evidence for the emergence and evolution of pathogenicity and niche preferences in the genus Campylobacter. Genome Biol Evol. 2014;6:2392–405.
Atif FA. Anaplasma marginale and Anaplasma phagocytophilum: Rickettsiales pathogens of veterinary and public health significance. Parasitol Res. 2015;114:3941–57.
Gemmell MR, Berry S, Mukhopadhya I, Hansen R, Nielsen HL, Bajaj-Elliott M, et al. Comparative genomics of Campylobacter concisus: analysis of clinical strains reveals genome diversity and pathogenic potential. Emerg Microbes Infect. 2018;7:116.
Acknowledgements
The authors of this study are greatly appreciated to staff members at Chungbuk Wildlife Center and the College of Veterinary Medicine, Chungbuk National University.
Funding
This work was supported by the Brain Pool program funded by the Ministry of Science and ICT through the National Research Foundation of Korea (2018H1D3A2002236) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (NRF-2016R1D1A1B03932312).
Author information
Authors and Affiliations
Contributions
SA and KJN conceived the study. SA, SK and YMY collected the samples and conducted the experiments. SA, YMY and KJN analyzed the data. SA and KJN prepared the report for publication. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Amer, S., Kim, S., Yun, Y. et al. Novel variants of the newly emerged Anaplasma capra from Korean water deer (Hydropotes inermis argyropus) in South Korea. Parasites Vectors 12, 365 (2019). https://doi.org/10.1186/s13071-019-3622-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3622-5