Abstract
Background
Cystic echinococcosis is a worldwide chronic zoonotic disease caused by infection with the larval stage of Echinococcus granulosus. Previously, we found significant accumulation of myeloid-derived suppressor cells (MDSCs) in E. granulosus infection mouse models and that they play a key role in immunosuppressing T lymphocytes. Here, we compared the long non-coding RNA (lncRNA) and mRNA expression patterns between the splenic monocytic MDSCs (M-MDSCs) of E. granulosus protoscoleces-infected mice and normal mice using microarray analysis.
Methods
LncRNA functions were predicted using Gene Ontology enrichment and the Kyoto Encyclopedia of Genes and Genomes pathway analysis. Cis- and trans-regulation analyses revealed potential relationships between the lncRNAs and their target genes or related transcription factors.
Results
We found that 649 lncRNAs were differentially expressed (fold change ≥ 2, P < 0.05): 582 lncRNAs were upregulated and 67 lncRNAs were downregulated; respectively, 28 upregulated mRNAs and 1043 downregulated mRNAs were differentially expressed. The microarray data was validated by quantitative reverse transcription-PCR. The results indicated that mRNAs co-expressed with the lncRNAs are mainly involved in regulating the actin cytoskeleton, Salmonella infection, leishmaniasis, and the vascular endothelial growth factor (VEGF) signaling pathway. The lncRNA NONMMUT021591 was predicted to cis-regulate the retinoblastoma gene (Rb1), whose expression is associated with abnormal M-MDSCs differentiation. We found that 372 lncRNAs were predicted to interact with 60 transcription factors; among these, C/EBPβ (CCAAT/enhancer binding protein beta) was previously demonstrated to be a transcription factor of MDSCs.
Conclusions
Our study identified dysregulated lncRNAs in the M-MDSCs of E. granulosus infection mouse models; they might be involved in M-MDSC-derived immunosuppression in related diseases.
Similar content being viewed by others
Background
Cystic echinococcosis is a worldwide chronic zoonotic disease caused by accidental ingestion of eggs of the genus Echinococcus and typically affects the liver and lungs [1]. It is endemic in pastoral regions around the world [2], causes a huge disease burden, and is characterized by long-term growth of hydatid cysts in humans and mammalian intermediate hosts. The cysts are filled with hydatid cyst fluid and protoscoleces [3].
Previously, we found significant accumulation of myeloid-derived suppressor cells (MDSCs) in mouse models infected with E. granulosus protoscoleces (Eg-psc) [4] and that they play a key role in downregulating the immune response of T lymphocytes. MDSCs are a heterogeneous population of myeloid cells composed of terminally differentiated macrophages, granulocytes, or dendritic cells. Various pathological conditions, such as cancer [5, 6], sepsis [7] and parasitic infection [8] result in aberrant MDSC expansion. MDSCs consist of two major subsets based on their phenotypic and morphological features: polymorphonuclear (PMN)-MDSCs and monocytic (M)-MDSCs. In mice, they are historically characterized by concurrent expression of the myeloid markers CD11b and Gr-1. The two major subsets can be identified more accurately based on the expression of Ly6G and Ly6C markers: PMN-MDSCs, CD11b+Ly6G+Ly6Clow; M-MDSCs, CD11b+Ly6G-Ly6Chi) [9, 10]. M-MDSCs and PMN-MDSCs inhibit immune function via different mechanisms. M-MDSCs suppress T cell function via both antigen-specific and nonspecific mechanisms by producing nitric oxide (NO) and cytokines [11, 12], and are more immunosuppressive than their counterparts when assessed on a per cell basis [13,14,15].
Long non-coding RNAs (lncRNAs) are commonly defined as transcribed RNAs of more than 200 nucleotides in length and lack protein-coding ability [16, 17]. Increasing evidence indicates that lncRNAs participate in several important biological processes, including carcinogenesis, cell differentiation, metabolism, and immunity responses [18,19,20], acting as signal molecules, decoys, guides, and scaffolds [21,22,23]. Although numerous lncRNAs have been discovered in recent years, only a limited number have been well characterized. At the same time, knowledge of the genome scale of lncRNAs and their underlying biological functions in MDSCs in parasitic infections remains limited. Moreover, MDSC functional plasticity via epigenetic modification leads to their characteristics reshaping [10].
In the present study, we used microarray analysis to investigate the lncRNA and mRNA expression profiles in the splenic M-MDSCs of normal and Eg-psc-infected mice, and performed bioinformatics analysis of the differentially expressed lncRNAs to explore the possible biological processes and pathways associated with M-MDSCs. The results demonstrate that aberrantly expressed lncRNAs may be new candidates for the immunosuppressive mechanism of M-MDSCs in parasitic diseases.
Methods
Mice, parasites, and infection
Female BALB/c mice (aged 6–8 weeks) were purchased from SLAC Laboratory. The Eg-psc were obtained from the hydatid cysts of naturally infected sheep livers under aseptic conditions, and washed three times using 0.9% NaCl containing 1000 mg/ml penicillin and 1000 U/ml streptomycin (Invitrogen, Frederick, MD, USA). Thirty BALB/c mice were intraperitoneally injected with a 200 μl suspension containing 2000 live Eg-psc in 0.9% NaCl; the controls were 30 BALB/c mice injected with 200 μl 0.9% NaCl. All mice were housed in specific pathogen-free conditions.
Cell isolation
Splenic M-MDSCs were isolated immediately after the BALB/c mice were sacrificed under sterile conditions at eight months after infection. Single-cell suspensions were enriched with magnetic cell sorting (MACS; Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s protocol. The M-MDSCs were separated on mini MACS columns (Miltenyi Biotec, Bergisch Gladbach, Germany) and yielded approximately 90% pure cells. Then, we randomly selected splenic M-MDSCs from three mice each in the infected and normal groups for the detection of lncRNA and mRNA arrays.
Microarray profiling
The lncRNA and mRNA expression patterns were detected in the splenic M-MDSCs of three Eg-psc-infected mice and three normal mice. The experiments were performed at OE BioTechCorporation (Shanghai, China). Agilent mouse lncRNA Microarray (4*180K, Design ID: 049801) was used in this experiment. Total RNA was extracted and purified with a RNeasy Mini Kit (Qiagen, p/n 74104, Boston, MA, USA) and quantified using NanoDrop ND-2000 (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Sample labeling, microarray hybridization, and washing were performed based on the manufacturer’s standard protocols. Briefly, total RNA was transcribed to double-stranded complementary DNA (cDNA), synthesized into complementary RNA (cRNA), and labeled with Cyanine 3-CTP. The labeled cRNA was hybridized onto the microarray. After washing, the arrays were scanned using an Agilent Scanner G2505C microarray scanner (Agilent Technologies).
Differential expression analysis
The raw data were analyzed using Feature Extraction software (version 10.7.1.1; Agilent Technologies) and then normalized using percentile normalization. Probes with least one of two conditions flagged in “P” were chosen for further data analysis. Differentially expressed lncRNAs were identified through fold change and the P-value as calculated with the t-test. Aberrantly expressed lncRNAs and mRNAs were defined as fold change ≥ 2.0 and P < 0.05. Subsequently, Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were used to explore the roles of the differentially expressed mRNAs. Hierarchical clustering was performed on six mouse splenic tissue samples using Cluster 3.0 (Stanford University School of Medicine, California, USA) and TreeView 2.0 (Baryshnikova Lab, Princeton University, New Jersey, USA) to distinguish the distinguishable gene expression pattern among the samples.
Co-expression network analysis
The co-expression of lncRNAs and the protein-coding genes was calculated using Pearson correlation coefficients with Cytoscape version 3.1.1 (US National Institute of General Medical Sciences). Correlations with P < 0.05 were considered statistically significant.
Functional enrichment analysis
The functions of lncRNA co-expressed mRNAs were analyzed using GO enrichment analysis which was divided into three functional categories: molecular function, biological process and cellular component. The pathways of the co-expressed mRNAs were analyzed using KEGG pathway analysis.
Quantitative reverse transcription-PCR (qRT-PCR)
qRT-PCR was performed to validate the microarray results according to the manufacturer’s protocol with a SYBR Green RT-PCR Kit (QuantiFast SYBR Green PCR Master Mix, Qiagen) on a Bio-Rad CFX96 system. The primers used for the qRT-PCR are shown in Additional file 1: Table S1. The lncRNA expression levels were quantified based on the threshold cycle (Ct) values. β-Actin served as the internal control. The relative gene expression was analyzed using the comparative Ct [2(-ΔΔCt)] method [24]. Three biological replicates were performed for each group.
Cis- and trans-regulation analysis
LncRNAs have been shown to enhance the expression of nearby genes through cis-regulation [25] and as the mRNA loci were within 300 kbp windows upstream and downstream of the given lncRNAs, we identified them as cis-regulated mRNAs of the corresponding lncRNAs.
Here, the lncRNAs identified showed high Pearson’s correlation with the neighboring protein-coding genes, suggesting that they act in cis on protein-coding genes to regulate M-MDSC functions. As it has been indicated that transcription factors regulate lncRNA production, we therefore used hypergeometric distribution testing to predict the potential transcription factors that may regulate the production of the differentially expressed lncRNAs.
Statistical analysis
Statistical analysis was performed using SPSS 19.0 (SPSS, New York, USA). All measurement data are reported as the mean ± standard deviation (SD). Differentially expressed lncRNAs were identified using t-tests or nonparametric tests. P < 0.05 was considered statistically significant.
Results
Differentially expressed lncRNAs in M-MDSCs
The raw data were analyzed using Feature Extraction software (version 10.7.1.1, Agilent Technologies) and were normalized with the quantile algorithm. GeneSpring (version 13.1, Agilent Technologies) was used for basic raw data analysis. Probes with at least 100% of the values in any one out of all conditions flagged as “Detected” were selected for further analysis. Differentially expressed mRNAs or lncRNAs were identified through fold change and the P-value as calculated with the t-test. The threshold set for upregulated or downregulated RNAs was fold change ≥ 2.0 and P < 0.05.
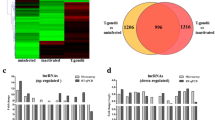
We detected 54,030 lncRNAs and 33,420 mRNAs in the M-MDSC samples. Microarray scanning and normalization determined that 649 lncRNAs and 1071 mRNAs were differentially expressed. Among them, 582 lncRNAs were upregulated and 67 lncRNAs were downregulated (Additional file 2: Table S2), and 28 upregulated and 1043 downregulated mRNAs were differentially expressed (Additional file 3: Table S3). Compared with the controls, FR208893 (fold change of 133.45992) was the most upregulated lncRNA, while FR325025 (fold change of 4.1526523) was the most downregulated lncRNA in the M-MDSCs.
Figure 1 shows a volcano plot of the differentially expressed lncRNAs, where red and green indicate significantly upregulated and downregulated lncRNAs, respectively. To identify gene expression patterns, hierarchical clustering was performed among samples using Cluster 3.0 and TreeView 2.0. Many lncRNAs were identified as differentially expressed between the two groups, indicating that the significantly altered expression of these lncRNAs may be involved in the immunoregulatory function of M-MDSCs (Fig. 2).
qRT-PCR validation
To verify the results of lncRNA microarray, we used qRT-PCR to detect the expression of 10 lncRNAs selected randomly from the differentially expressed lncRNA transcripts with splenic M-MDSCs samples with three biological replicates in each group. The qRT-PCR results were consistent with the lncRNA array analysis, and these lncRNAs are likely to play roles in response to the biological functions of MDSCs in parasitic infections (Fig. 3).
LncRNAs-mRNAs co-expression network
We constructed a co-expression network to investigate the correlation between each differentially expressed lncRNA in M-MDSCs and the target mRNAs. We ranked each lncRNA-mRNA correlation according to the P-value and selected the top 500 lncRNA functional terms for functional enrichment analysis, and counted the numbers of differently expressed lncRNAs enriched in the functional terms. These functional terms were used to predict the functions of the given lncRNAs based on GO enrichment and KEGG pathway analyses of the co-expressed mRNAs.
For GO analysis, differentially expressed lncRNAs were mostly enriched in nuclear envelope organization, positive regulation of target of rapamycin (TOR) signaling and positive regulation of protein complex assembly in biological processes (Fig. 4a); nuclear envelope, mitotic spindle, and trans-Golgi network in cellular component (Fig. 4b), and enzyme regulator activity, lipoteichoic acid binding, and NADH dehydrogenase activity in molecular functions (Fig. 4c).
It is widely believed that disorder of the signaling pathways under pathological conditions contributes to the development of suppressive myeloid cells [26]. KEGG pathway analysis indicated that the co-expressed mRNAs were mainly involved in regulating the actin cytoskeleton, Salmonella infection, leishmaniasis, and the vascular endothelial growth factor (VEGF) signaling pathway (Fig. 4d). MDSCs accumulate in lymphoid organs under parasitic infection (as in cancer), and can migrate to and invade the adjacent tissues and vasculature. Some key proteins involved in the actin cytoskeleton are linked to cancer cell invasion and metabolism [27]. As cystic echinococcosis is considered an infectious and inflammatory disease, the aberrantly expressed lncRNAs might regulate these processes.
Cis-regulation of lncRNAs
We found that 288 lncRNAs were considered cis-regulatory lncRNAs of their sense-overlapping genes, and we found that lncRNA NONMMUT021591 was predicted to cis-regulate the Rb1 (Additional file 4: Figure S1). Rb1 expression had been reported to be associated with abnormal M-MDSC differentiation [28].The lncRNA NONMMUT021591 may play a role in the immunosuppressive functions of MDSCs.
LncRNA-transcription factor network analysis
As it was indicated that transcription factors regulate lncRNA production, we used hypergeometric distribution testing to predict the transcription factors that could regulate the differentially expressed lncRNAs, and constructed a core network of the top 100 lncRNA-transcription factor pairs by ranking the P-value (Fig. 5). We predicted that 372 lncRNAs would interact with 60 transcription factors, forming an interaction network of 1746 connections. These lncRNAs were mostly regulated by IL6, FOSL1, YY1, PGR, TMEM37, PBX1, POU3F2, FOXF2, FOS, JUNB, JUND, CEBPD, RB1, MAX, E2F1, C/EBPβ, HERPUD1, E2F4 and ZBTB16. Among the transcription factors, C/EBPβ (CCAAT enhancer-binding transcription factor) [29] has been demonstrated to be a MDSC transcription factor. C/EBPβ regulates myeloid cell development and differentiation, and controls emergency granulopoiesis induced by cytokines and infections [7, 30]. Increased C/EBPβ expression is a characteristic biochemical feature of MDSCs.
LncRNA-target-transcription factor network analysis
For further identification of the functions of each dysregulated lncRNA in M-MDSCs, we analyzed the top 50 differentially expressed lncRNAs and their co-expressed mRNAs pairs according to the P-value to conduct the lncRNA-target-transcription factor network (Fig. 6). The network revealed several most likely transcription factors for these lncRNAs, and included FOSL1, YY1, IL6 and PGR. LncRNAs FR049933, FR291292, FR110455 and FR400826 are predicted to be mainly regulated by these transcription factors and participate in mitogen-activated protein kinase (MAPK) signaling pathway and VEGF signaling pathway, which are involved in MDSC function.
Discussion
Cystic echinococcosis is a worldwide chronic zoonotic disease in pastoral regions around the world [2]. Transmission is through predator-prey interactions between carnivore definitive hosts and herbivore intermediate hosts [31, 32]. Previously, we showed significant accumulation of CD11b+Gr-1+ myeloid cells in the spleen and peripheral blood in Eg-psc-infected mouse models [4] . Under infected conditions, myeloid cells are arrested in an immature state and possess a potential capacity to suppress immune cell responses by creating a suppressive environment; such cells are termed MDSCs. MDSCs have gained much attention due to their roles in immunosuppression and in the promotion of angiogenesis [33] and metastasis [34]. MDSCs exert immunosuppressive effects depending on the expanding subtype, the disease stage and the site where immunosuppression is occurring [35], and their mechanism may change with the disease progression or the affected organ. M-MDSCs suppress T-cell function via both antigen-specific and nonspecific mechanisms by producing high amounts of arginase 1, NO and immunosuppressive cytokines [11, 12]. Under infection conditions, the capacity of MDSCs to differentiate into macrophages and dendritic cells is inhibited. MDSCs are more immunosuppressive than their counterparts when assessed on a per cell basis [13,14,15]. The mechanism associated with MDSC regulation is becoming accepted as another means of regulating immune responses, and MDSCs are potential therapeutic targets in multiple infectious and inflammatory diseases. While the underlying molecular mechanisms of MDSCs and their sub-populations remain unclear, epigenetic modification of MDSC function could provide some molecular evidence concerning MDSC accumulation and immunosuppression. Studies have confirmed that a number of lncRNAs are crucial in biological processes, including regulating gene expression, cell development and metabolism, which are disordered in disease [18, 19, 36]. A global lncRNA expression profile specific to a functional MDSC population is not known. In the present study, we selected splenic M-MDSCs for lncRNA microarray analysis based on previous studies [4, 13, 15] to investigate and compare the lncRNA expression profile of M-MDSCs and to discover the role and significance of lncRNAs in M-MDSCs, which indicated that lncRNA is a mediator that recruits MDSCs in Eg-psc-infected models.
We compared the lncRNA expression profiles between splenic M-MDSCs from Eg-psc-infected mice and normal mice to identify the lncRNAs that may be important in M-MDSC differentiation and function. Compared with the normal mice, 582 lncRNAs were upregulated and 67 lncRNAs were downregulated, and 28 upregulated mRNAs and 1043 downregulated mRNAs were differentially expressed in the infected mice. The expression differences in the microarray detection were consistent with that of qRT-PCR detection. For further analysis of the differentially expressed lncRNAs, we constructed a co-expression network to investigate the correlation between each aberrant lncRNA and their target mRNAs. GO analysis and KEGG pathway analysis were carried out to investigate the biological functions of lncRNAs. The GO enrichment assay showed that the differentially expressed lncRNAs were mostly enriched in positive regulation of TOR signaling in biological processes. The TOR pathway is well recognized as being related to cell proliferation and metabolism. Moreover, mammalian TOR (mTOR) is an intrinsic factor essential for M-MDSC differentiation and immunosuppressive function [37]. KEGG pathway analysis showed that the mRNAs co-expressed lncRNAs were mainly involved in regulation of the actin cytoskeleton, leishmaniasis, and the VEGF signaling pathway, which have been widely researched and demonstrated to be associated with MDSCs [26]. These differentially expressed lncRNAs also participate in inflammatory signaling pathways, such as the MAPK signaling pathway, tumor necrosis factor (TNF) signaling pathway, and the nuclear factor kappa B (NF-κB) signaling pathway [38, 39]. Moreover, these inflammatory pathways have been widely researched and demonstrated to be associated with MDSCs function. The vascular endothelial growth factor (VEGF) is an important molecule involved in angiogenesis. Ostrand-Rosenberg et al. [38] demonstrated that inflammation could increase MDSCs levels by protecting MDSCs from Fas-mediated apoptosis through activation of the MAPK pathway. IL-33 [40] induced arginase-1 expression and activated the NF-κB and MAPK signaling pathways, augmenting the immunosuppressive ability of MDSCs. To further study the roles of specific lncRNAs in M-MDSCs, we predicted their corresponding mRNAs through cis- and trans-targeting. We found that 288 lncRNAs were considered cis-regulatory lncRNAs of their sense-overlapping genes; among them, NONMMUT021591 was predicted to cis-regulate Rb1. Rb1 expression is associated with abnormal M-MDSC differentiation. Youn et al. [28] demonstrated a novel regulatory mechanism of myeloid cells in cancer. Transcriptional silencing of the Rb1 gene altered M-MDSC differentiation into macrophages and dendritic cells to preferential differentiation towards PMN-MDSCs. Furthermore, M-MDSCs in tumor-bearing mice could acquire the phenotypic and morphological features of PMN-MDSCs. Three hundred and seventy-two lncRNAs were predicted to interact with 60 transcription factors; several among them, namely FOSL1, YY1, IL6 and PGR, were the most enriched terms. Among these transcription factors, C/EBPβ [27] has been demonstrated to be a MDSC transcription factor, and under inflammation and infection conditions, C/EBPβ could regulate myeloid cell development and differentiation and control emergency granulopoiesis [7, 30]. Increased expression of the transcriptional regulator C/EBPβ is a characteristic biochemical feature of MDSCs. In the present study, FR015378 was predicted to be regulated by C/EBPβ and participates in the VEGF signaling pathway in KEGG analysis. Therefore, it is reasonable to propose that the aberrantly expressed lncRNAs participate in parasitic infection induction of M-MDSCs by acting with their correlated mRNAs and transcription factors.
Conclusions
LncRNAs are critical in modulating the immune microenvironment and MDSCs; our findings provide a new understanding of M-MDSCs, and the data we present could guide the exploration of lncRNA-mediated immunosuppression in long-term parasitic infection. Although the sensitivity and specificity of lncRNA biomarkers in M-MDSCs should be further investigated, the functional lncRNAs can be explored as potential biomarkers or novel treatment strategies for immunoregulation in related diseases. Understanding the underlying mechanisms and functions of these immunosuppressive cell populations will pave the way for new parasite vaccine strategies.
Abbreviations
- C/EBPβ:
-
CCAAT/enhancer binding protein beta
- Eg-psc:
-
Echinococcus granulosus protoscoleces
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- lncRNA:
-
long non-coding RNA
- MAPK:
-
mitogen-activated protein kinase
- MDSCs:
-
myeloid-derived suppressor cells
- M-MDSCs:
-
monocytic myeloid-derived suppressor cells
- mTOR:
-
mammalian TOR
- NF-κB:
-
nuclear factor kappa B
- NO:
-
nitric oxide
- PMN-MDSCs:
-
polymorphonuclear myeloid-derived suppressor cells
- TNF:
-
tumor necrosis factor
- TOR:
-
target of rapamycin
- VEGF:
-
vascular endothelial growth factor
References
McManus DP, Gray DJ, Zhang W, Yang Y. Diagnosis, treatment, and management of echinococcosis. BMJ. 2012;344:e3866.
Budke CM, Carabin H, Ndimubanzi PC, Nguyen H, Rainwater E, Dickey M, et al. A systematic review of the literature on cystic echinococcosis frequency worldwide and its associated clinical manifestations. Am J Trop Med Hyg. 2013;88:1011–27.
Monteiro KM, de Carvalho MO, Zaha A, Ferreira HB. Proteomic analysis of the Echinococcus granulosus metacestode during infection of its intermediate host. Proteomics. 2010;10:1985–99.
Pan W, Zhou HJ, Shen YJ, Wang Y, Xu YX, Hu Y, et al. Surveillance on the status of immune cells after Echinnococcus granulosus protoscoleces infection in Balb/c mice. PLoS One. 2013;8:e59746.
Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. 2015;125:3356–64.
Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–506.
McClure C, McPeak MB, Youssef D, Yao ZQ, McCall CE, El Gazzar M. Stat3 and C/EBPbeta synergize to induce miR-21 and miR-181b expression during sepsis. Immunol Cell Biol. 2017;95:42–55.
Schmid M, Zimara N, Wege AK, Ritter U. Myeloid-derived suppressor cell functionality and interaction with Leishmania major parasites differ in C57BL/6 and BALB/c mice. Eur J Immunol. 2014;44:3295–306.
Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–802.
Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150.
Marigo I, Dolcetti L, Serafini P, Zanovello P, Bronte V. Tumor-induced tolerance immune suppression by myeloid derived suppressor cells. Immunol Rev. 2008;222:162–79.
Lindau D, Gielen P, Kroesen M, Wesseling P, Adema GJ. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138:105–15.
Dolcetti L, Peranzoni E, Ugel S, Marigo I, Fernandez Gomez A, Mesa C, et al. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. Eur J Immunol. 2010;40:22–35.
Movahedi K, Guilliams M, Van den Bossche J, Van den Bergh R, Gysemans C, Beschin A, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–44.
Haverkamp JM, Smith AM, Weinlich R, Dillon CP, Qualls JE, Neale G, et al. Myeloid-derived suppressor activity is mediated by monocytic lineages maintained by continuous inhibition of extrinsic and intrinsic death pathways. Immunity. 2014;41:947–59.
Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46.
Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46.
Liu YR, Tang RX, Huang WT, Ren FH, He RQ, Yang LH, et al. Long noncoding RNAs in hepatocellular carcinoma: novel insights into their mechanism. World J Hepatol. 2015;7:2781–91.
Moyo B, Nicholson SA, Arbuthnot PB. The role of long non-coding RNAs in hepatitis B virus-related hepatocellular carcinoma. Virus Res. 2016;212:103–13.
Chauhan R, Lahiri N. Tissue- and serum-associated biomarkers of hepatocellular carcinoma. Biomark Cancer. 2016;8:37–55.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom Proteom Bioinform. 2016;14:42–54.
Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods. 2001;25:402–8.
Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606.
Trikha P, Carson WE 3rd. Signaling pathways involved in MDSC regulation. Biochim Biophys Acta. 2014;1846:55–65.
Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–52.
Youn JI, Kumar V, Collazo M, Nefedova Y, Condamine T, Cheng P, et al. Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat Immunol. 2013;14:211–20.
Marigo I, Bosio E, Solito S, Mesa C, Fernandez A, Dolcetti L, et al. Tumor-induced tolerance and immune suppression depend on the C/EBPbeta transcription factor. Immunity. 2010;32:790–802.
Dai J, Kumbhare A, Youssef D, Yao ZQ, McCall CE, El Gazzar M. Expression of C/EBPβ in myeloid progenitors during sepsis promotes immunosuppression. Mol Immunol. 2017;91:165–72.
Ito A, Budke CM. The echinococcoses in Asia: the present situation. Acta Trop. 2017;176:11–21.
Craig P, Mastin A, van Kesteren F, Boufana B. Echinococcus granulosus: epidemiology and state-of-the-art of diagnostics in animals. Vet Parasitol. 2015;213:132–48.
Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6:409–21.
Talmadge JE, Gabrilovich DI. History of myeloid-derived suppressor cells. Nat Rev Cancer. 2013;13:739–52.
Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–20.
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39.
Wu T, Zhao Y, Wang H, Li Y, Shao L, Wang R, et al. mTOR masters monocytic myeloid-derived suppressor cells in mice with allografts or tumors. Sci Rep. 2016;6:20250.
Ostrand-Rosenberg S, Sinha P, Chornoguz O, Ecker C. Regulating the suppressors: apoptosis and inflammation govern the survival of tumor-induced myeloid-derived suppressor cells (MDSC). Cancer Immunol Immunother. 2012;61:1319–25.
Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res. 2011;9:133–48.
Xiao P, Wan X, Cui B, Liu Y, Qiu C, Rong J, et al. Interleukin 33 in tumor microenvironment is crucial for the accumulation and function of myeloid-derived suppressor cells. Oncoimmunology. 2016;5:e1063772.
Acknowledgements
The authors are grateful to Professor Junying Ma at the Qinghai Institute for Endemic Disease Prevention and Control for her help in collecting the parasite samples in Qinghai Province, China.
Funding
This work was supported by the National Natural Science Foundation of China (Nos: 81772224 and 81371842 to YS; 81501771 to JZ; 81601792 to YW; and 81702030 to JY), and the Fourth Round of Three-Year Public Health Action Plan of Shanghai, China (No. 15GWZK0101 to JC]. The funders had no role in the study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Author information
Authors and Affiliations
Contributions
YS and JC conceived and designed the study. AY, YS, JY, JZ, YW and SC performed the experiments and data analysis. YS, JC, YW, JZ and JY contributed reagents and materials. YA and YS wrote and JC revised the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology, China. The protocol was approved by the Laboratory Animals Welfare and Ethics Committee (LAWEC) of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention (Permit number: IPD 2013-006). A special effort was made to reduce the number of mice used in the study, to provide them with the most comfortable conditions, and to minimize animal suffering wherever possible.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Primers used in qPCR detection of selected lncRNAs. (XLSX 10 kb)
Additional file 2:
Table S2. Significantly and differentially expressed lncRNAs in M-MDSCs. (XLSX 49 kb)
Additional file 3:
Table S3. Significantly and differentially expressed mRNAs in M-MDSCs. (XLSX 83 kb)
Additional file 4:
Figure S1. The lncRNA NONMMUT021591 was predicted to cis-regulate the protein-coding gene Rb1. Red dots, genomic location of lncRNAs; blue dots, the corresponding genes; rho value, correlation coefficient. (TIF 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yu, A., Wang, Y., Yin, J. et al. Microarray analysis of long non-coding RNA expression profiles in monocytic myeloid-derived suppressor cells in Echinococcus granulosus-infected mice. Parasites Vectors 11, 327 (2018). https://doi.org/10.1186/s13071-018-2905-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-2905-6