Abstract
Background
Avian malaria occurs almost worldwide and is caused by Haemosporida parasites (Plasmodium, Haemoproteus and Leucocytozoon). Vectors such as mosquitoes, hippoboscid flies or biting midges are required for the transmission of these parasites. There are few studies about avian malaria parasites on Madagascar but none about suitable vectors.
Methods
To identify vectors of avian Plasmodium parasites on Madagascar, we examined head, thorax and abdomen of 418 mosquitoes from at least 18 species using a nested PCR method to amplify a 524 bp fragment of the haemosporidian mitochondrial cytochrome b gene. Sequences obtained were then compared with a large dataset of haemosporidian sequences detected in 45 different bird species (n = 686) from the same area in the Maromizaha rainforest.
Results
Twenty-one mosquitoes tested positive for avian malaria parasites. Haemoproteus DNA was found in nine mosquitoes (2.15%) while Plasmodium DNA was found in 12 mosquitoes (2.87%). Seven distinct lineages were identified among the Plasmodium DNA samples. Some lineages were also found in the examined bird samples: Plasmodium sp. WA46 (EU810628.1) in the Madagascar bulbul, Plasmodium sp. mosquito 132 (AB308050.1) in 15 bird species belonging to eight families, Plasmodium sp. PV12 (GQ150194.1) in eleven bird species belonging to eight families and Plasmodium sp. P31 (DQ839060.1) was found in three weaver bird species.
Conclusion
This study provides the first insight into avian malaria transmission in the Maromizaha rainforest in eastern Madagascar. Five Haemoproteus lineages and seven Plasmodium lineages were detected in the examined mosquitoes. Complete life-cycles for the specialist lineages WA46 and P31 and for the generalist lineages mosquito132 and PV12 of Plasmodium are proposed. In addition, we have identified for the first time Anopheles mascarensis and Uranotaenia spp. as vectors for avian malaria and offer the first description of vector mosquitoes for avian malaria in Madagascar.
Similar content being viewed by others
Background
The island of Madagascar is located approximately 400 km east of Africa in the Indian Ocean. Due to its isolation from mainland India and Africa it has many endemic species and is classified as an important biodiversity hotspot [1]. More than half of Madagascar′s breeding birds are endemic. This makes Madagascar an interesting area for the examination of the ecological and evolutionary dynamics of vector-host-parasite associations of avian malaria.
Avian malaria is caused by haemosporidian parasites including the genera Plasmodium, Haemoproteus and Leucocytozoon. In contrast to human malaria, avian malaria is distributed almost worldwide and has been detected in a wide range of species [2]. The three genera have similar life-cycles with differences during the asexual phase in the peripheral blood of their host [3]. The life-cycle involves the sexual phase and sporogony both occurring in the invertebrate host (vector) and the merogony and development of gametocytes which takes place in avian hosts [4]. For natural transmission of Plasmodium to the vertebrate host, the parasite undergoes a series of obligatory developmental, propagative and migrational processes inside a mosquito vector. These include zygote formation and ookinete development in the midgut lumen, oocyst formation and sporogony on the basal side of the midgut, sporozoite migration through the haemocoel, and invasion of the salivary glands [5]. Sporozoites, the infective stages, are then present in the salivary gland (thoracic part) of a mosquito and detectable for several weeks [6]. Since the end of the 20th Century, a number of field and laboratory experimental infection studies were used to identify dipterans as vectors transmitting haemosporidian parasites (e.g. [7, 8]). Recently, most studies use molecular methods to identify vector feeding preferences [9] and sporogonic stages of the parasites in salivary glands to identify potential vectors [10].
To date, few studies exist on blood parasites of Malagasy birds. Either blood samples were examined microscopically [11, 12] or just a small number was analyzed by PCR [13]. So far, data about vectors in Madagascar transmitting avian malaria parasites are lacking. To identify mosquitoes as suitable vectors for Plasmodium species, a demonstration of infective sporozoites in insects is essential. Due to predominant light infections and low parasite prevalence (often < 1%) in the majority of vector populations, the applicability of insect dissection and microscopic examination methods is limited in wildlife [6]. For parasite detection we therefore used a highly sensitive nested PCR. Positive results allowed us to determine significant links between mosquitoes and haemosporidian species.
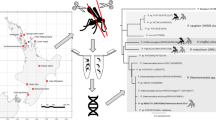
The purpose of our study was to assess the presence of avian malaria parasites in potential mosquito vectors and avian hosts using molecular techniques and to estimate infection rates among mosquito populations in the Maromizaha rainforest located in eastern Madagascar (Andasibe). In addition, Plasmodium spp. sequences isolated from mosquitoes were compared to those found in birds within the study area. A complete life-cycle with putative vector, host and parasite is proposed.
Methods
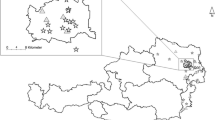
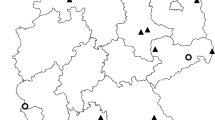
Mosquitoes and birds were caught in the Maromizaha rainforest located in the eastern part of Madagascar (18°56′49″S, 48°27′33″E), 30 km from Moramanga city, with an altitude above 943 m and a highest peak of 1,213 m. The area is protected and part of the Mantadia-Maromizaha-Zahamena rainforest corridor, a terrain that consists of hills with mountain ridges, valleys and small streams, dense and humid evergreen forest covers.
A total of 418 adult mosquitoes of at least 18 species (Table 1) were collected using twelve CDC (Center for Disease Control and Prevention) light traps [14] during one week (November 17–21, 2014). The sampling period was based on the knowledge of high mosquito diversity and density in forested areas of the central highland at the beginning of the rainy season (i.e. November) [15]. The traps were operated for 12 h periods from 6 pm to 6 am at each sampling point. To sample a representative mosquito population, traps were distributed along three sampling lines, thus, valley-floor, slope and ridge for each biotope, with four traps in each sampling point. Mosquitoes were anesthetized with chloroform vapor and identified using the keys of Ravaonjanahary [16] for Aedes, Grjebine [17] for Anopheles, Doucet [18] for Coquillettidia, Edwards [19] for Culex, Brunhes & Hervy [20] for Orthopodomyia and da Cunha Ramos [21] for Uranotaenia. After drying, mosquitoes were stored in ELISA plates to allow identification. DNA extractions were performed separately from head, thorax and abdomen of each mosquito in order to determine the infection rates and diversity of parasite lineages within the thoracic (including salivary glands) and abdominal parts, using the Zymo Research extraction kit (Quick-gDNA™ MiniPrep; Zymo Research Europe GmbH, Freiburg, Germany). The isolated DNA was stored at -20 °C until used.
A total of 686 blood samples were collected from 45 bird species mist-netted in the Maromizaha rainforest in the years 2006, 2007, 2010, 2012 and 2014. Sampling was done by taking a drop of blood from the brachial vein and stored in buffer [22]. For DNA extraction we used QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions. DNA was stored at -20 °C until further use.
To prevent contamination with target DNA, a negative control was included in each test run as well as a positive control to ensure PCR was working properly. Target sequence for amplification was a part of the haemosporidian mitochondrial cytochrome b gene. PCR was conducted in two steps whereby the primer pair HAEMNF and HAEMNR2 [23] was used to amplify a 580 bp fragment in the first PCR, and the internal primer pair HAEMF and HAEMR2 [24] used to amplify a 524 bp fragment in the second PCR. The reaction mixture consisted of 10 mM Tris-HCl, 50 mM KCl, 2 mM MgCl2, 20 pmol of each primer, 200 μM of each dNTP, 1.25 units Ampli-Taq (Applied Biosystems, Carlsbad, USA) and for the first PCR approximately 10–100 ng DNA in a total volume of 50 μl. For the second PCR 2 μl of the amplification product was used as template. Both PCRs were performed for 40 cycles with each cycle consisting of denaturation at 94 °C for 30 s, annealing at 50 °C for 60 s and elongation at 72 °C for 45 s. After amplification, 5 μl of the PCR products stained with GelRed™ (BIOTREND, Köln, Germany) were viewed on a 1.5% agarose gel. Amplification products were then purified using the PCR Product Purification Kit (Roche, Mannheim, Germany) and after sequencing (GATC Biotech AG) the resulting sequences were compared to sequences published in GenBank [25].
Results
A total of 418 mosquitoes were examined, representing at least 18 species of seven genera (Table 1). Culex (46.9%) and Uranotaenia (47.6%) were the two most frequent mosquito genera collected in the study. Other mosquito species found include those of the genera Aedes, Anopheles, Coquillettidia, Lutzia and Orthopodomyia. Using the two step PCR, we found 21 of the 418 mosquitoes (5.02%) to be haemosporidian DNA-positive.
These positive samples were of the mosquito species Anopheles mascarensis, Culex annulioris, C. pipiens, Uranotaenia alboabdominalis, Uranotaenia neireti, Uranotaenia n. sp. (Tantely, pers. Comm.) and Uranotaenia sp. The haemosporidian DNA detected include five Haemoproteus lineages, isolated from nine mosquitoes and seven Plasmodium lineages from 12 mosquitoes (Table 2).
Only mosquitoes with positive thoracic parts are considered to be putative vectors since infectious sporozoites are exclusively present in the salivary glands of arthropod vectors. The resulting haemosporidian DNA sequences of mosquitoes with positive thoracic parts therefore include Plasmodium sp. Uan1 (n = 1; from Uranotaenia n. sp.); Plasmodium sp. WA46 (EU810628.1) (n = 3; from Uranotaenia sp., Ur. neireti and Ur. alboabdominalis); Plasmodium sp. Ual1 (n = 1; from Ur. alboabdominalis); Plasmodium sp. Cp1 (n = 1; from Culex pipiens); Plasmodium sp. mosquito132 (AB308050.1) (n = 2; from Culex pipiens, Uranotaenia n. sp.); Plasmodium sp. PV12 (GQ150194.1) (n = 1; from Anopheles mascarensis); and Plasmodium sp. P31 (DQ659572.1) (n = 1; from Uranotaenia n. sp.). The sequences were compared to those found in bird blood samples collected in the years 2006, 2007, 2010, 2012 and 2014.
The Plasmodium sp. mosquito132 lineage was found in 51 bird blood samples from 15 bird species (Foudia omissa, Xanthomixis cinereiceps, Nesillas typica, Hypsipetes madagascariensis, Copsychus albospecularis, Zosterops maderaspatanus, Monticola sharpei, Ploceus nelicourvi, Foudia madagascariensis, Xanthomixis zosterops, Saxicola torquatus, Tylas eduardi, Philepitta castanea and Pseudobias wardi) belonging to eight bird families (Acrocephalidae, Bernieridae, Muscicapidae, Philepittidae, Ploceidae, Pycnonotidae, Vangidae and Zosteropidae).
Plasmodium sp. P31 was also found in 39 blood samples of weaver birds (Ploceidae), Foudia omissa (n = 35), Foudia madagascariensis (n = 3) and Ploceus nelicourvi (n = 1). The lineage Plasmodium sp. PV12 was found in 37 blood samples of 11 bird species (Foudia omissa, Saxicola torquatus, Philepitta castanea, Ploceus nelicourvi, Nesillas typica, Bernieria madagascariensis, Hypsipetes madagascariensis, Phedina borbonica, Foudia madagascariensis, Copsychus albospecularis, Motacilla flaviventris) belonging to eight bird families (Acrocephalidae, Bernieridae, Hirundinidae, Motacillidae, Muscicapidae, Philepittidae, Ploceidae and Pycnonotidae).
Plasmodium sp. WA46 was as well found in five Madagascar Bulbul (Hypsipetes madagascariensis, Pycnonotidae) blood samples.
Discussion
Blood-sucking arthropods are necessary for the life-cycle of avian malaria parasites. They act as definitive hosts and vectors of avian malaria parasites and are thus responsible for the transmission. Identification of such vector arthropods is therefore essential to unravel the transmission cycles of vector borne diseases like avian malaria [26]. For many parasite species of the genera Plasmodium, Haemoproteus or Leucocytozoon suitable arthropod vectors have already been identified [27]. Mosquitoes are the only known vectors for Plasmodium species.
To identify vectors of avian malaria on Madagascar, we examined 418 mosquitoes from at least 18 species individually using a part of the mitochondrial cytochrome b gene. Sequences found in the mosquitoes were compared to a large dataset of sequences isolated from 45 bird species (n = 686) of the same area. Twenty-one mosquitoes were found to contain DNA of avian Haemosporida.
We found Haemoproteus DNA in nine mosquitoes. Haemoproteus species develop oocysts in the head, thorax and midgut wall of mosquitoes but undergo abortive sporogonic development [6]. Therefore, mosquitoes are not competent vectors for this parasite. Haemosporidian parasites can be used to determine links between blood-sucking insects and their blood-source animals [6]. All identified Haemoproteus lineages in the present study were similar or highly homologous to sequences previously isolated from birds. This evidence therefore implicates that Uranotaenia alboabdominalis, Uranotaenia n. sp., Culex annulioris and Culex pipiens feed on birds in the study area and could be possible vectors for Plasmodium species.
Seven Plasmodium lineages were detected in 12 mosquito species. Of all examined mosquito species in this study containing Plasmodium DNA, only Culex pipiens has been previously known to be a suitable vector for avian Plasmodium species [27]. Culex pipiens was the most abundant mosquito in our study (39%) followed by the newly-described species Uranotaenia n. sp. (22.7%). We detected DNA of Plasmodium only in three Culex pipiens samples (2× thorax, 1× abdomen), indicating that this mosquito species may not play the most important role for avian Plasmodium transmission in the study area.
The endemic mosquito species Anopheles mascarensis is reported to act as a vector of Plasmodium falciparum, the causative agent of human malaria [28]. We found an avian Plasmodium lineage in the thorax of one sampled Anopheles mascarensis indicating that this endemic mosquito species plays an important role for both, human and avian malaria transmission.
Ten of the 12 (83.3%) Plasmodium lineages found were isolated from mosquitoes belonging to the genus Uranotaenia. This is the first report of Uranotaenia species acting as vectors for avian malaria. Although further studies are needed, the genus Uranotaenia might even be the major vector for avian malaria in this area.
The isolated Plasmodium lineages were compared to previously published sequences in GenBank and thus identified. We isolated the sequence Plasmodium sp. Ual1 from the thorax of one Uranotaenia alboabdominalis. The sequence was 95% homologous to the previously described lineage Plasmodium minuoviride haplotype CCA0640 (EU834703.1). This lineage was isolated by Perkins et al. [29] from Prasinohaema prehensicauda, a skink (Squamata) endemic to Papua New Guinea. Because species of the genus Uranotaenia are known to feed on cold-blooded animals [15] and due to the high homology we conclude that the sequence we found also stems from a Plasmodium species infecting reptiles. The lineages Plasmodium sp. Uan1 (isolated from thorax of Uranotaenia n. sp.) and Plasmodium sp. Cp1 (isolated from thorax of Culex pipiens) were highly homologous to sequences previously isolated from birds. Therefore, we conclude that these sequences represent two new Plasmodium lineages infecting birds. The other four sequences isolated were identical with lineages previously described and were also found in bird blood samples examined during this study. The presence of the identical haemosporidian cytochrome b sequence in birds and mosquitoes could be an indication of a vector-host relationship between the species described.
We isolated the lineage Plasmodium sp. WA46 [30] from four mosquitoes belonging to the genus Uranotaenia. The parasite sequence was found in the thoracic parts of Uranotaenia alboabdominalis, Ur. neireti and in one undescribed Uranotaenia species. We conclude that all three mosquito species are probably suitable vectors for the Plasmodium lineage WA46. The very same lineage was also found in five bird blood samples of the Madagascar bulbul (Hypsipetes madagascariensis, Pycnonotidae). The blood was sampled from different individuals in the years 2007, 2010, 2012 and 2014 suggesting a stable occurrence in the study area. The lineage Plasmodium sp. WA46 was originally isolated by Beadell et al. [30] from the common bulbul (Pycnonotus barbatus, Pycnonotidae). Our findings support the hypothesis, that the lineage WA46 is a specialist for the bird family Pycnonotidae. As Hypsipetes madagascariensis is the only bulbul species on Madagascar we assume that the life-cycle of Plasmodium sp. WA46 in our study area includes at least three vector mosquito species of the genus Uranotaenia and the host bird Hypsipetes madagascariensis.
The lineage Plasmodium sp. mosquito132 [26] was isolated from the thoracic parts of Culex pipiens and Uranotaenia n. sp. in this study. Ejiri et al. [26] originally isolated the lineage from Culex pipiens quinquefasciatus captured in Japan. We supported the role of Culex pipiens as vector for this lineage also on Madagascar but with Uranotaenia n. sp. we found a second vector mosquito. We isolated DNA of Plasmodium sp. mosquito132 also from 51 bird blood samples. The samples stem from individuals of 15 bird species belonging to eight bird families. The finding of the lineage in so many diverse bird species leads us to the conclusion that Plasmodium sp. mosquito132 is a generalist.
From the endemic mosquito Anopheles mascarensis we isolated the lineage Plasmodium sp. PV12 [31]. The lineage was originally isolated from the mosquito Coquillettidia aurites in Cameroon. We did not find the lineage in the four samples of Coquillettidia grandidieri examined in this study. Because of the small sample size we cannot exclude this mosquito species as a possible vector. Our findings suggest that Anopheles mascarensis is a vector of Plasmodium sp. PV12 in the study area. We also detected this lineage in 37 bird blood samples. The samples are from eleven 11 bird species belonging to eight families. The finding of the lineage in so many diverse bird species leads us to the conclusion that Plasmodium sp. PV12 is a generalist.
The Plasmodium lineage isolated from Uranotaenia n. sp. was identical to the previously described Plasmodium sp. P31 [32]. The lineage was originally isolated from Foudia madagascariensis (Ploceidae) captured on Madagascar. In the current study the same sequence was isolated from 39 blood samples of three weaver bird species, including Foudia omissa, Foudia madagascariensis and Ploceus nelicourvi. Therefore, Plasmodium sp. P31 seems to be specific to weaver birds (Ploceidae). Fodies were among the most frequently sampled birds. From observational studies they are known to move from the forest to the open, more degraded areas, and may therefore spread blood parasites into more pristine areas. The weaver bird lineage was detected in 39 samples from three different years (2006, 2007, 2010, 2012 and 2014). The high and apparently stable prevalence in the birds suggests that there is a stable life-cycle of Plasmodium sp. P31 on Madagascar with its intermediate host being weaver birds and Uranotaenia n. sp. as the first described vector.
To confirm the role of the genus Uranotaenia, Anopheles mascarensis and Culex pipiens in the transmission cycle of the reported avian malaria parasites on Madagascar, further analyses of blood-fed mosquitoes from infection experiments are required.
Conclusions
This study provides the first insight into avian malaria transmission in the Maromizaha forest in eastern Madagascar. Five Haemoproteus lineages and seven Plasmodium lineages were detected in the examined mosquitoes. Complete life-cycles for the specialist Plasmodium lineages WA46 and P31 and for the generalist Plasmodium lineages mosquito132 and PV12 are proposed. Plasmodium sp. WA46 is transmitted from mosquitoes of the genus Uranotaenia to birds belonging to the family Pycnonotidae. Plasmodium sp. P31 is transmitted from Uranotaenia n. sp. to birds belonging to the family Ploceidae. Plasmodium sp. mosquito132 is transmitted from Culex pipiens and Uranotaenia n. sp. to at least 15 different bird species on Madagascar. Plasmodium sp. PV12 is transmitted from Anopheles mascarensis to at least 11 different bird species on Madagascar.
This study identified Anopheles mascarensis and Uranotaenia spp. as putative vectors for avian malaria for the first time and offers the first description of vector mosquitoes for avian malaria on Madagascar.
References
Myers N, Mittermeier R, Mittermeier CG, da Fonseca G, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–8.
Atkinson CT, Van Riper C. Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Leucocytozoon and Haemoproteus. In: Bird-parasite interactions: ecology, evolution, and behavior. London: Oxford University Press; 1991. p. 19–48.
Valkiunas G. Avian malaria parasites and other Haemosporidia. Boca Raton: CRC Press; 2005.
Ishtiaq F, Guillaumot L, Clegg SM, Phillimore AB, Black RA, Owens IPF, et al. Avian haematozoan parasites and their associations with mosquitoes across Southwest Pacific Islands. Mol Ecol. 2008;17(20):4545–55.
Hillyer JF, Barreau C, Vernick KD. Efficiency of salivary gland invasion by malaria sporozoites is controlled by rapid sporozoite destruction in the mosquito haemocoel. Int J Parasitol. 2007;37:673–81.
Valkiunas G, Kazlauskiene R, Bernotiene R, Palinauskas V, Iezhova TA. Abortive long-lasting sporogony of two Haemoproteus species (Haemosporida, Haemoproteidae) in the mosquito Ochlerotatus cantans, with perspectives on haemosporidian vector research. Parasitol Res. 2013;112(6):2159–69.
Atkinson T, Greiner EC, Forrester J. Wild vectors in Florida of Haemoproteus meleagridis Levine from turkeys. J Wildl Dis. 1983;19:366–8.
Huff CG. Susceptibility of mosquitoes to avian malaria. Exp Parasitol. 1965;16:107–32.
Imura T, Sato Y, Ejiri H, Tamada A, Isawa H, Sawabe K, et al. Molecular identification of blood source animals from black flies (Diptera: Simuliidae) collected in the alpine regions of Japan. Parasitol Res. 2010;106(2):543–7.
Valkiūnas G, Santiago-Alarcon D, Levin II, Iezhova TA, Parker PG. A new Haemoproteus species (Haemosporida: Haemoproteidae) from the endemic Galapagos dove Zenaida galapagoensis, with remarks on the parasite distribution, vectors, and molecular diagnostics. J Parasitol. 2010;96(4):783–92.
Savage AF, Robert V, Goodman SM, Raharimanga V, Raherilalao MJ, Andrianarimisa A, et al. Blood parasites in birds from Madagascar. J Wildl Dis. 2009;45(4):907–20.
Barraclough R, Robert V, Peirce M. New species of haematozoa from the avian families Campephagidae and Apodidae. Parasite. 2008;15:105–10.
Ishtiaq F, Beadell JS, Warren BH, Fleischer RC. Diversity and distribution of avian haematozoan parasites in the western Indian Ocean region: a molecular survey. Parasitology. 2012;139(2):221–31.
Boyer S, Tantely ML, Randriamaherijaona S, Andrianaivolambo L, Cardinale E. Mosquitoes sampling strategy for studying West Nile virus vectors in Madagascar: abundance, distribution and methods of catching in high risk areas. Arch Inst Pasteur Madagascar. 2014;71(1):1–8.
Tantely M, Rakotoniaina J, Andrianaivolambo L, Tata E, Razafindrasata F, Fontenille D, Elissa N. Biology of mosquitoes that are potential vectors of Rift Valley fever virus in different biotopes of the central highlands of Madagascar. J Med Entomol. 2013;50:603–10.
Ravaonjanahary C. Les Aedes de Madagascar (Diptera-Culicidae). Paris: O.R.S.T.O.M; 1978.
Grjébine A. Insectes Diptères Culicidae Anophelinae. Paris: O.R.S.T.O.M; 1966.
Doucet J. Étude des Culicidae de la région de Vangaindrano (Diptera). Paris: Mémoires de l’Institut Scientifique de Madagascar; 1951.
Edwards FW. Mosquitoes of the Ethiopian Region. III.- Culicine adults and pupae. London: British Museum (Natural History); 1941.
Brunhes J, Hervy J. Insectes Diptères Culicidae Culicinae Genre Orthopodomyia de la sous-région malgacheet de la région afrotropicale. Paris: Muséum National d’Histoire Naturelle; 1995.
Da Cunha Ramos H, Brunhes J. Insecta, Diptera, Culicidae, Uranotaenia. Paris: IRD-CIRAD; 2004.
Wink M. Use of DNA markers to study bird migration. J Ornithol. 2006;147:234–44.
Waldenström AJ, Bensch S, Hasselquist D, Östman Ö. A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J Parasitol. 2004;90(1):191–4.
Bensch S, Stjernman M, Hasselquist D, Orjan O, Hannson B, Westerdahl H, Pinheiro RT. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc Biol Sci. 2000;267(1452):1583–9.
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:36–42.
Ejiri H, Sato Y, Sasaki E, Sumiyama D, Tsuda Y, Sawabe K, et al. Detection of avian Plasmodium spp. DNA sequences from mosquitoes captured in Minami Daito Island of Japan. J Vet Med Sci. 2008;70:1205–10.
Santiago-Alarcon D, Palinauskas V, Schaefer HM. Diptera vectors of avian haemosporidian parasites: Untangling parasite life cycles and their taxonomy. Biol Rev. 2012;87:928–64.
Fontenille D, Campbell G. Is Anopheles mascarensis a new malaria vector in Madagascar? Am J Trop Med Hyg. 1992;46:28–30.
Perkins SL, Austin CC. Four new species of Plasmodium from New Guinea Lizards: Integrating morphology and molecules. J Parasitol. 2009;95(2):424–33.
Beadell JS, Covas R, Gebhard C, Ishtiaq F, Melo M, Schmidt BK, et al. Host associations and evolutionary relationships of avian blood parasites from West Africa. Int J Parasitol. 2009;39(2):257–66.
Njabo KY, Cornel AJ, Sehgal RN, Loiseau C, Buermann W, Harrigan RJ, et al. Coquillettidia (Culicidae, Diptera) mosquitoes are natural vectors of avian malaria in Africa. Malar J. 2009. doi:10.1186/1475-2875-8-193.
Beadell JS, Ishtiaq F, Covas R, Melo M, Warren BH, Atkinson CT, et al. Global phylogeographic limits of Hawaii’s avian malaria. Proc Biol Sci. 2006;273(1604):2935–44.
Acknowledgements
We are indebted to the Malagasy authorities for granting all the relevant research and export permits and to GERP (Grouped'Etude et de Recherche sur les Primates de Madagascar), namely Jonah Ratsimbazafy and Rose Marie Randrianarison, for letting us work at Maromizaha. The work would not have been possible without the continued support over many years by Haja Rakotomanana and Daniel Rakotondravony from the University of Antananarivo, Department of Animal Biology. We thank Nicola Lillich, Jean-Robert Lekamisi and all the other Malagasy assistants for their help in the field. We would like to thank all field assistants, particulary Pia Reufsteck, and also Sonja Dumendiak and Katrin Fachet for their help in the laboratory.
Funding
Not applicable.
Availability of data and materials
Data generated or analysed during this study are partially included in this published article. The whole datasets generated during and/or analysed during the current study are not publicly available due to the use in further publications but are available from the corresponding author on reasonable request.
Authors’ contributions
SS carried out the molecular analysis and drafted the manuscript. AD and UM participated in the coordination of the study and helped drafting the manuscript. AD also carried out molecular analysis. SB facilitated the field study that was carried out by MLT and FJR involving capture and morphological identification of the mosquitoes in the field. FW had the idea for the study, participated in its design and coordination and helped to write the manuscript. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Schmid, S., Dinkel, A., Mackenstedt, U. et al. Avian malaria on Madagascar: bird hosts and putative vector mosquitoes of different Plasmodium lineages. Parasites Vectors 10, 6 (2017). https://doi.org/10.1186/s13071-016-1939-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1939-x