Abstract
Background
Of increasing importance to the medical and veterinary communities is the zoonotic filarioid nematode Onchocerca lupi. Onchocercosis, thus far found in wolves, dogs, cats and humans, is diagnosed via skin snips to detect microfilariae and surgical removal of adults from the eye of the host. These methods are time-consuming, laborious and invasive, highlighting the need for new tools for the diagnosis of O. lupi in susceptible hosts. Symptoms related to the presence of the adults in the eye can range from none apparent to severe, including blindness. No reliable chemotherapeutic protocols are available, as yet, to eliminate the infection. Paramyosin, an invertebrate-specific protein, has been well-studied as an allergen, diagnostic marker and vaccine candidate. The aim of this study, therefore, was to isolate and characterise paramyosin from O. lupi to assess its suitability for the development of a serological diagnostic assay.
Methods
The adult and microfilarial stages of O. lupi were isolated from the eyes and skin of a 3-year-old male dog. Total RNA was extracted and reverse transcribed into single stranded cDNA. Reverse-transcription PCR was used to isolate a full-length paramyosin cDNA from adult worms and to investigate the temporal expression patterns of this gene. All amplicons were sequenced using dideoxy chain termination sequencing. Bioinformatics was used to predict the amino acid sequence of the gene, to compare the DNA and protein sequences with those available in public databases and to investigate the phylogenetic relationship of all molecules. Antibody binding sites were predicted using bioinformatics and mapped along with published antigenic epitopes against the O. lupi paramyosin protein. The native protein, and three smaller recombinantly expressed peptides, were subjected to western blot using serum from dogs both positive and negative for O. lupi.
Results
Paramyosin of O. lupi was herein molecularly characterized, encoded by a transcript of 2,643 bp and producing a protein of 881 amino acids (101.24 kDa). The paramyosin transcript was detected, by reverse transcription PCR, in adults and microfilariae, but not in eggs. Phylogenetic analysis indicates that this molecule clusters with paramyosins from other filarioids to the exclusion of those from other taxa. A total of 621 unique antibody binding epitopes were predicted for this protein and another 28 were conserved in other organisms. This information was used to design three peptides, for recombinant expression, to identify the antibody binding epitope(s) and reduce potential cross-reactivity with serum from dogs infected with other filarioid nematodes. Native paramyosin, purified from microfilariae and adults, was detected by antibodies present in serum from dogs with known O. lupi infections.
Conclusions
Data provided herein may assist in the development of a serological diagnostic test, based on antibodies to O. lupi paramyosin, for the diagnosis of this infection, in order to gain more information on the real distribution of this little known filarioid of zoonotic concern.
Similar content being viewed by others
Background
Ocular parasitism in humans and animals may be caused by a range of zoonotic helminths, including strongylids, ascarids, thelaziids and filarioids [1]. The parasitic nematode Onchocerca lupi (Spirurida: Onchocercidae) localises to the connective tissue of the sclera of dogs, wolves and cats [2]. In dogs, subconjunctival granulomas are the most commonly reported manifestation of infection by this filarioid [3]; however, clinical signs may vary from none apparent [4] to severe (i.e. blindness) [2]. Affected dogs have periodically been reported from Hungary, Greece, Germany, Portugal and Spain [3, 5–10]. More recently, reports have also emerged of infections of dogs and cats in the United States of America [11, 12].
The suspected zoonotic potential of O. lupi [2, 8, 9, 11, 13] has only recently been confirmed [14–16]. To date, human ocular infection has been documented in Turkey [14, 17], Tunisia [15], Iran [18] and the United States of America [19]. Three additional cases, where localisation of O. lupi is to the cervical spine, have been reported in children in the USA [19–21], prompting an increased interest in this nematode by the medical and veterinary communities [22].
There is a paucity of information on the biology and epidemiology of O. lupi (e.g. vector insects and geographical distribution). The only report available, to date, on the prevalence of O. lupi came from apparently healthy dogs sampled in Greece (8.7 %) and Portugal (8.3 %) [4]. In addition, the DNA sequence data available are limited to single ribosomal and mitochondrial genes, or parts thereof, which have been used for molecular and phylogenetic analyses [23–25]. Importantly, the frequency, distribution and full zoonotic potential of this parasite has not yet been investigated, probably due to the difficulty in achieving a diagnosis of the infection, which is based on the detection of microfilariae in skin sediments or surgical removal and identification of the adult worm from the eye [26]. Some preliminary scientific evidence indicates that an ELISA, prepared with somatic paramyosin antigens from Onchocerca gibsoni (Og4C3), which infects cattle [27–29], cross-reacts with serum from dogs infected with O. lupi [30].
Paramyosin is a large structural protein (98–101 kDa) component of invertebrate muscle and found in endo- and ectoparasites of medical importance, but not in vertebrates [31]. By virtue of its major antigenic properties, this protein has been used for the development of diagnostic assays for filarioids of medical concern such as Wuchereria bancrofti and Brugia malayi [27, 32, 33]. Many studies of the paramyosin protein, as a potential vaccine candidate, have focussed on platyhelminths [34–39], with fewer studies on nematodes [40, 41] and ticks [42]. An immunomodulatory effect of this protein has also been identified, inhibiting components of the complement cascade [43, 44], binding of immunoglobulins and collagen [45, 46]. This suggests paramyosin is an important part of the mechanism by which a parasite invades and maintains itself in the host [31], making this protein of potential usefulness for serological diagnosis of, and protective immunity against, O. lupi.
Therefore, the research herein describes the isolation and characterisation of a paramyosin transcript from O. lupi, bioinformatic analysis of the predicted protein and preliminary assessment of the potential for the full protein and recombinant pieces for the development of a specific serological diagnostic assay.
Methods
Biological material
Three adult female and 2 adult male nematodes were isolated from the eye of an infected dog (3-year-old, male) that died accidentally, from an area (Algarve, southern Portugal) where O. lupi is known to occur [4]. Microfilariae were collected by sedimentation from the skin of the dorsal region of the dog and eggs by cutting the uterus of the adult females. All samples were preserved in RNAlater® (Life Technologies, California, USA) and stored at -80 °C until used. Serum samples were obtained from this dog and from O. lupi positive (n = 5) and negative dogs (n = 3) from previous research [30].
Nucleic acid isolation and single-stranded cDNA synthesis
Total RNA was isolated from adult and microfilarial O. lupi, following homogenization using a pestle and 400–600 μm glass beads (Sigma-Aldrich, Missouri, USA) employing the TriPure® isolation reagent according to the manufacturer's instructions (Roche Molecular Biochemicals, Basel, Switzerland). RNase inhibitor (RNasin®, Promega, Wisconsin, USA) was added to total RNA before quantification and storage. Due to the tiny amounts of RNA able to be extracted from these nematodes, no DNase treatment was performed. Nucleic acids were quantified using a Bioanalyzer (Agilent, California, USA) and all extracted material was stored at -80 °C until used. First-strand cDNA synthesis was performed using the SuperScript Reverse Transcriptase II kit (Invitrogen, California, USA), 0.5 g oligo dT primer (n = 12–18 primer, Promega, Wisconsin, USA) and 100 ng of total RNA from adult males and females, microfilariae and eggs according to the manufacturer’s instructions. Each completed reaction was diluted to 250 ng/l with TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) before use in reverse transcription PCR.
PCR of the paramyosin cDNA
PCR of the paramyosin cDNA, using 250 ng of single stranded cDNA from a single adult female, was performed using Phusion High-Fidelity DNA polymerase (Life Technologies, California, USA) and primers (PARAdegF 5′-ATG TCC GGT TCA TTG TAC CGT AG-3′; PARAdegR 5′-CTA TTG CTC ATC TTC GAG AAC GT-3′), designed from available filarial nematode paramyosin sequences (B. malayi NCBI Accession no. U77590.1, Onchocerca volvulus NCBI Accession no. M95813.1 and Acanthocheilonema viteae nAv_1_0_scaf01040, www.nematodes.org). Reactions consisted of 1× GC reaction buffer, 2 pmol of each primer, 0.2 mM of dNTPs and 1 U of Phusion Taq DNA polymerase in a volume of 50 μl. Cycling conditions were 98 °C/10 s for initial denaturation, then 35 cycles of 98 °C for10 s, 55 °C for 20 s and 72 °C for 3 min for DNA amplification and 72 °C for 10 min for the final extension. Products were resolved on 1 % agarose stained with 0.5× GelRed (Biotium, California, USA) and visualized on a GelLogic 100 gel documentation system (Kodak, New York, USA). Amplicon sequencing employed the PCR primers and Big Dye Terminator v.3.1 chemistry in a 3130 genetic analyzer (Applied Biosystems, California, USA).
Gene cloning and sequencing
The full-length cDNA isolated above was cloned into pGEM®-T Easy, employing T4 DNA ligase (3 U), and transformed into Escherichia coli (MACH-1 strain, Life Technologies, California, USA) using an established method [47]. Colonies were picked, resuspended in 20 μl of water, with 2 μl used as a PCR template for insert screening and the remaining grown overnight in Luria Bertani/ampicillin (100 g/ml) broth. Plasmid DNA from positive colonies was isolated using the PureYield™ Plasmid Miniprep System (Promega, Wisconsin, USA) and inserts were sequenced in both directions using the T7 and SP6 primers of pGEM®-T Easy [47].
Temporal expression of O. lupi paramyosin in adult males and females, microfilariae and eggs
The entire paramyosin (2,643 bp; PARA_degF, PARA_degR primers) and elongation factor 1α (as a control gene, designed from the O. volvulus ELF1α (1,776 bp; GenBank M64333.1, OvELF1α_F 5′-ATT GAG ATT TCG GGA TTA AGT GAA T-3′, OvELF1α_R 5′-TTC AGT GTA GCA GGA GCA TAT G-3′) genes were amplified using Phusion High-Fidelity DNA polymerase enzyme (Life Technologies, California, USA) according to the method used above for cDNA amplification (53 °C annealing for ELF1α). Reactions used 125 ng of single-stranded cDNA template for adults and eggs and 500 ng for microfilariae. Two reactions were performed for microfilariae using template from the first reaction, including the negative controls, in order to amplify both genes. The experiment was repeated 3 times (i.e. 3 replicates per gene) on 3 separate days. Products were sequenced using the PCR primers as described above.
Bioinformatic analyses and epitope mapping
The identity of the full-length O. lupi paramyosin cDNA was confirmed using BLASTx/n/p against nucleotides and proteins available in NCBI (blast.ncbi.nlm.nih.gov) with an E-value limit of 1e-05. The predicted O. lupi paramyosin protein was subjected additional analyses, including ProSite (prosite.expasy.org), the presence of signal peptides (http://www.cbs.dtu.dk/services/SignalP/; [48–50], transmembrane spanning domains (www.cbs.dtu.dk/services/TMHMM/) [51] and detection of conserved protein domains (ProSite; www.expasy.ch/tools/scnpsit1.html). Potential antigenic epitopes were predicted using algorithms based on published methods [52–55]. Epitopes reported in the literature for other organisms (Table 1) and bioinformatically predicted epitopes were mapped against the O. lupi protein using local BLAST+ (NCBI). Epitopes unique to O. lupi paramyosin were identified through comparisons with epitopes predicted for other filarial nematodes, namely A. viteae (bioinformatically predicted, see below), Ancylostoma caninum (NCBI Accession no. ABC86903.1), B. malayi (NCBI Accession no. Q01202.2), Dirofilaria immitis (NCBI Accession no. P13392.2), Loa loa (PantherDB Accession no. E1FX82), O. volvulus (NCBI Accession no. M95813.1) and W. bancrofti (NCBI Accession no. AEY79495.1). Paramyosin genes/proteins from other organisms were identified by BLASTp of non-redundant (NCBI, www.pantherdb.org/, www.metazoa.ensembl.org) and parasite-specific databases (www.broadinstitute.org/, www.nematodes.org/, www.nematode.net/, www.wormbase.org). Augustus gene prediction, using B. malayi as the training set, was used to predict the full-length genes and proteins from genomic data (http://bioinf.uni-greifswald.de/augustus/) [56]. These were confirmed via BLASTn/p against the NCBI database. All paramyosin molecules were aligned using MUSCLE [57] and full-length sequences subjected to phylogenetic analyses by Bayesian inference (MrBayes v 3.2.6) probabilities were calculated using 100,000 generations (ngen = 100,000, burnin = 20), employing 4 simultaneous tree-building chains (nchains = 4), saving every 100 th tree (samplefreq = 100). The heuristic neighbour-joining (NJ) analysis was performed employing MEGA 7.0 using the absolute difference model (2,000 replications) with the alignment gaps treated as missing data. The protein sequence from Meloidogyne hapla (www.wormbase.org; Accession number: Contig185) was assigned as the outgroup.
Protein expression and western blots
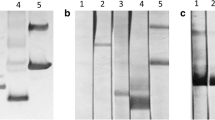
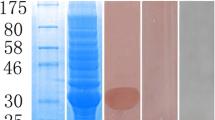
Native paramyosin was isolated from microfilariae and adults using a differential ammonium sulphate precipitation method [58]. Protein was measured using the Qubit® Protein Assay Kit according to the manufacturer’s instructions (Thermo Fisher Scientific, Massachusetts, USA). Subsequently, in order to define antigenic epitope(s) recognized by antibodies from dogs infected with O. lupi, bioinformatics was used to design 3 peptides, corresponding to both the presence of potential antigenic epitopes and natural methionine residues (to maintain the correct reading frame). These sections were designated pieces 1 (1,272 bp, 48.2 kDa), 2 (908 bp, 33.5 kDa) and 3 (513 bp, 19.65 kDa) from the N- to the C-terminal part of the protein respectively. Primers used were: Piece 1 - PARAdegF, Piece 1R 5′-CGC CTT CTC GTA CAA ATT CTT CAT-3′; Piece 2 - PIECE2F 5′-ATG AAG AAT TTG TAC GAG AAG GCG-3′, PIECE2R 5′-GAT TTC CTG AGA GCA TCA ATT TTC AT-3′; Piece 3 - PIECE3F 5′-ATG AAA ATT GAT GCT CTC AGG AAA TC-3′, PARAdegR using Phusion Taq polymerase under the conditions described above. All cDNAs were ligated into the SmaI restriction enzyme site of the pGEX3X glutathione S-transferase fusion protein expression vector (GE Healthcare Bio-Sciences, Pennsylvania, USA) using T4 DNA ligase (3 U) and transformed into E. coli (BL21 strain). Clones were induced using 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 3 h and then analyzed via SDS PAGE (10 % gels) using an established protocol [47]. Expressed peptides were purified using EZview™ Red Glutathione Affinity Gel (Sigma Aldrich, Missouri, USA) according to the manufacturer’s instructions, without elution of the peptide from the resin. Peptides were then cleaved from GST using Factor Xa protease (0.5 U, Promega, Wisconsin, USA) and the cleaved peptides isolated from the supernatant. Following SDS PAGE (10 % gels) of total nematode protein (positive control), purified native paramyosin (< 0.5 ng - microfilariae, 500 ng - adult), purified recombinant peptide (500 ng), uninduced bacterial culture (5 μl) and the Precision Plus Protein™ All Blue Prestained Protein Standards (BioRad, California, USA), proteins were transferred to PVDF membrane (0.5A/45 min; Roche Applied Sciences, Penzburg, Germany). Following blocking, membranes were incubated with serum from uninfected dogs (negative control) or those infected with O. lupi (1:1000 dilutions). Dog antibodies were detected using rabbit anti-dog IgG horseradish peroxidase conjugate (1:3000; Sigma Adrich, Missouri, USA) and detected using the femto-Chromo kit (G Biosciences, Missouri, USA). Documentation was via scanning on an Epson Perfection 2450 scanner.
Results
The full-length cDNA encoding paramyosin (henceforth named Ol-para) is 2,643 bp in length (NCBI Accession no. KJ699378). BLASTn analysis revealed the highest nucleotide identity to paramyosin from the related filarial nematode O. volvulus (98 %, EMBL Accession no. M95813.1), followed by L. loa (91 %, NCBI Accession no. JH712727.1) and B. malayi (90 %, NCBI Accession no. U77590.1). Isolation and alignment of the nucleotide sequences above, and from other organisms, and comparison with Ol-para revealed that the percentage identity with platyhelminths was, on average, 52 %. Non-filarioid nematodes showed an average nucleotide identity with Ol-para of 77 %, whilst the highest nucleotide identities were with the other filarioid nematodes, except W. bancrofti (52 %) (Table 2). Analysis of the nucleotide composition of Ol-para and other filarioid nematode sequences indicated that all are AT-rich (Ol-para, 60.23 %) (Table 2). The conceptually translated protein (Ol-PARA) was 881 amino acids in length, with a predicted molecular weight of 101.24 kDa (NCBI Accession no. AJO15918.1). Amino acid identities with O. volvulus, L. loa and B. malayi were 93 % for the first 2 and 90 %, for the latter. No signal peptides or transmembrane spanning domains were identified in Ol-PARA, consistent with the other nematode paramyosins analyzed. Conserved protein domain analysis revealed the presence of a proton donor site in Ol-PARA (and in some, but not all, of the other nematode proteins).
Reverse transcription PCR (RT-PCR) amplified Ol-para from adult males and females, microfilariae, but not from eggs, whereas ELF1α was amplified from all stages (Fig. 1). Sequencing and BLASTn analysis of O. lupi ELF1α indicated a 94 % nucleotide identity to O. volvulus ELF1α (NCBI Accession no. M64333.1).
Reverse transcription PCR of Onchocerca lupi paramyosin (2,643 bp) and of the control gene elongation factor 1α (1,776 bp, ELF1α). The 2.5 kb and 1.5 kb bands from the 1 kb DNA ladder used as the standard are indicated on the left for reference. Abbreviations: AF, adult female; AM, adult male; Mf, microfilariae; E, eggs
A total of 874 antibody binding epitopes, predicted using bioinformatic algorithms, and 14 published T-cell and 14 B-cell epitopes were mapped against Ol-PARA (Fig. 2 and Additional file 1: Figure S1). Of the 874 predicted antibody epitopes, when compared with those predicted for A. viteae, B. malayi, D. immitis, L. loa, O. volvulus and W. bancrofti, 621 were unique to O. lupi.
Immunologically reactive paramyosin epitopes reported in the literature for helminths and arthropods that are conserved in the Onchocerca lupi protein. Residues underlined indicate T-cell epitopes whilst those in bold indicate B-cell epitopes. The text in red indicates residues of the proton donor site. The paramyosin peptides designed using bioinformatics to detect the antibody binding epitopes are indicated using dashed line boxes
Forty-six full-length paramyosin molecules from a range of nematodes and platyhelminths were identified, some species having more than one form of paramyosin (Table 3). Alignment and phylogenetic analysis show distinct clusters corresponding to nematodes and platyhelminths (Fig. 3). In the nematode branch, free-living nematodes such as Caenorhabditis spp. grouped together, whilst the parasitic nematodes formed two separate groups. The Platyhelminthes consist of both digeneans and cestodes, each forming a separate group. Further phylogenetic analysis of the nematode paramyosin molecules separates the nematode proteins according to distinct taxonomic groups (Fig. 4).
Phylogenetic relationships among nematode and platyhelminth paramyosin proteins. Forty-six full-length paramyosin proteins from a range of nematodes and platyhelminths were aligned and subjected to phylogenetic analysis. The topology of the trees from Bayesian inference and maximum parsimony analyses were in consensus, the representative tree displayed here with bootstrap values assigned to each branch (Bootstrap cut-off of 50). The protein sequence from Meloidogyne hapla (www.wormbase.org; Accession number: Contig185) was assigned as the outgroup
Phylogenetic analysis of nematode proteins. Thirty-two full-length paramyosin proteins from a range of nematodes were aligned and subjected to phylogenetic analysis. The topology the of trees from Bayesian inference and maximum parsimony analyses were in consensus, the representative tree displayed here with bootstrap values assigned to each branch (Bootstrap cut-off of 50). The protein sequence from Meloidogyne hapla (www.wormbase.org; Accession number: Contig185) was assigned as the outgroup
Western blot analysis revealed that serum from dogs infected with O. lupi readily detected native paramyosin purified from microfilariae (< 0.5 ng, lower than the detection limit of the Qubit® Protein Assay Kit) and adults (< 1 μg) (Fig. 5). An additional co-purifying protein (~50 kDa) was detected, but was also detected by negative serum (data not shown). The microfilarial protein was detected, whilst the western blot of adults contained many additional proteins, most likely from the dog, as the adults are often embedded within the eye tissue, which cannot be completely removed. As DNA, RNA and protein are extracted from each sample simultaneously, enzyme digestion to facilitate removal of dog tissue was not performed due to the concurrent risk of RNA degradation. None of the purified, Factor Xa cleaved peptides, which were of the expected sizes, were detected by serum from dogs with O. lupi.
Discussion
Onchocerca lupi paramyosin (Ol-para, 2,643 bp cDNA) was herein isolated and characterised in adults, both males and females, and microfilariae. The predicted Ol-PARA protein (i.e. 881 amino acids, 101.24 kDa) represents a unique set of data for filarioids nematodes and, most specifically, for this little known vector-borne helminth. Indeed, although O. lupi is known to cause morbidity in both humans and animals, very little molecular information is available. The length of Ol-para is consistent with those of other filarioid nematodes, which range from the smallest 2,508 bp (L. loa, NCBI Accession no. XM_003141046.1) to 2,640 bp (B. malayi, NCBI Accession no. Q01202.2). The nucleotide identity levels also reflect the relatedness of Ol-para compared with paramyosin sequences from other organisms, with high homology to other filarioid nematodes (91–98 %) compared with non-filarioid nematodes (~77 %) and the platyhelminths (~52 %). The AT-richness of Ol-para (i.e. 60.23 %) was consistent with other filarioid nematode sequences, including O. volvulus (AT 61 %), L. loa (AT 61 %) and B. malayi (AT 61 %). Some, but not all, of the platyhelminth paramyosins (Schistosoma spp.) are also AT-rich (65–67 %), whilst the remaining platyhelminths and non-filarioid nematodes have percentages of AT nucleotides in their sequences of between 44 % and 54 % (Table 2). Whether there is any functional significance of this difference in AT proportions in paramyosin molecules is yet to be determined.
Unlike some other species of nematodes, however, only one transcript encoding paramyosin was detected in O. lupi adults and microfilariae, whilst no paramyosin transcript was detected in eggs. The detection of transcription of paramyosin in O. lupi microfilariae is consistent with what has been found in other nematodes. Indeed, two forms of paramyosin have been detected in B. malayi (NCBI Accession no. P13392.2), which differ in size by only three base pairs from adults (NCBI Accession no. P13392). Paramyosin transcripts in other species, such as the free-living nematode Caenorhabditis elegans, differ significantly in size (WormBase Accession no. F07A5.7a and b, 2,616 and 1,659 bp, respectively). In O. lupi it is possible that additional transcripts exist, but may be expressed at levels not able to be detected by the methods used. Upon deep transcriptomic sequencing of B. malayi life-cycle stages, paramyosin and ELF1α were identified across the life-cycle of the parasite [59]. Similarly, in Dictyocaulus viviparus (cattle lungworm) and Clonorchis sinensis (liver fluke) paramyosin is expressed throughout the life-cycle [60, 61]. Undoubtedly, next generation sequencing of the O. lupi transcriptome could be useful in determining the number of transcripts and confirming the expression patterns of paramyosin across the parasite life-cycle.
The Ol-PARA sequence is also consistent with that of other filarioid nematodes, with highest amino acid identity with the O. volvulus, B. malayi and L. loa, and within the range of lengths of proteins (90–93 % identity, 836–880 amino acids). Like other filarioid proteins, it has no signal peptides or transmembrane spanning domains. The Ol-PARA sequence, like five of 11 sequences available for other nematode species (A. caninum, B. malayi, C. elegans, D. viviparus and O. volvulus), has a proton donor site. This motif is commonly associated with enzyme catalysis of hydrolysis of 1,3- or 1,4- bonds in β-D-glucans (prosite.expasy.org), but the functional significance of this activity in paramyosin is not yet known and is, as yet, unreported in the available literature. There is evidence from some nematode species to suggest that there is a short and a long isoform of paramyosin, hence the differences in size (Table 3). However, there is not enough information available for many species to confirm this. Phylogenetic analysis of platyhelminth and nematode proteins indicated that, whilst these proteins share conserved T- and B-cell epitopes, they are distinctly different proteins. Based on the phylogenesis of paramyosin molecules, five groups of nematodes were recognized: the rhabditids, including Caenorhabditis spp., the strongylids (Haemonchus contortus and A. caninum), the metastrongylids (Dictyocaulus viviparous), the ascarids (Anisakis spp. and Ascaris suum) and the filarioids (Onchocherca spp., B. malayi, A. viteae, L. loa and D. immitis). The W. bancrofti paramyosin does not closely cluster with other filarioids (Fig. 4). It is possible that for W. bancrofti that only one of a number of transcripts has been identified.
Proteomic analysis of Ol-PARA has indicated that this protein, like other paramyosins, has the potential to elicit both humoral and cellular immune responses in the host [31]. The bioinformatic approach in this study, along with recombinant peptide expression, was however unable to identify the antibody binding epitope(s) of this protein. Nonetheless, 28 published epitopes (14 T- and 14 B-cell) experimentally determined in platyhelminths, ticks and O. volvulus, were conserved in Ol-PARA [38–43, 62–67].
In addition, paramyosin is a vaccine candidate, predominantly in platyhelminths and also in Trichinella spiralis [41], and it has been listed by the World Health Organization as one of six antigen candidates for vaccines against Schistosoma spp. [64]. This, including the positive antibody binding of very small amounts of microfilarial and adult native paramyosin detected herein, suggests that Ol-PARA should be tested as a potential vaccine candidate. Accordingly, Ol-para could also be tested as a DNA vaccine, particularly given the greater ease with which DNA can be manipulated when compared with proteins. To date, four studies have assessed paramyosin DNA-based vaccination (100 μg per rat/mouse) against Schistosoma japonicum [68, 69], B. malayi [70] and T. solium [71], with evidence of some protective effect. Nonetheless, for its high antigenicity, Ol-PARA could also be exploited for the development of a serological diagnostic test. Indeed, this antigen (Og4C3) has been used for the preparation of a number of diagnostic kits based on the detection of O. gibsoni paramyosin in cattle, which may also cross-react with filarioids infecting humans such as W. bancrofti and B. malayi [27–29, 32] and, to some extent, with O. lupi in serum from dogs that scored positive for microfilariae [30]. In addition, the identification of 621 antibody binding epitopes, unique to O. lupi when compared with other filarioid nematodes, provides a dataset invaluable for the development of a serological diagnostic assay through specific targeting of one or more of these epitopes. Antibody detection of O. lupi would not only assist in the detection of subclinically infected animals, gaining a better understanding of the true distribution and prevalence of O. lupi, but would also be amenable to the rapid screening of potential insect vectors. Vaccine studies would determine whether this molecule can provide a protective effect against infection by a zoonotic nematode for which the full zoonotic potential is yet to be determined. Future work will involve recombinant expression (eukaryotic) of the full protein, production of antibodies and testing of the serum against a range of filarial and other nematodes infecting humans and animals to ensure specificity. Serum from animals with infections of other filarioid nematodes will also be tested against the recombinant protein. If there is no cross-reactivity with serum from animals with other filarioid nematode infections this would provide a strong basis for the development of a specific serological diagnostic test.
Conclusions
This study indicates that paramyosin is a suitable candidate for the development of a specific diagnostic test and as a potential vaccine candidate, avoiding in the future a need for invasive, time-consuming tests. The development of a specific diagnostic test will enable further studies on the distribution and prevalence of this little known zoonotic filarioid.
References
Otranto D, Eberhard ML. Zoonotic helminths affecting the human eye. Parasit Vectors. 2011;4:41.
Sréter T, Széll Z. Onchocercosis: a newly recognized disease in dogs. Vet Parasitol. 2008;151(1):1–13.
Otranto D, Giannelli A, Trumble SN, Chavkin M, Kennard G, Latrofa MS, et al. Clinical case presentation and a review of the literature of canine onchocercosis by Onchocerca lupi in the United States. Parasit Vectors. 2015;8:89–96.
Otranto D, Dantas-Torres F, Giannelli A, Latrofa MS, Papadopoulos E, Cardoso L, Cortes H. Zoonotic Onchocerca lupi in dogs from Greece and Portugal. Emerg Infect Dis. 2013;9:2000–3.
Széll Z, Erdélyi I, Sréter T, Albert M, Varga I. Canine ocular onchocercosis in Hungary. Vet Parasitol. 2001;97:243–9.
Széll Z, Sréter T, Erdélyi I, Varga I. Ocular onchocercosis in dogs: aberrant infection in an accidental host or lupi onchocercosis? Vet Parasitol. 2001;101:115–25.
Hermosilla C, Hetzel U, Bausch M, Grübl J, Bauer C. First autochthonous case of canine ocular onchocercosis in Germany. Vet Rec. 2005;156:450–2.
Sallo F, Eberhard ML, Fok E, Baska F, Hatvani I. Zoonotic intravitreal Onchocerca in Hungary. Ophthalmology. 2005;112:502–4.
Faísca P, Morales-Hojas R, Alves M, Gomes J, Botelho M, Melo M, Xufre A. A case of canine ocular onchocercosis in Portugal. Vet Ophthalmol. 2010;13:117–21.
Miró G, Montoya A, Checa R, Gálvez R, Mínguez JJ, Marino V, Otranto D. First detection of Onchocerca lupi infection in dogs in southern Spain. Parasit Vectors. 2016;9:290–2.
Labelle AL, Daniels JB, Dix M, Labelle P. Onchocerca lupi causing ocular disease in two cats. Vet Ophthalmol. 2011;14:105–10.
Labelle AL, Maddox CW, Daniels JB, Lanka S, Eggett TE, Dubielzig RR, Labelle P. Canine ocular onchocercosis in the United States is associated with Onchocerca lupi. Vet Parasitol. 2013;193:297–301.
Burr Jr WE, Brown MF, Eberhard ML. Zoonotic Onchocerca (Nematoda: Filarioidea) in the cornea of a Colorado resident. Ophthalmology. 1998;105:1494–7.
Otranto D, Sakru N, Testini G, Gürlü VP, Yakar K, Lia RP, et al. Case report: First evidence of human zoonotic infection by Onchocerca lupi (Spirurida, Onchocercidae). Am J Trop Med Hyg. 2011;84:55–8.
Otranto D, Dantas-Torres F, Cebeci Z, Yeniad B, Buyukbabani N, Boral OB, et al. Human ocular filariasis: further evidence on the zoonotic role of Onchocerca lupi. Parasit Vectors. 2012;5:84.
Otranto D, Dantas-Torres F, Papadopoulos E, Petrić D, Ćupina AI, Bain O. Tracking the vector of Onchocerca lupi in a rural area of Greece. Emerg Infect Dis. 2012;18:1196–2000.
Ilhan HD, Yaman A, Morishima Y, Sugiyama H, Muto M, Yamasaki H, et al. Onchocerca lupi infection in Turkey: a unique case of a rare human parasite. Acta Parasitol. 2013;58:384–8.
Mowlavi G, Farzbod F, Kheirkhah A, Mobedi I, Bowman DD, Naddaf SR. Human ocular onchocerciasis caused by Onchocerca lupi (Spirurida, Onchocercidae) in Iran. J Helminthol. 2013;6:1–6.
Eberhard ML, Ostovar GA, Chundu K, Hobohm D, Feiz-Erfan I, Mathison BA, et al. Zoonotic Onchocerca lupi infection in a 22-month-old child in Arizona: first report in the United States and a review of the literature. Am J Trop Med Hyg. 2013;88:601–5.
Chen T, Moon K, de Mello DE, Feiz-Erfan I, Theodore N, Bhardwaj RD. Case report of an epidural cervical Onchocerca lupi infection in a 13-year-old boy. J Neurosurg Pediatr. 2015;16:217–21.
Dudley RWR, Smith C, Dishop M, Mirsky D, Handler MH, Suchitra R. A cervical spine mass caused by Onchocerca lupi. Lancet. 2015;386:1372.
Cantey PT, Weeks J, Edwards M, Rao S, Ostovar GA, et al. The Emergence of zoonotic Onchocerca lupi infection in the United States - a case-series. Clin Infect Dis. 2016;62:778–83.
Egyed Z, Sréter T, Széll Z, Beszteri B, Oravecz O, Márialigeti K, Varga I. Morphologic and genetic characterization of Onchocerca lupi infecting dogs. Vet Parasitol. 2001;102:309–19.
Egyed Z, Sréter T, Széll Z, Nyiro G, Márialigeti K, Varga I. Molecular phylogenetic analysis of Onchocerca lupi and its Wolbachia endosymbiont. Vet Parasitol. 2002;108:153–61.
Sréter-Lancz Z, Széll Z, Sréter T. Molecular genetic comparison of Onchocerca sp. infecting dogs in Europe with other spirurid nematodes including Onchocerca lienalis. Vet Parasitol. 2007;148:365–70.
Franchini D, Giannelli A, Di Paola G, Cortes H, Cardoso L, Lia RP, et al. Image diagnosis of zoonotic onchocercosis by Onchocerca lupi. Vet Parasitol. 2014;doi: 10.1016/j.vetpar.2014.03.007.
More SJ, Copeman DB. A highly specific and sensitive monoclonal antibody-based ELISA for the detection of circulating antigen in Bancroftian filariasis. Trop Med Parasitol. 1990;41:403–6.
Chanteau S, Moulia-Pelat JP, Glaziou P, Nguyen NL, Luquiaud P, Plichart C, et al. Og4C3 circulating antigen: a marker of infection and adult worm burden in Wuchereria bancrofti filariasis. J Infect Dis. 1994;170:247–50.
Langy S, Plichart C, Luquiaud P, Williams SA, Nicolas L. The immunodominant Brugia malayi paramyosin as a marker of current infection with Wuchereria bancrofti adult worms. Infect Immun. 1998;66:2854–8.
Giannelli A, Cantacessi C, Graves P, Becker L, Campbell BE, Dantas-Torres F, et al. A preliminary investigation of serological tools for the detection of Onchocerca lupi infection in dogs. Parasitol Res. 2014;113:1989–91.
Gobert GN, McManus DP. Update on paramyosin in parasitic worms. Parasitol Int. 2005;54:101–7.
Lammie PJ, Hightower AW, Eberhard ML. Age-specific prevalence of antigenemia in a Wuchereria bancrofti-exposed population. J Trop Med Hyg. 1994;51:348–55.
Abou-Elhakam HMA, Bauomy IR, El Deeb SO, El Amir AM. Immunodiagnosis of fasciolasis using sandwich enzyme-linked immunosorbent assay for detection of Fasciola gigantica paramyosin antigen. Tropical Parasitol. 2013;3:44–52.
Kalinna BH, McManus DP. A Vaccine against the Asian Schistosome, Schistosoma japonicum: an update on paramyosin as a target of protective immunity. Int J Parasitol. 1997;27:1213–9.
McManus DP, Liu S, Song G, Xu Y, Wong JM. The vaccine efficacy of native paramyosin (Sj-97) against Chinese Schistosoma japonicum. Int J Parasitol. 1998;28:1739–42.
Taylor MG, Huggins MC, Shi F, Lin J, Tian E, Ye P, et al. Production and testing of Schistosoma japonicum candidate vaccine antigens in the natural ovine host. Vaccine. 1998;16:1290–8.
Chen H, Nara T, Zeng X, Satoh M, Wu G, et al. Vaccination of domestic pig with recombinant paramyosin against Schistosoma japonicum in China. Vaccine. 2000;18:2142–6.
Hartmann S, Sereda MJ, Sollwedel A, Kalinna B, Lucius R. A nematode allergen elicits protection against challenge infection under specific conditions. Vaccine. 2006;24:3581–90.
Wang X, Chen W, Lu X, Tian Y, Men J, Zhang X, et al. Identification and characterization of paramyosin from cyst wall of metacercariae implicated protective efficacy against Clonorchis sinensis infection. PLoS ONE. 2012;7:e33703.
Strube C, Haake C, Sager H, Weber SS, Kaminsky R, et al. Vaccination with recombinant paramyosin against the bovine lungworm Dictyocaulus vivparus considerably reduces worm burden and larvae shedding. Parasit Vectors. 2015;8:119–30.
Yang J, Gu Y, Yang Y, Wei J, Wang S, et al. Trichinella spiralis: Immune response and protective immunity elicited by recombinant paramyosin formulated with different adjuvants. Exp Parasitol. 2010;124:403–8.
Hu Y, Zhang J, Yang S, Wang H, Zeng H, et al. Screening and molecular cloning of a protective antigen from the midgut of Haemaphysalis longicornis. Korean J Parasitol. 2013;51:327–34.
Deng J, Gold D, LoVerde P, Fishelson Z. Mapping of the complement C9 binding domain in paramyosin of the blood fluke Schistosoma mansoni. Int J Parasitol. 2007;37:67–75.
Zhang Z, Yang J, Wei J, Yang Y, Chen X, Zhao X, et al. Trichinella spiralis paramyosin binds to C8 and C9 and protects the tissue-dwelling nematode from being attacked by host complement. PLoS Negl Trop Dis. 2011;5:e1225.
Loukas A, Jones MK, King LT, Brindley PJ, McManus DP. Receptor for Fc on the surface of schistosomes. Infect Immun. 2001;69:3646–51.
Jiz M, Wu H-W, Meng R, Pond-Tor S, Reynolds M, et al. Pilot-scale production and characterization of paramyosin, a vaccine candidate for schistosomiasis japonica. Infect Immun. 2008;76(7):3164–9.
Campbell BE, Nisbet AJ, Mulvenna J, Loukas A, Gasser RB. Molecular and phylogenetic characterization of cytochromes c from Haemonchus contortus and Trichostrongylus vitrinus (Nematoda: Trichostrongylida). Gene. 2008;424:121–9.
Nielsen H, Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. In: Proceedings of the sixth international conference on intelligent systems for molecular biology (ISMB 6). Menlo Park, California, USA: AAAI Press; 1998. p. 122–30.
Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP. J Mol Biol. 2004;340:783–90.
Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 40: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6.
Sonnhammer ELL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Proceedings of the sixth international conference on intelligent systems for molecular biology. Menlo Park, California, USA: AAAI Press; 1998. p. 175–82.
Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148.
Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–9.
Karplus PA, Schulz GE. Prediction of chain flexibility in proteins. Naturwissenschaften. 1985;72:212–3.
Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–4.
Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Milstein CP. Isolation of Aulacomya Paramyosin. Biochem J. 1967;103:634–40.
Choi Y-J, Ghedin E, Berriman M, McQuillan J, Holroyd N, Mayhew GF, et al. A deep sequencing approach to comparatively analyze the transcriptome of life cycle stages of the filarial worm, Brugia malayi. PLoS Neg Trop Dis. 2011;5:e1409.
Park T-J, Kang J-M, Na B-K, Sohn W-M. Molecular cloning and characterization of a paramyosin from Clonorchis sinensis. Korean J Parasitol. 2009;47:359–67.
Strube C, Buschbaum S, von Samson-Himmelstjerna G, Schneider T. Stage-dependent transcriptional changes and characterization of paramyosin of the bovine lungworm Dictyocaulus viviparus. Parasitol Int. 2009;58:334–40.
Jenkins RE, Taylor MJ, Gilvary NJ, Bianco AE. Tropomyosin implicated in host protective responses to microfilariae in onchocerciasis. Proc Natl Acad Sci USA. 1998;95:7550–5.
Nara T, Tanabe K, Mahakunkijcharoen Y, Osada Y, Matsumoto N, Kita K, Kojima S. The B cell epitope of paramyosin recognized by a protective monoclonal IgE antibody to Schistosoma japonicum. Vaccine. 1997;15:79–84.
Fonseca CT, Cunha-Neto E, Goldberg AC, Kalil J, de Jesus AR, Carvalho EM, et al. Identification of paramyosin T cell epitopes associated with human resistance to Schistosoma mansoni reinfection. Clin Exp Immunol. 2005;142:539–47.
Gazarian KG, Gazarian TG, Solís CF, Hernández R, Shoemaker CB, Laclette JP. Epitope mapping on N-terminal region of Taenia solium paramyosin. Immunol Lett. 2000;72:191–5.
López-Moreno HS, Correa D, Laclette JP, Ortiz-Navarrete VF. Identification of CD4+ T cell epitopes of Taenia solium paramyosin. Parasite Immunol. 2003;25:513–6.
Wei J, Gu Y, Yang J, Yang Y, Wang S, Cui S, Zhu X. Identification and characterization of protective epitope of Trichinella spiralis paramyosin. Vaccine. 2011;29:3162–8.
Waine GJ, Yang W, Scott JC, McManus DP, Kalinna BH. DNA-based vaccination using Schistosoma japonicum Asian blood-fluke genes. Vaccine. 1997;15:846–8.
Zhou S, Liu S, Song G, Xu Y, Sun W. Protective immunity induced by the full-length cDNA encoding paramyosin of Chinese Schistosoma japonicum. Vaccine. 2000;18:3196–204.
Li B-W, Zhang S, Curtis KC, Weil GJ. Immune responses to Brugia malayi paramyosin in rodents after DNA vaccination. Vaccine. 2000;18:76–81.
Solís CF, Ostoa-Saloma P, Lugo-Martínez VH, Johnston A, Laclette JP. Genetic vaccination against murine cysticercosis by using a plasmid vector carrying Taenia solium paramyosin. Infect Immun. 2005;73:1895–7.
Yang W, Jackson DC, Zeng Q, McManus DP. Multi-epitope schistosome vaccine candidates tested for protective immunogenicity in mice. Vaccine. 2001;19:103–13.
Acknowledgments
The first author would like to take the opportunity to thank all past and present members of the Laboratorio di Parassitologia e Micologia e Infettive, Dipartimento di Medicina Veterinaria, Università degli Studi di Bari “Aldo Moro”, Italy. Special mentions for Rafael Antonio do Nascimento Ramos and Riccardo Paolo Lia. Maria João Vila-Viçosa (Victor Caeiro Laboratory of Parasitology, Universidade de Évora, Portugal) is thanked for her skilled technical support in the dissection of adult worms.
Funding
This study was partially funded by Merial.
Availability of data and material
The full-length paramyosin cDNA (Accession no. KJ699378) and protein (Accession no. AJO15918.1) sequences are available in the NCBI database. All predicted antibody binding epitopes are available in Additional file 1: Figure S1.
Authors’ contributions
BC, AG and DO conceived the research. HC, LC, BC and AG collected the samples. BC and GA performed the experiments. AP performed the DNA sequencing. BC performed the data analysis and wrote the manuscript, with contributions from DO for interpretation of the data and revision of the manuscript. AG, SL and FDT contributed to revision of the manuscript. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was conducted according to the Guideline on Good Clinical Practices (The European Agency for the Evaluation of Medicinal Products, Veterinary Medicines and Information Technology Unit, VICH Topic GL9; www.emea.eu.int/pdfs/vet/vich/059598en.pdf) and procedures were approved by the ethical commission at the University of Évora (identification number: AE02Fila2013), complying with Portuguese legislation for the protection of animals (Decree-Law no. 113/2013). An owner consent agreement was obtained before sample collection.
Author information
Authors and Affiliations
Corresponding author
Additional file
Additional file 1: Figure S1.
Epitopes predicted by bioinformatics and those reported in the literature for other organisms mapped to Onchocerca lupi paramyosin. Residues underlined indicate T-cell epitopes whilst those in bold indicate B-cell epitopes. Of the 874 predicted antibody epitopes, when compared with those predicted for Acanthocheilonema viteae, Brugia malayi, Dirofilaria immitis, Loa loa, Onchocerca volvulus and Wuchereria bancrofti, 621 were unique to Onchocerca lupi. (DOCX 32 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Campbell, B., Cortes, H., Annoscia, G. et al. Paramyosin of canine Onchocerca lupi: usefulness for the diagnosis of a neglected zoonotic disease. Parasites Vectors 9, 493 (2016). https://doi.org/10.1186/s13071-016-1783-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1783-z