Abstract
Background
The intestine of hookworms contains enzymes and proteins involved in the blood-feeding process of the parasite and is therefore a promising source of possible vaccine antigens. One such antigen, the hemoglobin-digesting intestinal aspartic protease known as Na-APR-1 from the human hookworm Necator americanus, is currently a lead candidate antigen in clinical trials, as is Na-GST-1 a heme-detoxifying glutathione S-transferase.
Methods
In order to discover additional hookworm vaccine antigens, messenger RNA was obtained from the intestine of male hookworms, Ancylostoma ceylanicum, maintained in hamsters. RNA-seq was performed using Illumina high-throughput sequencing technology. The genes expressed in the hookworm intestine were compared with those expressed in the whole worm and those genes overexpressed in the parasite intestine transcriptome were further analyzed.
Results
Among the lead transcripts identified were genes encoding for proteolytic enzymes including an A. ceylanicum APR-1, but the most common proteases were cysteine-, serine-, and metallo-proteases. Also in abundance were specific transporters of key breakdown metabolites, including amino acids, glucose, lipids, ions and water; detoxifying and heme-binding glutathione S-transferases; a family of cysteine-rich/antigen 5/pathogenesis-related 1 proteins (CAP) previously found in high abundance in parasitic nematodes; C-type lectins; and heat shock proteins. These candidates will be ranked for downstream antigen target selection based on key criteria including abundance, uniqueness in the parasite versus the vertebrate host, as well as solubility and yield of expression.
Conclusion
The intestinal transcriptome of A. ceylanicum provides useful information for the identification of proteins involved in the blood-feeding process, representing a first step towards a reverse vaccinology approach to a human hookworm vaccine.
Similar content being viewed by others
Background
Human hookworm infection remains one of the leading neglected tropical diseases (NTDs), affecting more than 400 million people living in developing countries and causing the loss of about 3.2 million disability adjusted life years (DALYs) [1, 2]. Hookworm is a blood-feeding nematode that hooks onto the host intestinal mucosa using sharp teeth or cutting plates, causing significant host blood loss. As a consequence, hookworm infection is the major cause of iron-deficiency anemia in endemic regions [3]. Current control of hookworm infection mainly relies on mass drug administration with a single annual dose of an anthelmintic such as albendazole or mebendazole. However, a recent systematic analysis revealed that anthelmintic treatment with mebendazole had no impact on the improvement of anemia in hookworm-infected regions [4] and is associated with a low cure rate overall [5, 6], while the cure rates with albendazole were highly variable [7]. Therefore, development of a vaccine has emerged as a practical and feasible alternative technology to control hookworm infection or complement anthelmintic drug treatment [1, 8]. A human hookworm vaccine is considered both cost-effective and cost-saving relative to mass drug administration [9].
Hookworm infection begins when infective larvae penetrate the skin of the host and migrate through the circulatory system and the lungs before reaching the intestine, where they develop into adult worms and start blood-feeding [3]. The hookworm’s survival exclusively depends on blood-feeding and the digestion of blood proteins including hemoglobin and serum proteins as a major source of nutrition [10]. During the past decade, the molecular basis of this blood-feeding process and the digestion has been partially identified as a cascade of hemoglobinases [11]. After the worm takes up blood into the intestine, the red blood cells are lysed by hemolysin and hemoglobin is released. The released hemoglobin is degraded and digested by a series of hemoglobinases, initiating cleavage of the hemoglobin molecule by an aspartic protease (APR) [12, 13], followed by further digestion with several cysteine proteases [10, 14] and metalloproteinases [15]. These proteases are expressed in the brush border membrane of the parasite's intestine [10, 15, 16]. A vaccine development strategy is in process that focuses on interfering with the blood-feeding process of the hookworm. It targets proteins, including enzymes, found in the gut of the major human hookworm, Necator americanus, and involved in the degradation of hemoglobin and the detoxification of breakdown products of the process [1]. Vaccination elicits anti-enzyme antibodies that can reach the hookworm intestine through blood feeding and subsequently inactivate their target enzymes in the hookworm’s digestive tract and other organs. Among the lead candidate antigens are Na-APR-1, a cathepsin-D aspartic protease required for hemoglobin digestion and Na-GST-1, a glutathione-S transferase involved in detoxification of toxic heme derived from hemoglobin digestion – both molecules induce significant protective immunity in vaccinated animals against hookworm infection [16, 17]. These two antigens have been selected as the leading hookworm vaccine antigens for product development and are currently in phase 1 clinical trials [1, 8]. Ac-CP-2, another cysteine protease of the canine hookworm Ancylostoma caninum, was also determined to be expressed on the intestine brush-border membrane. Like Na-APR-1, laboratory animals vaccinated with recombinant Ac-CP-2 also produced antibodies that stunted worm development and reduced egg count after being challenged with hookworm larvae [18]. In addition to their efficacy in preclinical studies, a second rationale for choosing hookworm gut antigens is that antigens from the hookworm intestine are not directly exposed to the host immune system during natural infection, thereby reducing the likelihood of eliciting host IgE responses and allergic antibodies. This issue had previously thwarted other vaccine development efforts for infective larval stage-derived antigens [19].
In an effort to identify additional antigens in the hookworm intestine that might be suitable for the development of novel vaccine antigens and therapeutics, we undertook an antigen discovery program. Based on the success of previous reverse vaccinology programs for selecting bacterial vaccine antigens [20], we are interested in initiating a similar approach for the human hookworm vaccine based on the gut transcriptome. Such antigens could be added to, or considered alternatives to, the current candidates, depending on the progression of the clinical trials. We believe that the complete profile of the hookworm intestine’s gene expression pattern could significantly facilitate this selection process. For this study we used Ancylostoma ceylanicum as a model, a hookworm that infects both animals (e.g. dogs, hamsters) and humans (in focal areas of Malaysia and elsewhere in Southeast Asia) and expresses proteins sharing high homology with the two major human hookworms N. americanus and A. duodenale [21] which only infect humans and cannot properly infect laboratory animals without the use of immunosuppressant [22]. The full transcriptome of the A. ceylanicum adult intestine was sequenced and analyzed. The results reveal some new macromolecules likely involved in parasite survival in the mammalian host with the potential to serve as future vaccine candidates or drug targets for controlling hookworm infections.
Methods
Preparation of A. ceylanicum intestinal RNA
Hookworm A. ceylanicum was maintained in hamsters according to protocol AN-5762 approved by Baylor College of Medicine’s Institutional Animal Care and Use Committee (IACUC). Fresh A. ceylanicum adult worms were recovered from the intestines of hamsters 21 days after infection with A. ceylanicum larvae [23]. In order to avoid contamination from the worm’s reproductive organs and eggs, only male worms were collected. Total hookworm intestinal tissue was isolated from 50 male adult worms. The anterior esophagus and any contaminating reproductive organs were removed. The isolated intestines were preserved immediately in RNAlater (Qiagen, Hilden, Germany) to prevent degradation of RNA. Total RNA was extracted from the hookworm intestines using Trizol reagent (Invitrogen, Carlsbad, CA, USA), then digested with Ambion Turbo DNase (Ambion/Applied Biosystems, Austin, TX, USA) to remove any contaminated chromosomal DNA. The quality and yield of the purified RNA were assessed using an Agilent 2100 bioanalyzer (Agilent Technologies, Cedar Creek, TX, USA) using the ratio of 28S/18S rRNA and a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The integrity of the purified RNA was determined using the RNA Integrity Number [24].
RNA-seq library construction and Illumina sequencing
Messenger RNA (mRNA) was prepared from the purified total RNA using the MicroPoly(A)Purist™ Kit (Ambion/Applied Biosystems, Austin, TX, USA), and then converted to cDNA using Ovation®’s RNA-Seq V2 kit (NuGen Technologies, Inc., San Carlos, CA, USA) with both a poly-A and a random primer. An Illumina sequencing library was constructed according to the manufacturer's instructions (Illumina Inc., San Diego, CA, USA). Briefly, 1 μg cDNA was sheared using a Covaris S220 DNA Sonicator (Covaris Inc., Woburn, MA, USA) and fragments with a size range of 200–400 bp were collected. Single Primer Isothermal Amplification (SPIA) adapters were added to those fragments, followed by PCR amplification to enrich adaptor-ligated fragments. Quantitative PCR (Kapa Biosystems Inc., Woburn, MA, USA) was used to determine the concentration of the resulting library and to produce the cluster counts appropriate for the Illumina platform. The library was finally loaded into the flow cells of the HiSeq2000 sequencer to generate paired end reads of approximately 100 bp in length [21].
The mRNA-sequence reads were processed using an in-house Perl script to remove adaptor sequences as well as those with low complexity or derived from non-target sources (golden hamster genome database, GCF_000349665.1, and GenBank Bacterial Sequences database, GBBCT). The processed reads were mapped to the unpublished A. ceylanicum genome (available at http: nematode.net [25]), using Tophat 2.0.8 [26] with the parameter mate-inner-distance set to 300. Breadth of coverage and read count of individual A. ceylanicum gene transcripts were calculated using Refcov, version 0.3 (http://gmt.genome.wustl.edu/packages/refcov/) [27] and HTSeq-count (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html) [28], respectively. Genes with at least 50 % breadth of coverage were considered to be expressed in the male A. ceylanicum intestine. The corresponding counts were used to represent the expression abundances of those genes. The significantly abundant gene transcripts in A. ceylanicum intestine were identified with the Fisher’s exact test in Bioconductor package edgeR on the basis of comparison of the gene transcript counts in the intestinal transcriptome with the whole male worm transcriptome. A corrected false discovery rate (FDR) cutoff of 0.05 was used.
Quantitative reverse transcription PCR (qRT-PCR)
To validate the results from the RNA-seq experiments, five genes with higher expression levels in the intestine than in the whole worm were chosen for quantitative reverse transcription PCR (qRT-PCR). Total RNA was freshly extracted from whole worms and isolated intestines of A. ceylanicum. Total cDNA was reverse-transcribed using the SuperScript III First-Strand Synthesis System per the manufacturer’s protocol (Invitrogen, Waltham, USA). Quantitative PCR was performed using a PowerUp™ SYBR® Green (Life Technologies, Carlsbad, USA) cocktail in a ViiA7 real-time PCR detection system (Applied Biosystems). The PCR conditions were 95 °C for 1 min, followed by 35 cycles of 95 °C for 25 s, 55 °C for 20 s, 72 °C for 1 min, and concluded by a 10 min incubation step at 72 °C. The 18S rRNA of A. ceylanicum (DQ464371.1) was selected as a standard comparison control gene.
Bioinformatics analysis
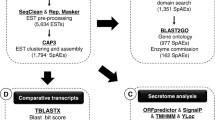
The functional annotations based on primary sequence level was done using Interproscan (https://www.ebi.ac.uk/interpro/), and further classified into different Gene Ontologies (GO). We further added various sub-categories within each GO-ontology and weighted them with FPKM (Fragments Per Kilobase of transcript per Million mapped reads, a measure of the relative expression of an individual transcript) values for each gene. Those transcripts highly expressed in the worm’s intestine were determined through comparison with data from the whole adult male transcripts available from the SRA for A. ceylanicum (accession numbers: SRX1116908 and SRX1116915) downloaded and employed in this study. The Merops database (https://merops.sanger.ac.uk/), a manually curated peptidase and inhibitor database, was used to annotate and classify peptidases in order to identify which proteases or peptidases are highly expressed in the hookworm intestine. Pepunit.lib sequences from release 9.12 were downloaded from the website (http://merops.sanger.ac.uk/download/) and used to create a custom Blastp database using BLAST command line tools and an e-value of less than 10-6. The hits were weighted by using their FPKM values and then summed to plot using proportional values. Molecules involved in transmembrane transport were identified using the TrSSP dataset (http://bioinfo.noble.org/TrSSP/?dowhat=Datasets). The transporter sequences were downloaded from the Transporter Substrate Specificity Prediction Server (TrSSP) and used for BlastP searches against the hookworm intestinal RNA-Seq genes obtained in this study by setting an e-value of less than 10-6. Phylogenetic trees were generated for those functional protein homologues identified in this study using Phylogeny.fr [29] (http://www.phylogeny.fr/index.cgi). Signal peptides were predicted using Phobius Server (http://phobius.sbc.su.se/) [30]. The transmembrane domains were predicted using TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) [31].
Results and discussion
Isolation of hookworm intestines
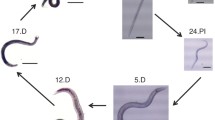
The intestines of hookworms were isolated from living adult male A. ceylanicum worms collected from infected hamsters under microscopy (Fig. 1a). Total RNA was extracted from the isolated intestine and the total RIN for the hookworm intestinal RNA was 8.6 on a scale that extends from 10 (intact) to 2 (degraded), indicating that most of the RNA had remained intact throughout the purification process (Fig. 1b).
The intestines were isolated from A. ceylanicum male worms avoiding esophagus and reproductive organs (a), and total RNA was extracted and treated with DNase. The integrity of the total intestinal RNA was analyzed by electrophoresis and the RNA Integrity Number (RIN) for the purified RNA was 8.6 (b)
Hookworm intestinal transcriptome analysis
The RNA-seq library constructed using the sample obtained above was subjected to Illumina RNA sequencing. The RNA-seq data generated from the adult male intestine of A. ceylanicum is available from NCBI under BioProject ID PRJNA72583 with SRA accession number of SRX1127457. The transcriptional profile analysis revealed that a total of 8,495 genes, versus a total of 15,892 transcripts in the whole genome (BioProject ID PRJNA72583), had been expressed in our sample of the hookworm A. ceylanicum intestine. Among the 8,495 A. ceylanicum intestinal genes, 6,559 have predicted IPR domains according to the InterPro scan analysis. A total of 7.1 % of intestinal transcripts possessed a signal peptide that is statistically higher than that in the whole worm (4.1 %) (Chi-square test, χ 2 = 104.988, P-value < 0.0001), suggesting that more transcripts in intestine encode proteins that secret into the intestine. Hookworm intestine contained 19.0 % of transcripts that contain transmembrane domain, which is similar to that in the whole worm (19.8 %). Secreted or membrane-associated proteins are expected to be involved in the worm’s digestive process. Comparison of the FPKM value for the intestine expressed transcripts with the corresponding gene FPKM-derived from the transcriptome of the whole A. ceylanicum male worm, resulting in 964 genes being expressed at significantly higher level in the A. ceylanicum intestine than in the whole worm based on the Fisher’s exact test in Bioconductor package edgeR [32] (Table 1).
Genes highly expressed in the hookworm intestine
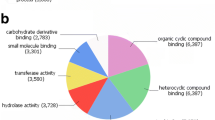
The genes highly expressed in the hookworm intestine are likely involved in blood-feeding and therefore constitute important targets for hookworm vaccine or therapeutic drug development [11]. The 100 genes with the highest FPKM in the intestine are listed in Additional file 1: Table S1, together with a putative functions or characterization. The higher expression levels in the intestine for five selected genes were also confirmed by qRT-PCR using independently prepared batches of RNA samples from whole worms and intestines of A. ceylanicum, with 12.0–68.8 % increase in intestine compared to the whole worm calculated by 2(-ΔΔCt) (Table 2). After comparison of the relative transcript abundance to the whole worm, a gene ontology analysis was carried out on those transcripts highly expressed in the intestine. The analysis determined that those proteins with hydrolytic functions or binding/transporting activities are the most highly expressed genes in the intestine compared to the whole worm, especially those involved in protein digestion and the transport of digested macromolecules such as amino acids, fatty acid, sugar and ions. The top ten most abundant groups of genes expressed in intestine belong to the categories of proteolysis [FPKM intestine/whole worm (ratio) = 37,125/2,971 (12.5)], lipid-binding/transporters [30,077/78 (385.6)], protein-binding [6,317/476 (13.3)], ATP-binding [5,681/435 (13.1)], DNA-binding [4,456/276 (16.1)], membrane [3,289/232 (14.2)], zinc ion-binding [3,163/289 (11)], integral component of membrane [3,061/219 (14)], transferase activity [2,959/212 (14)], other transporters [2,732/164 (16.7)] (Fig. 2). These most abundantly expressed genes and their categorization are also shown in Additional file 1: Table S1.
We further explored various subcategories within the main gene ontologies and weighted those using FPKM values. Those protein categories with the highest expression levels in the intestine are analyzed below.
Proteases
With a total FPKM of 37,125 in the intestine, compared to 2,971 FPKM in the whole worm, proteases constitute the most highly expressed proteins in the worm’s intestinal transcriptome. The Merops database was used to run BLASTP on 8,495 protein sequences from RNA-seq using an e-value of less than 10-6. We obtained 3,473 hits, of which 674 were unique. We determined that 518 of those unique hits were classified as peptidases and 156 of them as peptidase inhibitors.
When peptidases were further sub-classified into aspartic peptidases, cysteine peptidases, metallopeptidases, serine peptidases, threonine peptidases and unknown peptidases, we found that the most common peptidase in the intestine were the cysteine peptidases (2.6 % of the total FPKM), followed by serine peptidases (2.1 %), metallopeptidase (1.4 %), aspartic peptidases (0.7 %) and threonine peptidases (0.2 %). Compared to the whole worm transcripts, the cysteine peptidase, serine peptidase, aspartic peptidase and threonine peptidase were preferentially expressed in the intestine (Fig. 3), indicating their possible involvement in blood digestion.
Searching the intestinal protease profile identified in this study, the aspartic peptidase ANCCEY_13850 shares 89 % amino acid sequence identity with Na-APR-1. Its expression level is high in the intestine, with the ninth highest FPKM level observed in the protease category (Table 3, FPKM 579.3). Moreover, a cysteine protease, ANCCEY_09304, was identified that shares 55.0 % sequence identity with Ac-CP-2 (Table 3). In addition to the homologues of Na-APR-1 or Ac-CP-2 in the A. ceylanicum intestine involved in the hemoglobin digestion, there are additional proteases that may contribute to the proteolysis of hemoglobin or other serum proteins adapted to the blood feeding of hookworm. There were a total of 26 proteases, including serine proteases, cysteine proteases, aspartic proteases, metalloproteases, and aminopeptidases, which were expressed with an at least 8 times higher FPKM in the intestine than in the adult worm except for ANCCEY_13850 with 1.5 times higher (Table 3). These proteases could be the important targets for therapeutic or preventive vaccine or drug development.
Transporters
The hookworm intestine expresses different transporters with functions related to the absorption of digested macromolecules and the maintenance of homeostasis between the worm and outside environment [21, 33, 34]. The specific binding proteins, transporters or pumps on the gut membrane are necessary for the uptake of metabolites such amino acids, glucose, lipids, ions and water after blood feeding digestions [35]. A search against the TrSSP database revealed that virtually all of the categories of transporters are expressed in the intestine at much higher levels than in the whole worm. These highly expressed transporters include amino acid transporters, protein/mRNA transporters, sugar transporters, ion transporters and other transporters. The latter two categories of transporters (cation and other) may indicate their importance in ion exchange and other material transport, such as for lipids, ATP, or other nucleotides (Fig. 4). Amino acids and small peptides derived from blood likely play a key role in hookworm nutrition. However, their uptake and absorption rely on the amino acids/peptide transporters produced by hookworm intestine. Deletion of an intestinal peptide transporter from free-living nematode Caenorhabditis elegans reduced the uptake of digested peptides from the gut lumen, resulting in delayed development, reduced progeny and body size [36].
In addition to the high expression of amino acid/peptide transporters, the genes involved in lipid-binding and transportation are also highly expressed in the hookworm gut (Figs. 2 and 4). Vitellogenin and fatty acid and retinol-binding (FAR) proteins represent two major categories of lipid-binding proteins involved in cross-membrane transport. More than three vitellogenins (ANCCEY_09758, ANCCEY_15699, ANCCEY_01522) are exclusively expressed in the hookworm gut with high FPKM from 4298.3–7970.4 (Additional file 1: Table S1; Table 4). Vitellogenin is a lipid transport and storage protein consisting of four domains including lipovitellin-1, phosvitin, lipovitellin-2, and a von Willebrand factor type D domain (vWFD) [37, 38]. The 3-D structure of vitellogenin reveals that phosvitin and lipovitellin domains form amphipathic structures of a lipid pocket to receive their lipid cargo [39]. The vWFD is located at the cysteine-rich C-terminus of vitellogenin with homology to human von Willebrand factor type D domains. Unlike human vWFD though, metazoan vWFDs do not contain RGD motifs, which are associated with extracellular matrix binding. The conservation of cysteine-rich vWFDs throughout the metazoan, including nematodes, implicates functional importance, possibly protecting vitellogenin from being digested by proteases [39]. The knockdown of vitellogenin in C. elegans had previously been shown to result in reduced survival after bacterial infection, indicating the crucial role of vitellogenin in nematode’s life-cycle [40].
Another category of highly expressed lipid-binding proteins identified in this study includes fatty acid and retinol-binding proteins (FAR). Lipids and retinoids are relatively insoluble in water and, in their oxidative free form, can be toxic to membranes. Consequently, parasitic nematodes secrete a structurally unique class of FAR proteins to sequester fatty acids and retinoids from the host necessary for the worms’ essential biological processes [41–43]. Crystallization of Na-FAR-1 reveals its large and more complex internal ligand-binding cavity for binding to lipid [43]. In this study, we have identified at least three FAR proteins (ANCCEY_04371, ANCCEY_00853 and ANCCEY_04372) with high FPKM values (132.7–411.2).
Glutathione-S transferases (GSTs)
After digestion of hemoglobin, free heme is released into the hookworm intestine. Due to its oxidative iron, free heme is a potent enzyme inhibitor and source of reactive oxygen species that may damage the parasite’s structure and DNA [44]. As in other blood-feeding parasites such as Plasmodium spp. and Schistosoma spp. [35, 45, 46], hookworm has evolved mechanisms to detoxify the free heme by expressing nematode-specific Nu-class GSTs that have higher affinity heme-binding sites through homodimerization of two GST molecules [47–49]. Due to the importance of hookworm GSTs involved in the heme binding and detoxification, vaccination with hookworm GSTs produced protective immunity in laboratory animals against hookworm larval challenge [17, 47, 48], and thus Na-GST-1 from N. americanus has been selected as the leading vaccine candidate for human hookworm infection [48]. To date, three GSTs have been cloned from N. americanus adult worms. Here a total of 13 new GSTs were identified in the transcriptome of the A. ceylanicum adult intestine (Fig. 5). ANCCEY_00737 shares the highest level of similarity with Na-GST-1 (60.0 % amino acid identity). Possibly some of them are involved in the detoxification of heme and carrying of reduced non-toxic heme to functional sites required by this parasite, because heme is a co-factor of some essential enzymes and hookworm is considered as heme auxotrophic [50]; therefore hookworm gut-expressed GSTs could be candidates for vaccine or drug development.
ASP/CAP proteins
Upon stimulation by a host signal during penetration into the skin, arrested hookworm infective larvae assume feeding and development, and simultaneously secrete many proteins related to migration and parasitism in the host. Some of those activated secreted proteins are called Ancylostoma-secreted proteins or activation-associated secreted proteins (ASPs) [51, 52]. This group of proteins exists in a wide range of eukaryotic organisms, including plants, vertebrates and invertebrates including helminths, and belongs to the large family of cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins (CAPs) [53], also known as sperm-coating protein (SCP)-like extracellular proteins, or SCP/Tpx-1/Ag5/PR-1/Sc7 (SCP/TAPS) proteins [54]. Except for their association with establishment of parasitism in the host, the exact function of ASPs remains mostly unknown. Recent studies have demonstrated that Na-ASP-2 of the human hookworm N. americanus binds to the human B-cell antigen receptor complex CD79A and downregulated about 1,000 B-cell messenger RNAs including factors involved in leukocyte transendothelial migration pathways and the B-cell signaling receptor pathway, indicating nematode-secreted ASP/CAP proteins may cause immunomodulation as a strategy of immune evasion [55]. Vaccination with recombinant ASP-1 or ASP-2 secreted by hookworm larvae produced significant protective immunity in different animal models [17, 23, 56]. Unfortunately, the larvae-secreted ASPs induced an IgE response during natural infection and pre-existing anti-Na-ASP-2 IgE caused a significant allergic response upon vaccination with recombinant Na-ASP-2, which has stopped the development of Na-ASP-2 as a vaccine in endemic areas [19]. However, it has been shown that ASP’s fusion to human IgG Fc fragment could significantly reduce the anti-Na-ASP-2 IgE-triggered histamine release [57].
The A. ceylanicum genome contains a total of 432 ASP genes, more than any other hookworm species described so far [21, 34, 58, 59]. Except for those secreted by activated larvae, some well-studied ASPs are only expressed in adult worms [60]. In this study, we identified 62 ASP genes expressed in the hookworm intestine, of which 11 are within the 100 most abundant gene products in the intestine, second only to the protease group (Table 5; Additional file 1: Table S1). Phylogenetic analysis suggests that these ASPs are closely related to the adult worm expressed proteins, Ac-ASP-3, Ac-ASP-4, Ac-ASP-5 and Ac-ASP-6 [60], but less related to the larvae-secreted Ac-ASP-1 [51] and Ac-ASP-2 [52] (Fig. 6). Due to their specific expression in the adult intestine, these proteins are not expected to be exposed to the host immune system, and therefore there is less concern regarding the induction of an IgE response during natural infection, possibly making them suitable vaccine antigen candidates.
C-type lectins
C-type lectins are sugar-binding proteins mediating both pathogen recognition and cell-cell interactions using structurally related Ca++-dependent carbohydrate-recognition domains (C-type CRDs) [61]. Nematodes secrete certain CTLs homologous to some key receptors of the mammalian host immune system, such as CD23 and CD206 [62–64]. It is conceivable that nematode-secreted CTL-like products interfere with the immunological recognition or effector function, suggesting a potential role at the host-parasite interface, possibly as a strategy to evade the host immune response [65]. In C. elegans more than 278 CTL-like genes have been found [66], with some of them induced upon bacterial infection [67]. The A. ceylanicum intestine also expresses an abundant number of C-type lectins. In this study 8 CTLs were identified among the 100 most abundant proteins in the A. ceylanicum intestine (Table 6). Two of them (ANCCEY_05930 and ANCCEY_10784) contain a von Willebrand factor type A domain at their N-terminus, which might make these proteins important in adhesion processes involving platelets, collagen and other proteins. This is especially true for ANCCEY_05930 which is extremely highly abundant in the hookworm intestine (FPKM 20518.0).
Heat shock proteins (HSPs)
HSPs are a family of highly conserved proteins induced upon different kinds of environmental stress such as heat, infection, inflammation, toxins, starvation, hypoxia, pH extremes, and nutrient deprivation [68]. Not only acting as chaperones by stabilizing intracellular proteins to maintain correct folding or by helping those proteins damaged to refold, HSPs also play significant roles in immunomodulation such as antigen presentation, activation of lymphocytes and maturation of dendritic cells [69]. Due to their important roles in parasite survival, HSPs have been chosen as vaccine candidates and the protective efficacy of HSP vaccinations has been shown against various parasitic infections including Plasmodium yoelii [70], Brugia malayi [71], Leishmania donovani [72] and Trichinella spiralis [73]. In this study, we identified 8 HSPs within the 100 most abundant proteins expressed in hookworm intestine, including HSP20, 40 and 70 (Table 7). The high expression in intestine and conserved functions for worm’s survival suggest these hookworm intestine-expressed HSPs could be the targets for preventive vaccines or pharmaceutical drugs.
Conclusions
Whereas reverse vaccinology approaches have yielded promising results in terms of identifying vaccine targets against bacterial pathogens, so far this approach has had limited success for more complex eukaryotic organisms. Among the reasons why anthelmintic vaccines have not benefited from reverse vaccinology approaches include the large genome of parasitic helminths and difficulties in shaping moderate to high throughput approaches for protein expression and preclinical testing in laboratory animal models. To address the first component, we have narrowed our antigen selection program for the human hookworm vaccine to the parasite’s gut transcriptome.
The hookworm intestine expresses a wide range of proteins or enzymes involved in blood-feeding and homeostasis between the parasite and its host. The functional antigens expressed in the hookworm gut have become major targets for vaccine and drug development. In order to discover additional hookworm vaccine antigens, the A. ceylanicum intestinal transcriptome was generated and analyzed. Ancylostoma ceylanicum was selected because of its relative convenience for vaccine testing in a hamster laboratory animal model. Analysis of the full transcriptome of the adult male A. ceylanicum intestine reveals that two categories of proteins were highly expressed in the hookworm intestine, namely proteolytic enzymes involved in blood digestion and transporter proteins involved in the absorption of nutrient metabolites and the maintenance of homeostasis between the parasite and its environment. Glutathione S-transferases involved in the binding and detoxification of oxidative heme derived from blood-feeding were also highly expressed in the intestine. In addition, other proteins including a family of cysteine-rich/antigen 5/pathogenesis-related proteins (CAP); C-type lectins; and heat shock proteins all also upregulated in the hookworm intestine and possibly contribute to the survival of worm in the host. Thus at least two classes of potential hookworm antigens are related to adult blood-feeding, whereas the function of the CAP proteins, C-type lectins, and heat shock proteins in the host-parasite relationship are still under investigation. More hookworm-specific intestinal proteins involved in blood-feeding may further be identified by comparing other available intestinal transcriptomes of nematodes such as Ascaris suum (non-blood-feeding) [74], Trichuris suis (tissue-feeding) and Haemonchus contortus (blood-feeding) [75]. In subsequent studies, the vaccine candidates identified here will be ranked for downstream antigen target selection based on criteria including abundance, possible functions related to survival, uniqueness in the parasite versus the vertebrate host, and ultimately solubility and yield of expression.
Abbreviations
- APR:
-
Aspartic protease
- ASP:
-
Ancylostoma-secreted protein
- CAP:
-
Cysteine-rich/antigen 5/pathogenesis-related 1 proteins
- CP:
-
Cysteine protease
- CTL:
-
C-type lectin
- DALY:
-
Disability adjusted life years
- FAR:
-
Fatty acid and retinol-binding protein
- FDR:
-
False discovery rate
- FPKM:
-
Fragments per kilobase of transcript per million mapped reads
- GO:
-
Gene ontologies
- GST:
-
Glutathione-S transferase
- HSP:
-
Heat shock protein
- IACUC:
-
Institutional animal care and use committee
- NTD:
-
Neglected tropical disease
- SPIA:
-
Single primer isothermal amplification
- TrSSP:
-
Transporter substrate specificity prediction server
- vWFD:
-
von Willebrand Factor type D domain
References
Hotez PJ, Beaumier CM, Gillespie PM, Strych U, Hayward T, Bottazzi ME. Advancing a vaccine to prevent hookworm disease and anemia. Vaccine. 2016;34(26):3001–5.
Hotez PJ, Alvarado M, Basanez MG, Bolliger I, Bourne R, Boussinesq M, et al. The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PLoS NTD. 2014;8(7):e2865.
Hotez PJ, Pritchard DI. Hookworm infection. Sci Am. 1995;272(6):68–74.
Smith JL, Brooker S. Impact of hookworm infection and deworming on anaemia in non-pregnant populations: a systematic review. Trop Med Int Health. 2010;15(7):776–95.
Keiser J, Utzinger J. Efficacy of current drugs against soil-transmitted helminth infections: systematic review and meta-analysis. JAMA. 2008;299(16):1937–48.
De Clercq D, Sacko M, Behnke J, Gilbert F, Dorny P, Vercruysse J. Failure of mebendazole in treatment of human hookworm infections in the southern region of Mali. Am J Trop Med Hyg. 1997;57(1):25–30.
Soukhathammavong PA, Sayasone S, Phongluxa K, Xayaseng V, Utzinger J, Vounatsou P, et al. Low efficacy of single-dose albendazole and mebendazole against hookworm and effect on concomitant helminth infection in Lao PDR. PLoS NTD. 2012;6(1):e1417.
Hotez PJ, Diemert D, Bacon KM, Beaumier C, Bethony JM, Bottazzi ME, et al. The human hookworm vaccine. Vaccine. 2013;31 Suppl 2:B227–32.
Bartsch SM, Hotez PJ, Hertenstein DL, Diemert DJ, Zapf KM, Bottazzi ME, et al. Modeling the economic and epidemiologic impact of hookworm vaccine and mass drug administration (MDA) in Brazil, a high transmission setting. Vaccine. 2016;34(19):2197–206.
Ranjit N, Zhan B, Hamilton B, Stenzel D, Lowther J, Pearson M, et al. Proteolytic degradation of hemoglobin in the intestine of the human hookworm Necator americanus. J Infect Dis. 2009;199(6):904–12.
Hotez PJ, Bethony JM, Diemert DJ, Pearson M, Loukas A. Developing vaccines to combat hookworm infection and intestinal schistosomiasis. Nat Rev Microbiol. 2010;8(11):814–26.
Williamson AL, Brindley PJ, Abbenante G, Datu BJ, Prociv P, Berry C, et al. Hookworm aspartic protease, Na-APR-2, cleaves human hemoglobin and serum proteins in a host-specific fashion. J Infect Dis. 2003;187(3):484–94.
Williamson AL, Lecchi P, Turk BE, Choe Y, Hotez PJ, McKerrow JH, et al. A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J Biol Chem. 2004;279(34):35950–7.
Ranjit N, Zhan B, Stenzel DJ, Mulvenna J, Fujiwara R, Hotez PJ, Loukas A. A family of cathepsin B cysteine proteases expressed in the gut of the human hookworm, Necator americanus. Mol Biochem Parasitol. 2008;160(2):90–9.
Jones BF, Hotez PJ. Molecular cloning and characterization of Ac-mep-1, a developmentally regulated gut luminal metalloendopeptidase from adult Ancylostoma caninum hookworms. Mol Biochem Parasitol. 2002;119(1):107–16.
Loukas A, Bethony JM, Mendez S, Fujiwara RT, Goud GN, Ranjit N, et al. Vaccination with recombinant aspartic hemoglobinase reduces parasite load and blood loss after hookworm infection in dogs. PLoS Med. 2005;2(10):e295.
Xiao S, Zhan B, Xue J, Goud GN, Loukas A, Liu Y, et al. The evaluation of recombinant hookworm antigens as vaccines in hamsters (Mesocricetus auratus) challenged with human hookworm, Necator americanus. Exp Parasitol. 2008;118(1):32–40.
Loukas A, Bethony JM, Williamson AL, Goud GN, Mendez S, Zhan B, et al. Vaccination of dogs with a recombinant cysteine protease from the intestine of canine hookworms diminishes the fecundity and growth of worms. J Infect Dis. 2004;189(10):1952–61.
Diemert DJ, Pinto AG, Freire J, Jariwala A, Santiago H, Hamilton RG, et al. Generalized urticaria induced by the Na-ASP-2 hookworm vaccine: implications for the development of vaccines against helminths. J Allergy Clin Immunol. 2012;130(1):169–e176. e166.
Delany I, Rappuoli R, Seib KL. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb Perspect Med. 2013;3(5):a012476.
Schwarz EM, Hu Y, Antoshechkin I, Miller MM, Sternberg PW, Aroian RV. The genome and transcriptome of the zoonotic hookworm Ancylostoma ceylanicum identify infection-specific gene families. Nat Genet. 2015;47(4):416–22.
Fujiwara RT, Geiger SM, Bethony J, Mendez S. Comparative immunology of human and animal models of hookworm infection. Parasite Immunol. 2006;28(7):285–93.
Goud GN, Zhan B, Ghosh K, Loukas A, Hawdon J, Dobardzic A, et al. Cloning, yeast expression, isolation, and vaccine testing of recombinant Ancylostoma-secreted protein (ASP)-1 and ASP-2 from Ancylostoma ceylanicum. J Infect Dis. 2004;189(5):919–29.
Sonntag KC, Tejada G, Subbujaru S, Berretta S, Benes FM, Woo TW. Limited predictability of postmortem human brain tissue quality by RNA integrity numbers. J Neurochem. 2016;138(1):53–9.
Martin J, Rosa BA, Ozersky P, Hallsworth-Pepin K, Zhang X, Bhonagiri-Palsikar V, et al. Helminth.net: expansions to Nematode.net and an introduction to Trematode.net. Nucleic Acids Res. 2015;43(Database issue):D698–706.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36.
Tekes G, Hofmann-Lehmann R, Stallkamp I, Thiel V, Thiel HJ. Genome organization and reverse genetic analysis of a type I feline coronavirus. J Virol. 2008;82(4):1851–9.
Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9.
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–9.
Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338(5):1027–36.
Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Wang Z, Abubucker S, Martin J, Wilson RK, Hawdon J, Mitreva M. Characterizing Ancylostoma caninum transcriptome and exploring nematode parasitic adaptation. BMC Genomics. 2010;11:307.
Ranjit N, Jones MK, Stenzel DJ, Gasser RB, Loukas A. A survey of the intestinal transcriptomes of the hookworms, Necator americanus and Ancylostoma caninum, using tissues isolated by laser microdissection microscopy. Int J Parasitol. 2006;36(6):701–10.
Skelly PJ, Da'dara AA, Li XH, Castro-Borges W, Wilson RA. Schistosome feeding and regurgitation. PLoS Pathog. 2014;10(8):e1004246.
Meissner B, Boll M, Daniel H, Baumeister R. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans. J Biol Chem. 2004;279(35):36739–45.
Yamamura J, Adachi T, Aoki N, Nakajima H, Nakamura R, Matsuda T. Precursor-product relationship between chicken vitellogenin and the yolk proteins: the 40 kDa yolk plasma glycoprotein is derived from the C-terminal cysteine-rich domain of vitellogenin II. Biochim Biophys Acta. 1995;1244(2-3):384–94.
Baker ME. Invertebrate vitellogenin is homologous to human von Willebrand factor. Biochem J. 1988;256(3):1059–61.
Finn RN. Vertebrate yolk complexes and the functional implications of phosvitins and other subdomains in vitellogenins. Biol Reprod. 2007;76(6):926–35.
Fischer M, Regitz C, Kull R, Boll M, Wenzel U. Vitellogenins increase stress resistance of Caenorhabditis elegans after Photorhabdus luminescens infection depending on the steroid-signaling pathway. Microbes Infect. 2013;15(8-9):569–78.
Garofalo A, Kennedy MW, Bradley JE. The FAR proteins of parasitic nematodes: their possible involvement in the pathogenesis of infection and the use of Caenorhabditis elegans as a model system to evaluate their function. Med Microbiol Immunol. 2003;192(1):47–52.
Basavaraju SV, Zhan B, Kennedy MW, Liu Y, Hawdon J, Hotez PJ. Ac-FAR-1, a 20 kDa fatty acid- and retinol-binding protein secreted by adult Ancylostoma caninum hookworms: gene transcription pattern, ligand binding properties and structural characterisation. Mol Biochem Parasitol. 2003;126(1):63–71.
Rey-Burusco MF, Ibanez-Shimabukuro M, Gabrielsen M, Franchini GR, Roe AJ, Griffiths K, et al. Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus. Biochem J. 2015;471(3):403–14.
Brophy PM, Pritchard DI. Metabolism of lipid peroxidation products by the gastro-intestinal nematodes Necator americanus, Ancylostoma ceylanicum and Heligmosomoides polygyrus. Int J Parasitol. 1992;22(7):1009–12.
Jani D, Nagarkatti R, Beatty W, Angel R, Slebodnick C, Andersen J, et al. HDP-a novel heme detoxification protein from the malaria parasite. PLoS Pathog. 2008;4(4):e1000053.
Deponte M, Becker K. Glutathione S-transferase from malarial parasites: structural and functional aspects. Method Enzymo. 2005;401:241–53.
Zhan B, Liu S, Perally S, Xue J, Fujiwara R, Brophy P, et al. Biochemical characterization and vaccine potential of a heme-binding glutathione transferase from the adult hookworm Ancylostoma caninum. Infect Immun. 2005;73(10):6903–11.
Zhan B, Perally S, Brophy PM, Xue J, Goud G, Liu S, et al. Molecular cloning, biochemical characterization, and partial protective immunity of the heme-binding glutathione S-transferases from the human hookworm Necator americanus. Infect Immun. 2010;78(4):1552–63.
Asojo OA, Homma K, Sedlacek M, Ngamelue M, Goud GN, Zhan B, et al. X-ray structures of Na-GST-1 and Na-GST-2 two glutathione S-transferase from the human hookworm Necator americanus. BMC Struct Biol. 2007;7:42.
Rao AU, Carta LK, Lesuisse E, Hamza I. Lack of heme synthesis in afree-living eukaryote. Proc Natl Acad Sci USA. 2005;102(12):4270–5.
Hawdon JM, Jones BF, Hoffman DR, Hotez PJ. Cloning and characterization of Ancylostoma-secreted protein. A novel protein associated with the transition to parasitism by infective hookworm larvae. J Biol Chem. 1996;271(12):6672–8.
Hawdon JM, Narasimhan S, Hotez PJ. Ancylostoma secreted protein 2: cloning and characterization of a second member of a family of nematode secreted proteins from Ancylostoma caninum. Mol Biochem Parasitol. 1999;99(2):149–65.
Gibbs GM, Roelants K, O'Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins - roles in reproduction, cancer, and immune defense. Endocr Rev. 2008;29(7):865–97.
Cantacessi C, Campbell BE, Visser A, Geldhof P, Nolan MJ, Nisbet AJ, et al. A portrait of the "SCP/TAPS" proteins of eukaryotes - developing a framework for fundamental research and biotechnological outcomes. Biotechnol Adv. 2009;27(4):376–88.
Tribolet L, Cantacessi C, Pickering DA, Navarro S, Doolan DL, Trieu A, et al. Probing of a human proteome microarray with a recombinant pathogen protein reveals a novel mechanism by which hookworms suppress B-cell receptor signaling. J Infect Dis. 2015;211(3):416–25.
Sen L, Ghosh K, Bin Z, Qiang S, Thompson MG, Hawdon JM, et al. Hookworm burden reductions in BALB/c mice vaccinated with recombinant Ancylostoma secreted proteins (ASPs) from Ancylostoma duodenale, Ancylostoma caninum and Necator americanus. Vaccine. 2000;18(11-12):1096–102.
Zhan B, Santiago H, Keegan B, Gillespie P, Xue J, Bethony J, et al. Fusion of Na-ASP-2 with human immunoglobulin Fcgamma abrogates histamine release from basophils sensitized with anti-Na-ASP-2 IgE. Parasite Immunol. 2012;34(8-9):404–11.
Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, et al. Genome of the human hookworm Necator americanus. Nat Genet. 2014;46(3):261–9.
Abubucker S, Martin J, Yin Y, Fulton L, Yang SP, Hallsworth-Pepin K, et al. The canine hookworm genome: analysis and classification of Ancylostoma caninum survey sequences. Mol Biochem Parasitol. 2008;157(2):187–92.
Zhan B, Liu Y, Badamchian M, Williamson A, Feng J, Loukas A, et al. Molecular characterisation of the Ancylostoma-secreted protein family from the adult stage of Ancylostoma caninum. Int J Parasitol. 2003;33(9):897–907.
Weis WI, Taylor ME, Drickamer K. The C-type lectin superfamily in the immune system. Immunol Rev. 1998;163:19–34.
Harcus Y, Nicoll G, Murray J, Filbey K, Gomez-Escobar N, Maizels RM. C-type lectins from the nematode parasites Heligmosomoides polygyrus and Nippostrongylus brasiliensis. Parasitol Int. 2009;58(4):461–70.
Loukas A, Maizels RM. Helminth C-type lectins and host-parasite interactions. Parasitol Today. 2000;16(8):333–9.
Loukas A, Doedens A, Hintz M, Maizels RM. Identification of a new C-type lectin, TES-70, secreted by infective larvae of Toxocara canis, which binds to host ligands. Parasitology. 2000;121(Pt 5):545–54.
Loukas A, Mullin NP, Tetteh KK, Moens L, Maizels RM. A novel C-type lectin secreted by a tissue-dwelling parasitic nematode. Curr Biol. 1999;9(15):825–8.
Schulenburg H, Hoeppner MP, Weiner 3rd J, Bornberg-Bauer E. Specificityof the innate immune system and diversity of C-type lectin domain(CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiol.2008;213(3-4):237–50.
Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12(14):1209–14.
Colaco CA, Bailey CR, Walker KB, Keeble J. Heat shock proteins: stimulators of innate and acquired immunity. Biomed Res Int. 2013;2013:461230.
Corigliano MG, Fenoy I, Sander V, Maglioco A, Goldman A, Clemente M. Plant heat shock protein 90 as carrier-adjuvant for immunization against a reporter antigen. Vaccine. 2013;31(49):5872–8.
Sanchez GI, Sedegah M, Rogers WO, Jones TR, Sacci J, Witney A, et al. Immunogenicity and protective efficacy of a Plasmodium yoelii Hsp60 DNA vaccine in BALB/c mice. Infect Immun. 2001;69(6):3897–905.
Dakshinamoorthy G, Samykutty AK, Munirathinam G, Shinde GB, Nutman T, Reddy MV, Kalyanasundaram R. Biochemical characterization and evaluation of a Brugia malayi small heat shock protein as a vaccine against lymphatic filariasis. PLoS ONE. 2012;7(4):e34077.
Kaur T, Sobti RC, Kaur S. Cocktail of gp63 and Hsp70 induces protection against Leishmania donovani in BALB/c mice. Parasite Immunol. 2011;33(2):95–103.
Fang L, Sun L, Yang J, Gu Y, Zhan B, Huang J, Zhu X. Heat shock protein 70 from Trichinella spiralis induces protective immunity in BALB/c mice by activating dendritic cells. Vaccine. 2014;32(35):4412–9.
Rosa BA, Jasmer DP, Mitreva M. Genome-wide tissue-specific gene expression, co-expression and regulation of co-expressed genes in adult nematode Ascaris suum. PLoS NTD. 2014;8(2):e2678.
Wang Q, Rosa BA, Jasmer DP, Mitreva M. Pan-Nematoda transcriptomic elucidation of essential intestinal functions and therapeutic targets with broad potential. EBioMedicine. 2015;2(9):1079–89.
Acknowledgements
The unpublished A. ceylanicum reference genome and the whole worm RNA-seq data used in this study were produced by the parasite genomics group at the McDonnell Genome Institute in collaboration with the laboratory of Dr. John Hawdon at George Washington University. The genome data are available from GenBank under BioProject ID PRJNA72583, and from http://nematode.net and http://nematode.net/NN3_frontpage.cgi?navbar_selection=speciestable&subnav_selection=Ancylostoma_ceylanicum. The unpublished RNA-seq data are available from the SRA sequence data archives.
Funding
The research described herein is supported by the Michelson Medical Research Foundation via the Sabin Vaccine Institute. The analyses performed in Dr. Mitreva’s lab were supported by the National Institute of General Medicine Science Grant R01GM097435 to MM.
Availability of data and materials
The RNA-seq data generated from the adult male intestine of A. ceylanicum are available from NCBI under BioProject ID PRJNA72583: (http://www.ncbi.nlm.nih.gov/bioproject/PRJNA72583), or from WormBase database under BioProject ID PRJNA72583: (http://parasite.wormbase.org/Ancylostoma_ceylanicum_prjna72583/Info/Index).
Authors’ contributions
BZ conceived the study. JW performed the experimental work. AD analyzed the sequence data. XG, MM performed sequencing and interpreted that data. ZL performed some laboratory experiments. RM performed qPCR. BZ, US, MEB and PJH interpreted the results and prepared the manuscript. All authors read and approved the final manuscript.
Authors’ information
JW AD ZL RM US MEB PJH and BZ are researchers at Baylor College of Medicine, National School of Tropical Medicine and the section of Pediatric Tropical Medicine, as well as the Texas Children’s Hospital and Sabin Vaccine Institute Center for Vaccine Development (www.bcm.edu/tropicalmedicine), Houston, USA. XG and MM are researchers in McDonnel Genome Institute, Washington University School of Medicine, St. Louis, USA.
Competing interests
Some of the authors (BZ, PJH, MEB) are inventors on patents filed for the human hookworm vaccine.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Hookworm A. ceylanicum was maintained in hamsters according to protocol AN-5762 approved by Baylor College of Medicine’s Institutional Animal Care and Use Committee, Houston, USA.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Table S1.
The 100 most abundant proteins in A. ceylanicum intestine. (PDF 326 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wei, J., Damania, A., Gao, X. et al. The hookworm Ancylostoma ceylanicum intestinal transcriptome provides a platform for selecting drug and vaccine candidates. Parasites Vectors 9, 518 (2016). https://doi.org/10.1186/s13071-016-1795-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1795-8