Abstract
Background
Long-lasting insecticidal nets (LLINs) are the principal tool for malaria control in Africa and are presently treated with a single class of insecticide; however, increasing levels of insecticide resistance threaten their success. In response to this threat nets have been developed that incorporate the synergist, piperonyl butoxide (PBO), which inhibits the activity of cytochrome P450s which is one main mechanisms of insecticide resistance, allowing resistance to pyrethroids to be reversed. However, data on the value and cost effectiveness of these nets is lacking. A large-scale cluster randomised trial of conventional LLINs and PBO-LLINs was conducted in Uganda in 104 health sub-districts (HSDs) in 2017–2019. Prior to the mass distribution of LLINs, a baseline entomological survey was carried out, the results of which are reported herein. Ten households from each HSD were randomly selected for entomological surveillance at baseline which included household mosquito collections.
Results
Prior to LLIN distribution entomological collections were carried out in 1029 houses across the 104 HSDs. Anopheles gambiae (s.l.) was the principal vector in all but 9 of the 71 HSDs that yielded vector species. Molecular analysis found An. gambiae (s.s.) to be the predominant vector collected. Plasmodium falciparum was detected in 5.5% of An. gambiae (s.s.) and in 4.0% of An. funestus (s.s.) examined. Infection rates of other plasmodium species (P. vivax, P. ovale and P. malariae) were lower with infection rates of 1.2% and 1.7% for An. gambiae (s.s.) and An. funestus (s.s.), respectively. The knockdown resistance (kdr) mutation Vgsc-L1014S was found at very high frequency in An. gambiae (s.s.) with the Vgsc-L1014F mutation at low frequency and the wild-type allele virtually absent. In An. arabiensis the wild-type allele was predominant. The resistance-associated alleles, Cyp4j5-L43F and Coeae1d were found at moderate frequencies which varied across the study site. Vgsc-N1575Y mutation was not found in any samples examined.
Conclusions
No significant differences between planned intervention arms was observed in vector densities, sporozoite infection rate or insecticide resistance marker frequency across the study site prior to the distribution of LLINs. Very high levels of kdr resistance were observed in all areas; however, the resistance-associated markers Cyp4j5-L43F and Coeae1d were found at varying frequencies across the study site which may have implications for the effectiveness of standard LLINs.
Trial registration This study is registered with ISRCTN, ISRCTN17516395. Registered 14 February 2017, http://www.isrctn.com/ISRCTN17516395
Similar content being viewed by others
Background
The burden of malaria in sub-Saharan Africa has reduced markedly in the last 20 years. Malaria control has been focused on scaling up of insecticide treated net coverage (ITNs), indoor residual spraying (IRS), and treatment with artemisinin-based combination therapy (ACT) which together have reduced the incidence of clinical disease by 40% [1]. Increased coverage of vector control is estimated to have been the major driver behind the decline in malaria mortality between 2000–2015 [1, 2]. Long-lasting insecticidal nets (LLINs) are currently the principal tool for malaria control in Africa and are presently treated with a single class of insecticide; however, there is growing concern that the rapid spread of resistance to pyrethroids, could render LLINs ineffective [3, 4]. Moreover, the most recent data from 2016, suggest that after years of falling annual malaria mortality, advances may have stalled in Africa. There are therefore justified concerns about the emergence and spread of insecticide resistance and the impact this may have on the continued effectiveness of insecticide-based interventions [2, 3, 5, 6]. A recent WHO co-ordinated trial across a range of transmission settings in five countries concluded that users of LLINs still benefited from reduced rates of infection and morbidity even in areas where vectors show high levels of resistance to pyrethroids [5]. This earlier study was unable to determine whether the community effect had been lost, and as modelling data suggest increasing resistance will adversely affect malaria vector control [4] there is a pressing need for improved anti-vector interventions. One promising intervention is the development of LLIN technology which incorporates the synergist, piperonyl butoxide (PBO), which is intended to overcome one of the most important insecticide resistance mechanisms circulating in malaria vector populations. PBO inhibits the activity of cytochrome P450s and thus could reverse resistance to pyrethroids caused by these enzymes. Gene expression studies have demonstrated that P450s are differentially expressed in numerous populations of the primary African malaria vectors (Anopheles gambiae, An. funestus and An. arabiensis) [7,8,9,10]. Additional studies show that these overexpressed P450s are capable of metabolising pyrethroids and conferring elevated levels of insecticide resistance when expressed transgenically [7, 9, 11].
Resistance to pyrethroids in malaria vectors is widespread and intense in Uganda [6, 12, 13], and has been implicated in the limited epidemiological impact of both IRS- [14] and LLIN-based control campaigns [15]. P450-associated resistance has been observed in four districts in the East and North of the country [13] and a DNA diagnostic has been developed for a non-synonymous change in the p450, Cyp4j5, which has been reproducibly associated with pyrethroid resistance in several sites in Uganda and western Kenya [11]. Uganda is therefore an appropriate location to test whether combination PBO-LLINs are more effective than conventional LLINs.
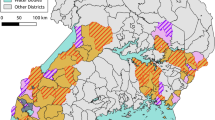
A cross-sectional community survey was conducted in Uganda in 2017–2018 in 104 health sub-districts (HSDs) (48 districts) at baseline, prior to the mass distribution of LLINs by the Ministry of Health in 2017 and 2018 (Staedke et al. unpublished data). This mass distribution of nets was undertaken as a cluster randomised trial of conventional LLINs (Permannet 2.0 and Olyset) and PBO-LLINs (Permanent 3.0 and Olyset Plus) with the four different net types constituting the four arms of the study (registered trial ISRCTN 17516395). Of the 104 HSDs included in the study, 38 were located in the eastern region and the remaining 66 in the western region (Fig. 1a). The study region encompasses approximately half the total area of Uganda and includes a wide range of epidemiological settings with varying levels of malaria transmission and insecticide resistance [15, 16]. Prior to the survey, 50 households were randomly selected in each HSD for epidemiological surveys and a subset of 10 of these were selected at random for entomological surveillance which included household mosquito collections using mechanical aspirators and a questionnaire focusing on vector control at the household level. Full details of the epidemiological survey are presented in (Staedke et al. unpublished data). In this paper we present results from the entomological surveys.
In brief, mosquitoes were identified phenotypically to genus and a subset of Anopheles mosquitoes were identified to species and screened for malaria infection using molecular methods. Insecticide resistance of the primary malaria vector, An. gambiae (s.s.), was assessed by screening mosquitoes for the genetic signatures of insecticide resistance rather than traditional phenotypic methods. Previous studies have shown that resistance measured by phenotypic methods is not predicative of LLIN failure [5] and this approach can only detect large changes in resistance levels within a population. High levels of pyrethroid resistance across Uganda have been previously observed [6, 12, 13, 17], meaning that phenotypic screening may not be sensitive enough to detect small differences in resistance level temporally and geographically. Moreover, phenotypic assays are logistically difficult to perform on a large scale, and maybe unreliable if testing conditions are not rigorously controlled across all sites. Molecular screening of resistance-associated markers allows small changes in resistance to be observed and permits detailed geographical mapping of resistance across all 104 health sub-districts. Previous work in Uganda and Kenya has shown that several mutations in the P450, Cyp4j5, and the carboxylesterase gene, Coeae1d, are reliably associated with pyrethroid resistance and can be used as markers of resistance in this location [11]. These and other insecticide resistance-associated variants were genotyped in the primary malaria vector, An. gambiae (s.s.). Together these data (i) provide a comprehensive picture of malaria vector and insecticide resistance distribution across a broad swathe of Uganda; and (ii) enable us to determine if there were significant differences in vector distribution and insecticide resistance between planned intervention arms before the mass distribution of LLINs.
Methods
Household selection
A detailed sampling schema is given in (Staedke et al., unpublished data). In brief, a sample of 50 households from each of the 104 clusters was randomly selected using a two-stage cluster sampling procedure (Staedke et al., unpublished data). The randomization process was stratified by region, with 66 clusters in the western region, and 38 clusters in the eastern region (Fig. 1a), because whilst insecticide resistance is relatively well characterised in eastern Uganda comparatively little is known about patterns in the west. Enumeration areas (EAs) identified in the 2014 national census by the Uganda Bureau of Statistics formed the primary sampling unit [18]. Ten EAs were randomly selected in each of the 104 HSDs using probability proportionate to size (PPS) sampling. Households within each selected enumeration area were mapped and enumerated by the study team and a randomized list of households was produced in each EA. Households were approached for inclusion into the study by the study team until five households that had at least one child aged between 2–10 years present had been enrolled [19]. From the five households enrolled into the community survey in each EA, one household was randomly selected for enrolment into the entomological survey. Therefore, up to ten households were surveyed from each of the 104 HSDs included in the study, giving a maximum of 1040 households for entomological surveillance. Mosquito collections were made in the early morning and a questionnaire completed. Consent was obtained the previous day. Entomological data were collected on paper forms and later transferred to a Microsoft Access database. Internal consistency checks were carried out.
Collections
Mosquitoes were collected from houses using mechanical (‘Prokopack’) aspirators [20]. Household collections were carried out by one person per house for a duration of ten minutes before 10:00 h. All mosquitoes collected were counted and morphologically identified to genus. Anopheles mosquitoes were identified to species group [21] and blood-fed status recorded. Anopheles mosquitoes were stored individually in pierced 0.2 ml tubes and sealed in ziplock bags containing silica gel. Up to 50 Anopheles gambiae (s.l.) and 50 An. funestus (s.l.) mosquitoes per HSD were selected for molecular analysis. DNA extractions were carried out on the head and thorax using Nexttec Biotechnologie DNA extraction plates (Nexttec Biotechnologie GmbH, Hilgertshausen, Germany) according to the manufacturer’s instructions.
Molecular analysis
Mosquitoes identified morphologically as An. gambiae (s.l.) were identified to species level by SINE PCR [22]. Mosquitoes identified phenotypically as An. funestus (s.l.) were identified to species by PCR of the ITS2 region on the rDNA [23]. Presence of the 2La chromosome inversion in An. gambiae (s.l.) specimens was assessed by PCR [24]. Sporozoite rates in An. gambiae (s.l.) and An. funestus (s.l.) were calculated using the Taqman assay for detection of Plasmodium falciparum, P. vivax, P. ovale and P. malariae [25]. Anopheles gambiae (s.l.), the dominant vector group, were screened for a number of mutations known to be associated with pyrethroid resistance. The Vgsc-L1014F and Vgsc-L1014S mutations were assayed using the TaqMan assay [26] or LNA-kdr assay [27]. Screening for the Vgsc-N1575Y, Cyp4J5-L43F and Coeae1d mutations used TaqMan assays following standard protocols [11, 28]. All Taqman and LNA assays were analysed using AriaMX V1.5 or MXPro software and the ΔR threshold adjusted manually for each dye, if necessary. Results for “ΔR last”, the final baseline-corrected fluorescence reading as measured in the last cycle, were then exported into Microsoft Excel for analysis and the genotype at each locus determined.
Data from household mosquito collections and questionnaires was compiled in Microsoft Access. Molecular data was compiled in Microsoft Excel. All data analyses were carried out using R statistical software version 3.3.1. Analysis of mosquito density used Generalized Linear Mixed Effect Models (GLMMs) using a Type 2 Negative Binomial distribution with model fit determined by Akaike information criterion (AIC) to account for repeated measures (up to ten households per HSD). Analysis of frequency data (sporozoite infections and molecular markers) used General Linear Models with logit link function for a binomial dependent variable.
Results
Entomological collections were carried out from 1029 houses. A total of 4703 female anopheline mosquitoes were collected of which 4009 were identified morphologically as An. gambiae (s.l.) and 694 as An. funestus (s.l.) (Fig. 1a, b). Female individuals were collected in 71 of 104 HSDs and in all but 9 of the 71 HSDs An. gambiae (s.l.) was predominant (Fig. 1c). The density of female An. gambiae (s.l.) (Fig. 1d) and An. funestus (s.l.) (Additional file 1: Figure S1) was analysed as a function of planned net distribution arm with HSD as random effect using GLMM based on a Type 2 Negative Binomial Model. The best-fit model for both species, was determined by AIC, a method which allows the relative fit of different models to be estimated for a dataset, with lower values indicating a better relative fit than higher values. The best-fit model was found to include region (East vs West) but did not include planned intervention arm showing that there was no significant difference in the densities of either An. gambiae (s.l.) or An. funestus (s.l.) by planned intervention arm (Table 1).
Heads and thoraces from 1942 Anopheles mosquitoes were separated from abdomens, and DNA extracted for molecular analysis. Of these, 1368 were An. gambiae (s.s.), 88 An. arabiensis, 470 An. funestus (s.s.) and 1 An. parensis. Morphological identification was accurate for 86% of mosquitoes identified phenotypically as An. funestus (s.l.) and 94% of mosquitoes identified as An. gambiae (s.l.)
Molecular identification was not possible for 15 specimens using standard PCR for An. gambiae and An. funestus species groups. Successful amplification and sequencing of the mitochondrial cytochrome c oxidase subunit 1 (mtDNA cox1) gene (using primers TL2-N-3014 and CI-J-2183) [29] was achieved for 11 out of these 15 specimens. A BLAST search against the NCBI database [30] was carried out to identify similarity to known anopheline sequences. The observed percentage identity ranged between 92–99%, with a query coverage of 96–100%. The sequences of the top matches were collated, and redundant sequences removed, before alignment using multiple sequence alignment by MUSCLE [31] and a phylogenetic tree constructed using PHYML algorithm [32] (Additional file 2: Figure S2). Three samples were clustered in the An. gambiae (s.l.) group, and three within the An. funestus (s.l.) group. Two specimens, one of which was positive for P. falciparum, clustered with An. pharoensis (percentage identity 93%); one sample was most likely An. rufipes (percentage identity of 95%), whilst two specimens were of uncertain status clustering with Anopheles minimus and Anopheles culicifacies, both vectors not found in sub-Saharan Africa.
Sporozoite infection rates varied across the study site (Fig. 2a, b, Additional file 3: Figure S3). Plasmodium falciparum was detected in 5.5% of An. gambiae (s.s.) and in 4.0% of An. funestus (s.s.) examined. No Plasmodium spp. were observed in the limited sample of An. arabiensis. Infection rates of other Plasmodium species (P. vivax, P. ovale and P. malariae) were lower with infection rates of 1.2% and 1.7% for An. gambiae (s.s.) and An. funestus (s.s.), respectively. Plasmodium infection rates in An. gambiae (s.s.) (Fig. 2c, d) and An. funestus (s.s.) (Fig. 2e, f) were analysed as a function of net distribution arm with HSD as random effect using a logistic GLMM Binomial Model. The best-fit model, as determined by AIC, did not include intervention arm showing that there was no significant difference in infection rates by planned intervention arm.
Sporozoite infection rates. a P. falciparum sporozoite infection rate in Anopheles. b Sporozoite infection rate for combined P. vivax, P. ovale and P. malariae in Anopheles. c P. falciparum sporozoite infection rate in An. gambiae (s.s.) by net distribution arm. d Sporozoite infection rate for the combined P. vivax, P. ovale and P. malariae (P.OVM) in An. gambiae (s.s.) by net distribution arm. e P. falciparum sporozoite infection rate in An. funestus (s.s.) by net distribution arm. f Sporozoite infection rate for combined P. vivax, P. ovale and P. malariae (P. OVM) in An. funestus (s.s.) by net distribution arm
Resistance screening of the primary vector group, An. gambiae (s.l.) was carried out, and health sub-district specific resistance marker frequency data were generated for the resistance variants Vgsc-L1014S, Vgsc-L1014F, Vgsc-N1575Y, Cyp4j5-L43F and Coeae1d mutations, using PCR based approaches. The kdr mutations, Vgsc-L1014S and Vgsc-L1014F, that cause alterations to the target site of the voltage-gated sodium channel and are associated with resistance to pyrethroids and DTT, were both detected in our sample. In An. gambiae (s.s.) the L1014S was at a frequency of 0.94, whilst Vgsc-L1014F was detected at a frequency 0.06 (Figs. 3a, 4a-c, Additional file 4: Figure S4). The wild-type allele was detected at extremely low levels (< 0.01). In An. arabiensis the wild-type allele was detected at a frequency of 0.96, whilst resistance-associated alleles were detected at a frequency of 0.01 and 0.03 for L1014S and L1014F respectively.
Samples were screened for the Vgsc-N1575Y mutation, which is also located in the voltage-gated sodium channel and is known to have a synergistic effect on pyrethroid and DDT resistance when combined with the Vgsc-L1014F mutation [28]. The mutation, which has been found across West and Central Africa, including the neighbouring country of DRC in An. gambiae (s.s.), An. coluzzii and An. arabiensis [27, 28, 33], was not found in any of the specimens examined. This is perhaps unsurprising given the low frequency of the L1014F mutation in the population. The pyrethroid metabolic resistance-associated variant, Cyp4j5-L43F [11], located within a p450 gene in the 2La inversion was found at a frequency of 0.60 in An. gambiae (s.s.) (Figs. 3b, 4d, Additional file 4: Figure S4). The Coeae1d resistance-associated allele [11] located in a carboxylesterase gene and highly associated with pyrethroid resistance in East African An. gambiae populations, was found at a frequency of 0.54 in An. gambiae (s.s.) (Figs. 3c, 4f, Additional file 4: Figure S4). The 2La chromosome inversion, associated with adaptation to aridity and humidity [34, 35], biting and resting behaviour [36], and increased susceptibility to Plasmodium [37] was found at a frequency of 0.60 in An. gambiae (s.s.) (Figs. 3d, 4e, Additional file 4: Figure S4).
Resistance marker frequencies for Anopheles gambiae (s.s.) were analysed as a function of net distribution arm and region (East vs West) using GLM. No evidence for a significant difference in resistance marker frequency by planned intervention arm or region was observed (Table 2 and Additional file 5: Table S1).
Discussion
This study provides a comprehensive description of the distribution of major malaria vector species in Uganda, reporting data from 48 of 121 administrative districts. Anopheles gambiae (s.s.) was the dominant endophilic vector across the majority of sites in Uganda with An. funestus and An. arabiensis generally observed at lower densities. Plasmodium falciparum infection rates of 5.5% in An. gambiae (s.s.) and 4.0% in An. funestus (s.s.) were higher than previously observed in Uganda [38]. This may partially reflect differences in the assay methods used in this work compared to the previous study, ELISA vs Taqman, with the latter having increased sensitivity [25]. Whilst the Prokopak-based collection method precludes the estimation of entomological inoculation rates, these data suggest high rates of transmission across Uganda despite widespread conventional LLIN use [19, 39]. These data corroborate early work which has revealed a relatively limited impact of both DDT and pyrethroid based vector control on epidemiological indices [14, 15] thereby highlighting the need for alternative vector control technologies.
Three additional potential vector species were detected: An. parensis, An. pharoensis and An. rufipes. All three species have been recorded in Uganda and in parts of their range they have been found to be sporozoite-positive [40]. It is possible that these species may be malaria vectors in some of the study locations, and whilst the single specimen of An. pharoensis was positive for P. falciparum DNA, these species were captured at very low densities in our indoor-collections. It is possible that these species might have a role in the maintenance of residual transmission and in the transmission of filarial and viral infections [40].
Three known pyrethroid resistance-associated variants were widely distributed across the country. The knockdown resistance mutations (Vgsc-1014F/S) were near or at fixation in An. gambiae (s.s.) populations as expected from earlier studies [6, 11, 41, 42]. The presence of the Vgsc-1014F mutation has been reported sporadically in the region [41, 43] and despite evidence that it may confer higher rates of resistance to pyrethroids that the Vgsc-1014S mutation (reviewed in [44]) there is to date no conclusive evidence of allelic replacement. The two metabolic resistance-associated variants Cyp4j5-L43F and Coeae1d were found across the country at intermediate frequencies. The importance of these mutations on the effectiveness of PBO-LLINs in the field is not yet fully understood however it is hoped that by screening for these resistance markers at six monthly intervals (6, 12 and 18 months post-baseline) it will be possible to track the differential impact of the interventions in areas of differing insecticide resistance.
Conclusions
Importantly for the purposes of the cluster randomized trial there were no significant differences observed between planned the intervention arms at baseline for any of the key variables; vector density, Plasmodium infection rates or insecticide resistance marker frequency. In the principal vector, An. gambiae (s.s.), extreme levels of kdr resistance were observed in all areas with Vgsc-L1014S predominating. The resistance-associated markers, Cyp4j5-L43F and Coeae1d were found at differing frequencies across the study site which may have consequences for the effectiveness of standard and PBO impregnated LLINs.
References
Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–11.
WHO. Global report on insecticide resistance in malaria vectors: 2010–2016. Geneva: World Health Organization; 2018.
Hemingway J, Ranson H, Magill A, Kolaczinski J, Fornadel C, Gimnig J, et al. Averting a malaria disaster: will insecticide resistance derail malaria control? Lancet. 2016;387:1785–8.
Glunt KD, Coetzee M, Huijben S, Koffi AA, Lynch PA, N’Guessan R, et al. Empirical and theoretical investigation into the potential impacts of insecticide resistance on the effectiveness of insecticide-treated bed nets. Evol Appl. 2018;11:431–41.
Kleinschmidt I, Bradley J, Knox TB, Mnzava AP, Kafy HT, Mbogo C, et al. Implications of insecticide resistance for malaria vector control with long-lasting insecticidal nets: a WHO-coordinated, prospective, international, observational cohort study. Lancet Infect Dis. 2018;18:640–9.
Mawejje HD, Wilding CS, Rippon EJ, Hughes A, Weetman D, Donnelly MJ. Insecticide resistance monitoring of field-collected Anopheles gambiae s.l. populations from Jinja, eastern Uganda, identifies high levels of pyrethroid resistance. Med Vet Entomol. 2013;27:276–83.
Müller P, Warr E, Stevenson BJ, Pignatelli PM, Morgan JC, Steven A, et al. Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. PLoS Genet. 2008;4:e1000286.
Ibrahim SS, Riveron JM, Stott R, Irving H, Wondji CS. The cytochrome P450 CYP6P4 is responsible for the high pyrethroid resistance in knockdown resistance-free Anopheles arabiensis. Insect Biochem Mol Biol. 2016;68:23–32.
Edi CV, Djogbénou L, Jenkins AM, Regna K, Muskavitch MAT, Poupardin R, et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 2014;10:e1004236.
Mitchell SN, Stevenson BJ, Müller P, Wilding CS, Egyir-Yawson A, Field SG, et al. Identification and validation of a gene causing cross-resistance between insecticide classes in Anopheles gambiae from Ghana. Proc Natl Acad Sci USA. 2012;109:6147–52.
Weetman D, Wilding CS, Neafsey DE, Müller P, Ochomo E, Isaacs AT, et al. Candidate-gene based GWAS identifies reproducible DNA markers for metabolic pyrethroid resistance from standing genetic variation in East African Anopheles gambiae. Sci Rep. 2018;8:2920.
Abeku TA, Helinski MEH, Kirby MJ, Ssekitooleko J, Bass C, Kyomuhangi I, et al. Insecticide resistance patterns in Uganda and the effect of indoor residual spraying with bendiocarb on kdr L1014S frequencies in Anopheles gambiae s.s. Malar J. 2017;16:156.
Okia M, Hoel DF, Kirunda J, Rwakimari JB, Mpeka B, Ambayo D, et al. Insecticide resistance status of the malaria mosquitoes: Anopheles gambiae and Anopheles funestus in eastern and northern Uganda. Malar J. 2018;17:157.
Kigozi R, Baxi SM, Gasasira A, Sserwanga A, Kakeeto S, Nasr S, et al. Indoor residual spraying of insecticide and malaria morbidity in a high transmission intensity area of Uganda. PLoS ONE. 2012;7:e42857.
Katureebe A, Zinszer K, Arinaitwe E, Rek J, Kakande E, Charland K, et al. Measures of malaria burden after long-lasting insecticidal net distribution and indoor residual spraying at three sites in Uganda: a prospective observational study. PLoS Med. 2016;13:e1002167.
Uganda Bureau of Statistics (UBOS) and ICF International. Uganda malaria indicator survey 2014–15. Kampala, Uganda, and Rockville, Maryland: UBOS and ICF International; 2015.
Rubaihayo J, Tukesiga E, Abaasa A. Reduced susceptibility to pyrethroid insecticide treated nets by the malaria vector Anopheles gambiae s.l. in western Uganda. Malar J. 2008;7:92.
Steinhardt LC, Yeka A, Nasr S, Wiegand RE, Rubahika D, Sserwanga A, et al. The effect of indoor residual spraying on malaria and anemia in a high-transmission area of northern Uganda. Am J Trop Med Hyg. 2013;88:855–61.
Gonahasa S, Maiteki-Sebuguzi C, Rugnao S, Dorsey G, Opigo J, Yeka A, et al. LLIN Evaluation in Uganda Project (LLINEUP) - factors associated with ownership and use of long-lasting insecticidal nets in Uganda: a cross-sectional survey of 48 districts. Malar J. 2018;17:421.
Vazquez-Prokopec GM, Galvin WA, Kelly R, Kitron U. A new, cost-effective, battery-powered aspirator for adult mosquito collections. J Med Entomol. 2009;46:1256–9.
Gillies MT, Coetzee M. A supplement to the anophelinae of Africa south of the Sahara (Afrotropical region). Johannesburg: Publications of the South African Institute for Medical Research; 1987.
Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163.
Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;66:804–11.
White BJ, Santolamazza F, Kamau L, Pombi M, Grushko O, Mouline K, et al. Molecular karyotyping of the 2La inversion in Anopheles gambiae. Am J Trop Med Hyg. 2007;76:334–9.
Bass C, Nikou D, Blagborough AM, Vontas J, Sinden RE, Williamson MS, et al. PCR-based detection of Plasmodium in Anopheles mosquitoes: a comparison of a new high-throughput assay with existing methods. Malar J. 2008;7:177.
Bass C, Nikou D, Donnelly MJ, Williamson MS, Ranson H, Ball A, et al. Detection of knockdown resistance (kdr) mutations in Anopheles gambiae: a comparison of two new high-throughput assays with existing methods. Malar J. 2007;6:111.
Lynd A, Oruni A, van’t Hof AE, Morgan JC, Naego LB, Pipini D, et al. Insecticide resistance in Anopheles gambiae from the northern Democratic Republic of Congo, with extreme knockdown resistance (kdr) mutation frequencies revealed by a new diagnostic assay. Malar J. 2018;17:412.
Jones CM, Liyanapathirana M, Agossa FR, Weetman D, Ranson H, Donnelly MJ, et al. Footprints of positive selection associated with a mutation (N1575Y) in the voltage-gated sodium channel of Anopheles gambiae. Proc Natl Acad Sci USA. 2012;109:6614–9.
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701.
Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–6.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704.
Edi A, N’Dri B, Chouaibou M, Kouadio F, Pignatelli P, Raso G, et al. First detection of N1575Y mutation in pyrethroid resistant Anopheles gambiae in Southern Côte d’Ivoire. Wellcome Open Res. 2017. https://doi.org/10.12688/wellcomeopenres.12246.1.
Fouet C, Gray E, Besansky NJ, Costantini C. Adaptation to aridity in the malaria mosquito Anopheles gambiae: chromosomal inversion polymorphism and body size influence resistance to desiccation. PLoS One. 2012;7:e34841.
Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–97.
Coluzzi M, Sabatini A, Petrarca V, Di Deco MA. Behavioural divergences between mosquitoes with different inversion karyotypes in polymorphic populations of the Anopheles gambiae complex. Nature. 1977;266:832–3.
Riehle MM, Bukhari T, Gneme A, Guelbeogo WM, Coulibaly B, Fofana A, et al. The Anopheles gambiae 2La chromosome inversion is associated with susceptibility to Plasmodium falciparum in Africa. eLife. 2017;6:e25813.
Kilama M, Smith DL, Hutchinson R, Kigozi R, Yeka A, Lavoy G, et al. Estimating the annual entomological inoculation rate for Plasmodium falciparum transmitted by Anopheles gambiae s.l. using three sampling methods in three sites in Uganda. Malar J. 2014;13:111.
Yeka A, Gasasira A, Mpimbaza A, Achan J, Nankabirwa J, Nsobya S, et al. Malaria in Uganda: challenges to control on the long road to elimination: I. Epidemiology and current control efforts. Acta Trop. 2012;121:184–95.
Kyalo D, Amratia P, Mundia CW, Mbogo CM, Coetzee M, Snow RW. A geo-coded inventory of anophelines in the Afrotropical Region south of the Sahara: 1898–2016. Wellcome Open Res. 2017;2:57.
Verhaeghen K, Bortel WV, Roelants P, Okello PE, Talisuna A, Coosemans M. Spatio-temporal patterns in kdr frequency in permethrin and DDT resistant Anopheles gambiae s.s. from Uganda. Am J Trop Med Hyg. 2010;82:566–73.
Wilding CS, Weetman D, Rippon EJ, Steen K, Mawejje HD, Barsukov I, et al. Parallel evolution or purifying selection, not introgression, explain similarity in the pyrethroid detoxification linked GSTE4 of Anopheles gambiae and An. arabiensis. Mol Genet Genomics. 2015;290:201–15.
Ochomo E, Subramaniam K, Kemei B, Rippon E, Bayoh NM, Kamau L, et al. Presence of the knockdown resistance mutation, Vgsc-1014F in Anopheles gambiae and An. arabiensis in western Kenya. Parasit Vectors. 2015;8:616.
Donnelly MJ, Isaacs AT, Weetman D. Identification, validation, and application of molecular diagnostics for insecticide resistance in malaria vectors. Trends Parasitol. 2016;32:197–206.
Acknowledgements
The authors would like to thank all the staff and administration at IDRC for their considerable efforts in facilitating this project. In particular Martin Mugote, Violet Tuhaise, Jackson Asiimwe and Maxwell Kilama for their supervision of field activities; Peter Mutungi, Simon Peter Kigozi and Geoff Lavoy for their considerable assistance in data management; Lillian Taaka, Christine Nabirye and Nicholas Wendo for assistance in logistics and procurement; Susan Nayiga, Isiko Joseph, Erias Muyanda and Henry Opolot for coordination and support. We are extremely grateful to the entomology field workers for collecting and identifying mosquitoes, and to the community survey teams and team leaders who facilitated their work. We would also like to acknowledge and thank the members of the Uganda National Malaria Control Program and the Liverpool School of Tropical Medicine for logistical and other support provided as we carried out these surveys, and the district health, administrative, and political leadership teams for all their support and guidance during community entry in the 48 districts of the study area.
Funding
This project was primarily supported by The Against Malaria Foundation. Additional funding was provided by Award Number R01AI116811 from the National Institute of Allergy and Infectious Diseases (NIAID) and a Wellcome Trust MSc Fellowship in Public Health and Tropical Medicine (Oruni-203511/Z/16/Z). The content of the manuscript is solely the responsibility of the authors.
Availability of data and materials
Data supporting the conclusions of this article are included within the article and its additional files. The datasets reported herein will be made publicly available on completion of the study but are available from the corresponding author on reasonable request.
Authors’ contributions
SGS, MRK, GD and MJD conceived of the study, with input from JO, AY and JH. SGS, GD and MJD developed the procedures and drafted the protocol with MRK and JH. AL led the data collection in the field, with oversight from SG, and support from CMS, JO, MRK, AK, MK and SGS. AL and AO performed all laboratory analyses. AL and MJD managed the data and led the data analysis. AL, AO and MJD interpreted the data and drafted the manuscript, with input from SGS. All authors reviewed the manuscript and gave permission for publication. AL is the corresponding author, and MJD had full access to all the data in the study and had final responsibility for the decision to submit for publication. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ugandan National Council for Science and Technology (UNCST Ref HS 2176), Makerere University School of Medicine Research & Ethics Committee (SOMREC 2016-133), London School of Hygiene & Tropical Medicine Ethics Committee (LSHTM Ref 12019), and the Liverpool School of Tropical Medicine (Ref 16-072), which sponsored the study. Written informed consent to participate in the study was obtained by the head of household (or their designate) for all participating households.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Figure S1.
The density of female An. funestus (s.l.) was analysed as a function of net distribution arm with HSD as random effect using GLMM based on a Type 2 negative Binomial Model. The best-fit model, as determined by AIC, did not include planned intervention arm showing that there was no significant difference in the densities of An. funestus (s.l.) by intervention.
Additional file 2: Figure S2.
Phylogenetic tree constructed from mitochondrial cox1 gene sequence of the unknown mosquito samples (in red boxes) were compared with known sequences of major anopheline vectors from the NCBI database. A mid-rooted phylogenetic tree was plotted using PHYML (Maximum Likelihood relationship) algorithm, following multiple sequence alignment by MUSCLE. The black dots are the tree nodes which represent a common ancestor. The figures show the branch length which represents the amount of change in-terms of mutations that has occurred with time between members.
Additional file 3: Figure S3.
Sporozoite infection rates by sub region. a P. falciparum sporozoite infection rate in An. gambiae (s.s.). b Combined sporozoite infection rate for P. vivax, P. ovale and P. malariae in An. gambiae (s.s.). c P. falciparum sporozoite infection rate in An. funestus (s.s.). d Combined sporozoite infection rate for P. vivax, P. ovale and P. malariae in An. funestus (s.s.).
Additional file 4: Figure S4.
Resistance and polytene chromosome allele frequencies in Anopheles gambiae (s.s.) by sub-region. a Vgsc 1014F/S. b Cyp4j5-L43F. c Coeae1d. d 2La inversion.
Additional file 5: Table S1.
Resistance and polytene chromosome allele frequencies in Anopheles gambiae (s.s.) analysed as a function of sub-region using a generalized linear model.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lynd, A., Gonahasa, S., Staedke, S.G. et al. LLIN Evaluation in Uganda Project (LLINEUP): a cross-sectional survey of species diversity and insecticide resistance in 48 districts of Uganda. Parasites Vectors 12, 94 (2019). https://doi.org/10.1186/s13071-019-3353-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3353-7