Abstract
Background
Trypanosoma cruzi infection via oral route results in outbreaks or cases of acute Chagas disease (ACD) in different Brazilian regions and poses a novel epidemiological scenario. In the Espírito Santo state (southeastern Brazil), a fatal case of a patient with ACD led us to investigate the enzootic scenario to avoid the development of new cases. At the studied locality, Triatoma vitticeps exhibited high T. cruzi infection rates and frequently invaded residences.
Methods
Sylvatic and domestic mammals in the Rio da Prata locality, where the ACD case occurred, and in four surrounding areas (Baia Nova, Buenos Aires, Santa Rita and Todos os Santos) were examined and underwent parasitological and serological tests. Triatomines were collected for a fecal material exam, culturing and mini-exon gene molecular characterization, followed by RFLP-PCR of H3/Alul. Paraffin-embedded cardiac tissue of a patient was washed with xylene to remove paraffin and DNA was extracted using the phenol-chloroform method. For genotype characterization, PCR was performed to amplify the 1f8, GPI and 18S rRNA genes. In the case of V7V8 SSU rRNA, the PCR products were molecularly cloned. PCR products were sequenced and compared to sequences in GenBank. Phylogenetic analysis using maximum likelihood method with 1000 bootstrap replicates was performed.
Results
None of the animals showed positive hemocultures. Three rodents and two dogs showed signs of infection, as inferred from borderline serological titers. T. vitticeps was the only triatomine species identified and showed T. cruzi infection by DTUs TcI and TcIV. The analysis of cardiac tissue DNA showed mixed infection by T. cruzi (DTUs I, II, III and IV) and Trypanosoma dionisii.
Conclusions
Each case or outbreak of ACD should be analyzed as a particular epidemiological occurrence. The results indicated that mixed infections in humans may play a role in pathogenicity and may be more common than is currently recognized. Direct molecular characterization from biological samples is essential because this procedure avoids parasite selection. T. dionisii may under certain and unknown circumstances infect humans. The distribution of T. cruzi DTUS TcIII and TcIV in Brazilian biomes is broader than has been assumed to date.
Similar content being viewed by others
Background
The genus Trypanosoma (Trypanosomatidae, Kinetoplastida), which includes the subgenus Schizotrypanum, is composed of numerous species that are distributed worldwide. Humans or other mammals can serve as suitable hosts. With the exception of Trypanosoma cruzi, other species of this subgenus are restricted to bats. Due to their morphological similarity, these other species have been classically described as T. cruzi-like [1, 2]. The biological cycles of Schizotrypanum trypanosomes are similar, differing only in the identity of their mammalian hosts and their hemipteran vectors. Trypanosoma cruzi marinkellei is transmitted by triatomine insects of the genus Cavernicola, and Trypanosoma dionisii is transmitted by Cimicidae. Species of Schizotrypanum are the only trypanosomes described thus far that infect mammalian cells and multiply inside them as amastigotes [2–4]. There is still much to study regarding T. dionisii and T. c. marinkellei. Furthermore, despite being the subject of intensive study for more than 100 years, there are still several unanswered questions pertaining to the biology of T. cruzi.
Trypanosomiasis by T. cruzi is primarily a sylvatic enzooty. This flagellate species is widely distributed in nature, occurring from the southern United States (USA) through southern Argentina and Chile [5]. Trypanosoma cruzi circulates among 150 mammal species and is capable of colonizing almost any tissue of its mammalian hosts. It can also be transmitted by dozens of triatomine species [6]. The parasite transmission cycle is complex in nature because, in addition to its tremendous host species diversity, T. cruzi is highly genetically diverse [7]. Currently, six Discrete Typing Units (DTUs), TcI to TcVI, in addition to TcBat, are recognized [8–10]. Correlations among DTUs/geographical distribution/host species and pathogenicity are still controversial. Classically, TcII, TcV and TcVI were related to severe human diseases and TcI, TcIII and TcIV were related to the sylvatic cycle [10], but in the Amazon region, Colombia and Venezuela, reports have described human disease by TcI, TcIII and TcIV [11–16]. Although diverse studies have proposed these and other correlations, this topic still requires further clarification. Trypanosoma cruzi populations can be selected when they are grown under laboratory conditions or even when natural infections lead to erroneous conclusions regarding DTU variety and putative associations [17, 18]. Similarly, due to the undersampling of hosts and habitats, the ecology of the DTUs of T. cruzi is far from well understood.
In Brazil, the efficient control of Chagas disease (CD) due to intra-domiciliary transmission of T. cruzi by Triatoma infestans has been largely achieved. However, human infection by T. cruzi is re-emerging as a food-borne disease in previously non-endemic areas, such as the Amazon region, where it is associated with the ingestion of Açai juice [19–21]. The oral route transmission has been demonstrated to be a highly efficient mechanism of infection [22, 23]. Acute Chagas disease (ACD) cases and outbreaks involving triatomines, which were not previously considered as the main vector species for the contaminative vectorial route, demonstrate that any triatomine can act as a vector when it is related to oral transmission [24]. Moreover, in sylvatic environments, this mechanism is likely the primary means of infection between animals [25].
In the Espírito Santo State (ES), the invasion of domiciles by infected triatomines (mainly Triatoma vitticeps but also Panstrongylus geniculatus) is frequently reported in rural areas, primarily in mountainous regions that have irregular terrain [26]. Triatoma vitticeps is the more prevalent species in ES and can be found in Rio de Janeiro, Minas Gerais and Bahia states [27, 28]. Triatoma vitticeps occasionally forms colonies associated with opossum nests in peridomiciles and has high infection rates by flagellates, such as T. cruzi [29, 30]. In a study conducted between 2010 and 2012 (Dario, unpublished data), 55 T. cruzi isolates derived from T. vitticeps and P. geniculatus collected in ES subjected to molecular characterization, demonstrated the transmission of four T. cruzi DTUs (TcI, TcII, TcIII and TcIV).
Despite the high T. cruzi infection rates, T. vitticeps has always been considered a secondary vector of CD due to the long time interval between feeding and defecation, reducing the success for the classical triatomine-human route transmission [31]. Since 2007, according to the Espírito Santo state Health Department (Sesa/ES), only three cases of CD were reported in ES. The last case, in 2012, led to the death of a 2-year-old patient, and epidemiological investigation showed the cause to be ACD acquired by the oral route due to the manipulation of a recently dead (and infected) T. vitticeps.
This study had the following objectives: (i) to determine the T. cruzi DTU that was related to the fatal case that occurred in ES and (ii) to study the ecology of the transmission cycle of T. cruzi in the area where the ACD occurred and in nearby locations where triatomines continuously invade residences. The aim of these objectives is to contribute to the development of local control measures to prevent additional cases of CD.
Methods
Cardiac tissue sample
A cardiac fragment was collected during the post mortem patient’s exam, embedded in paraffin and sent to Sesa/ES. The cardiac fragment was kindly donated by Dr Janaina A. Shineider Casotti from Sesa/ES.
Study area
The Guarapari municipality is located in the Central coast of the ES state. It is 594,487 km2 in size and has a population of 105,286; of these, 4758 live in rural areas. The rural area in which the case occurred (and where we examined the domestic and wild animals) is located mostly in mountainous areas, which contain remnants of Atlantic rainforest. The residents of the rural area report banana and coffee agriculture to be the main source of their income.
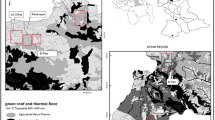
The study was conducted at five localities: Rio da Prata, where the patient's case occurred; Baia Nova; Buenos Aires; Todos os Santos; and Santa Rita (Fig. 1), which, according to Zoonosis Control Center (ZCC), have registered a higher number of infected triatomines invading residences in recent years.
Representative map of the locality of the serological results of small sylvatic and domestic mammals. The positive mammals are framed in yellow. Only one rodent was positive at the Buenos Aires location and two were positive at the Baia Nova location. Two dogs were positive at the Baia Nova (n = 1) and Santa Rita (n = 1) locations
Small wild mammal capture
Fieldwork was conducted in June 2012, just a few months after the occurrence of the fatal case. Small wild mammals were captured using the following protocol: linear transects were designed in which capture points were established (each point 10 m apart) using alternating Shermann® (H. B. Sherman Traps, Tallahassee, FL, USA) and Tomahawk® (Tomahawk Live Traps, Tomahawk, WI, USA) live traps baited with a mixture of banana, peanut butter, bacon and sardine. The traps were placed near houses and in wild habitats. Seven transects with 10 Sherman and 10 Tomahawk traps each were placed in the field for five nights, with a total capture effort of 840 traps-night.
All captured animals were manipulated according to the safety manual for the use of wild mammals in research [32] and were anesthetized (9:1 ketamine chlorhydrate 10 % and acepromazine 2 %) for blood sample collection (by cardiac puncture) for parasitological and serological analyses. Only those mammals that required taxonomical confirmation obtained by karyotyping were sacrificed [33].
Dog survey
An active search for dogs was performed at the following locations: Rio da Prata, Baia Nova and Santa Rita. At all of these locations, with the owner’s consent, a blood sample was collected by puncturing the cephalic vein under aseptic conditions with Vacutainer® tubes with anticoagulant for serological and parasitological tests. A canine questionnaire was given to the owner that requested the following information: name of the dog, age, sex and the dog’s primary function (protection, hunting or company). We considered dogs to be juveniles when they were less than one year of age and adults when they were more than one year of age. Each dog was considered a single event, even if they lived at the same house.
Trypanosoma cruzi survey
To survey for T. cruzi, parasitological and serological tests were performed for both small wild mammals and dogs. The following parasitological tests were conducted: (i) fresh blood examination to visualize T. cruzi flagellates and (ii) hemoculture, which involved the inoculation of 0.6 ml of blood into two tubes containing Novy Mc Neal Nicole (NNN) medium with Liver Infusion Tryptose (LIT) overlay. The tubes were examined every two weeks for a total of three (for seronegative animals) or five months (for seropositive animals). When positive, the parasites were amplified in LIT, cryopreserved and deposited in the Coleção de Trypanosoma de Mamíferos Silvestres, Domésticos e Vetores, Fiocruz - COLTRYP (Oswaldo Cruz Foundation, Rio de Janeiro - RJ/Brazil). Positive hemoculture results also showed that the animal exhibited notable parasitemia levels.
A serological survey for the detection of anti-T. cruzi IgG antibodies was performed using an Indirect Immunofluorescent Antibody Test (IFAT), as described by [34]. The antigens used in the reaction were an equal mixture of parasites derived from the strains I00/BR/00 F (TcI) and MHOM/BR/1957/Y (TcII). The sera of Murinae rodents were tested with anti-rat IgG, while the sera of dogs were tested with anti-dog IgG. All sera were conjugated to fluorescein isothiocyanate (Sigma, St Louis, MO, USA). Echimyidae rodents and marsupials sera were tested using intermediary anti-IgG antibodies for Thrichomys spp. and anti-IgG for Didelphidae, respectively, both of which were raised in rabbits. The reaction was revealed using anti-rabbit IgG antibodies conjugated with fluorescein (Sigma, St Louis, MO, USA). The cutoff values for serological results were 1:40 for dogs and marsupials and 1:10 for rodents [35].
To avoid possible cross-reactions with other trypanosomatids, small mammals and dogs were screened to detect anti-Leishmania IgG antibodies through IFAT, as described above, using antigens derived from a mixture of Leishmania infantum and L. braziliensis. Animals were considered positive for Leishmania spp. when the serological titers for this parasite were higher than for T. cruzi by at least two dilutions and were considered to present both infections when titers were > 1:40 in each assay. Animals were considered to present T. cruzi infection when the serological titer was higher that the cutoff value analysis and/or when hemoculture were positive.
In-house Enzyme-Linked Immunosorbent Assays (ELISA) were performed to confirm infections in dogs by T. cruzi and Leishmania sp. The mean negative control optical density, which added 20 % to this value via a dog serum panel, established the cutoff values in each plate. For each serological reaction, two T. cruzi and Leishmania sp. positive and negative control sera were added.
Trypanosoma cruzi survey in triatomines
Triatomines were collected inside houses by residents and delivered to the ZCC. Triatomine identification was performed according to the method of Lent & Wygodzinsky [36]. The intestinal contents were removed using scissors and forceps, and examined on a microscope slide with a cover slip under an optical microscope to search for flagellar forms similar to T. cruzi. When the exam was positive, the sample was cultured in NNN with LIT overlay and was examined every two weeks for up to five months [37, 38]. In addition, the culture was amplified, cryopreserved and deposited in COLTRYP, as described previously.
Trypanosoma cruzi molecular characterization
DNA extraction
T. cruzi DNA derived from epimastigotes in the logarithmic phase of the cultures and DNA from the cardiac tissue embedded in the paraffin was extracted using the standard phenol-chloroform method [39]. Prior to this step, the cultures were washed with phosphate-buffered saline (PBS) solution and incubated with proteinase K (100 μg/ml) and 0.5 % sodium dodecyl sulfate (SDS) at 56 °C for two hours. For paraffin removal, the cardiac tissue was washed using a previously described method [40, 41]. DNA from the cardiac tissue was quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, San Jose, CA, USA), and the final concentration was adjusted to 50, 100, 150, 200 and 250 ng/μl. To avoid contamination, only unused aerosol-resistant pipette tips were used, and PCR was conducted in a separate room free of any T. cruzi or T. dionisii DNA (we do not have T. dionisii isolates in our laboratory). Irradiation with ultraviolet (UV) light was also performed on all materials, such as pipets, filter tips, PCR tubes and the cabinet area where the PCR was carried out.
Culture characterization
The parasite characterization of epimastigotes from positive cultures was performed as follows. First, multiplex-PCR was performed to amplify the non-transcribed spacer of the mini-exon gene [42] for the identification of TcI (DTU I), TcII (DTU II/V/VI), zymodeme 3 (DTU III/IV) and T. rangeli fragments of 200 bp, 250 bp, 150 bp and 100 bp [43], respectively, as well as mixed infections. Positive samples, except for TcI, were amplified by PCR for the histone 3 (H3) gene [44] followed by restriction fragment length polymorphism (RFLP) analysis. The fragments were digested by the AluI enzyme for discrimination of Z3 (DTUs III or IV).
Electrophoresis of PCR products was carried out in a 2 % agarose gel, which was stained with ethidium bromide solution and visualized under UV light. All reactions included distilled water as a negative control. Trypanosoma cruzi strains, representing all DTUs (TcI-SylvioX/10cl1; TcII-Esmeraldocl3; TcIII-M5631cl5; TcIV-92122102R and TcV/VI-SC43cl1), and T. rangeli (Choco) samples were used as positive controls.
Cardiac tissue characterization
For DTU identification, DNA extracted from cardiac tissue was used to amplify three nuclear markers: 1f8 (950 bp), glucose-phosphate isomerase (GPI) (652 bp) and the third portion of variable regions 7 and 8 (V7V8) of 18S rRNA gene (650 bp) (45), according to previous studies [46–48]. PCR products were purified using the Illustra GFX PCR DNA and gel band purification kit (GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK). In addition, the V7V8 region of SSU rRNA (750–800bp) was amplified as described [49].
V7V8 SSU rRNA PCR products were purified with a Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA) and cloned using the pGEM-T Easy Vector System (Promega, Madison, WI, USA) per the manufacturer’s protocol. Sixteen colonies were randomly collected and minipreps were performed with Invisorb Spin Plasmid Mini Two kits (STRATEC Biomedical AG, Germany).
All of the samples were sequenced for both strands of DNA with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster city, CA, USA) on an ABI 3730 DNA sequencer available on the PDTIS/FIOCRUZ sequencing platform (Fig. 2). Two clones generated poor sequences and were excluded from the analysis.
Sequence and phylogenetic analysis
The sequences were edited, aligned and corrected using the BioEdit software. The sequences were compared with nucleotide sequences deposited in GenBank using the BLAST (Basic Local Alignment Search Tool) algorithm. Phylogenetic tree construction was performed using Mega 5 software [50]. We used the maximum likelihood (ML) method, employing the best DNA model. The best substitution model was identified as having the lowest Bayesian Information Criterion score (BIC): Hasegawa-Kishino-Yano for the 1f8 gene, Tamura 3+ G (a gamma-distributed rate of variation among sites) parameter for the GPI gene, Kimura 2-parameter for the 18S rRNA gene, and the Kimura 2 + G parameter for V7V8 SSU rRNA, with bootstrapping at 1000 replicates. We used T. cruzi (TcI to TcVI), T. c. marinkellei, T. rangeli and T. dionisii sequences from GenBank as references. All sequences analyzed were deposited in the GenBank database under the accession numbers KR905432–KR905446 for the 18S rRNA marker, KT737478 for GPI and KT983981 for 1f8. The GenBank accession numbers can be viewed in Additional file 1.
Results
Identification of Trypanosoma cruzi DTUs and T. dionisii in cardiac tissue
In this study, we decided to use three nuclear markers to genotype the DNA obtained from the cardiac tissue: 1f8, GPI and 18S rRNA genes. Nuclear markers cluster separately with T. cruzi DTUs (TcI to TcIV) [44, 51–53]. For the 18S rRNA gene, which exists as thousands of copies in the genome, we used two different regions for characterization: a variable region (V7V8) and a third portion of this variable region [45, 48]. 18S rRNA gene allows identification of different species within the subgenus Schizotrypanum and is considered a reliable marker to distinguish between T. cruzi DTUs [45, 54–57]. The 1f8 gene allows discrimination between the DTUs TcI and TcIV [44, 46]. The GPI gene, which is also considered a suitable target to distinguish between DTUs, revealed that TcI, TcII, TcIII and TcIV are robust monophyletic groups [5, 13, 52, 58, 59].
We demonstrated via a PCR method the occurrence of four sympatric T. cruzi DTUs (TcI, TcII, TcIII and TcIV) in the cardiac tissue of a patient who died in the acute phase of Chagas disease. This is the first time that we observed such a diversity of DTUs in a human case. Furthermore, we also detected T. dionisii, a Trypanosoma species that has only been described in bats until now, by phylogenetic and additional alignment analyses (Additional file 2).
The DTU TcIV was detected in cardiac tissue by employing 1f8 and GPI as molecular targets. The sequence obtained by amplification of the 1f8 gene was subjected to BLAST algorithm and shown to be similar to both TcIII (CM17 and M5631) and TcIV (CANIIII and M4167) strains (96–97 %). According to the ML phylogenetic analysis, the DNA sequence clustered together with the TcIV reference strain CANIII (Fig. 3a). To confirm this result, we sought to determine the occurrence of this DTU based on GPI. With this target, the nucleotide sequence showed 99 % identity to TcIV (Ep272, Saimiri3cl1 and CANIII) strains. In addition, the phylogenetic analysis clustered the sequence with the TcIV reference strain Ep272 (Fig. 3b). DNA from the cardiac tissue was subjected to PCR and molecular cloning of the V7V8 SSU rRNA region. The BLAST algorithm showed that one clone presented a similarity of 100 % to TcI (Dm28c strain), one clone presented a similarity of 99 % to TcII/VI (isolate TCC873, Tulahuencl2, TCC2558), three clones presented similarities of 99 % and 100 % to TcIII (MT3663 strain, isolates TryCC1356 and 1078) and six clones presented similarities of 99–100 % to TcIV (Jose Julio, MT4167 and CanIII strains). The phylogenetic analysis clustered the clones with TcI (n = 01), TcII/TcVI (n = 01), TcIII (n = 03) and TcIV (n = 06) (Fig. 4).
Furthermore, three clones were identified as T. dionisii via phylogenetic analysis using the ML method. The identification of T. dionisii also occurred via sequence analysis of the 18S rRNA gene. The sequence obtained from 18S rRNA nested-PCR was subjected to BLAST algorithm. This sequence showed 100 % identity with T. dionisii isolates CBT 63, 64 and 69. In the phylogenetic analysis, the sequence clustered with T. dionisii isolates, confirming the presence of T. dionisii in this human cardiac tissue sample (Figs. 3c and 4).
Molecular phylogenetic analysis by maximum likelihood methods. a for 1f8 gene with HKY parameter and bootstrap value of 1000 replicates; b for GPI gene with T92 + G parameter and bootstrap value of 1000 replicates; and c for 18S rRNA K2 parameter and bootstrap value of 1000 replicates. The 1f8 and GPI DNA sequences clustered with DTU TcIV reference strains CANIII (A) and Ep272 (B), and 18S rDNA sequence clustered together with T. dionisii isolates. T. rangeli isolates RGB, PG and San Augustin were used as the outgroup, except for 1f8 gene, because no sequence is available on GenBank
Molecular phylogenetic analysis by maximum likelihood of the V7V8 SSU rRNA gene. K2 + G parameter and bootstrap value of 1000 replicates. The clones from cardiac tissue clustered with Jose Julio and CANIII strains (TcIV); SO3cl5 clone1 and clone2, M6241cl6 clone2 and MT3663 strains (TcIII/TcV); Dm28c and Colombiana strains (TcI); Esmeraldocl3 clone1 and clone2, and CLBR (TcII/TcVI); and T. dionisii isolates TCC/USP:495 and CBT 59. T. rangeli isolates PG and San Agustin were used as the outgroup
Small mammal capture and Trypanosoma cruzi infection
The study area clearly had a reduced mammalian fauna density and diversity. Despite an extensive capture effort involving 840 traps for five nights, only one species of Rodentia (Trinomys paratus (n = 5)) and four species of Didelphimorphia [Didelphis aurita (n = 1), Philander frenatus (n = 1), Metachirus nudicaudatus (n = 2) and Marmosops incanus (n = 2)] were captured. Additionally, we examined four synanthropic rodents (Rattus rattus) that were collected from the peridomicile area. The relative abundance of mammals captured was higher for Didelphimorphia, which represented 54.5 % of the mammals captured, whereas the percentage of Rodentia represented 45.5 %.
None of the sylvatic animals had parasites based on the examination of fresh blood or hemoculture. In serological tests, only three specimens of T. paratus, two from Baia Nova and one from Buenos Aires locations, were found to be infected with T. cruzi (Fig. 1). The three positive rodents presented only borderline serological titers (1:20) (Table 1).
Dogs and Trypanosoma cruzi infection
Dogs are considered sentinel hosts [60], signaling that T. cruzi cycle is occurring in a peridomicile area. Fifteen dogs were examined from the following locations: Rio da Prata -house of the infected patient (n = 10), Baia Nova (n = 2) and Santa Rita (n = 3). Of this total, only two, one from Baia Nova and the other from Santa Rita, displayed only borderline IFAT tests for T. cruzi in IFAT (both 1:40) and ELISA (Table 1). All of the dogs (n = 10) from the house where the patient lived were negative for T. cruzi based on serological and parasitological tests. This finding means that the dogs from Rio de Prata have not been exposed to T. cruzi infection.
Triatomine infection and molecular characterization
Five triatomine specimens were delivered to the ZCC during our expedition. All of the specimens were identified as T. vitticeps. These specimens were from São Miguel (n = 1), Rio da Prata (n = 2) and Baia Nova (n = 2). Four T. vitticeps (75 %) had T. cruzi based on the intestinal content exam. Only one sample from Rio da Prata (LBT 3214) was negative.
Four positive samples were subjected to culture and three samples presented epimastigote forms. Molecular characterization using the non-transcribed spacer of the mini-exon gene was performed, which classified the samples as TcI (DTU TcI) - LBT 3211 and Z3 (DTU TcIII/TcIV) - LBT 3198 and LBT 3210 (Fig. 5a). To discriminate between TcIII and TcIV, which is not possible with the mini-exon gene, the LBT 3198 and LBT 3210 samples were further characterized at the DTU level using the H3 marker, resulting in their classification as TcIV (Fig. 5b).
Trypanosoma cruzi DTU molecular characterization of T. vitticeps isolates LBT 3198, LBT 3210 and LBT 3211. a Products amplified by the mini-exon gene and analyzed by electrophoresis on 2 % agarose gel stained with ethidium bromide. The sample LBT 3198 and LBT 3210 presented an amplicon size of 150 bp and was identified as zimodeme 3 (DTU TcIII-TcIV). The sample LBT 3211 was identified as TcI, as it presented an amplicon size of 200 bp. b PCR-RFLP with histone 3 gene/AluI restriction enzyme for T. cruzi DTU genotype on 3 % agarose gel stained with ethidium bromide. The samples LBT 3198 and LBT 3210 presented amplicons sizes of 210 bp and 440 bp, genotyping these samples as DTU TcIV. DTU reference strains: TcI-SylvioX/10cl1, TcII-Esmeraldocl3, TcIII-M5631cl5, TcIV-9212102R, TcV/TcVI-SC43cl1, T. rangeli-Choco, NC:negative control
Discussion
Another fatal ACD case acquired via the oral route has been reported. Studies investigating this mechanism have attracted attention due to the high number of cases and outbreaks in Brazil, especially in the Amazon region, in addition to other South American countries [61–63]. This new/ancient epidemiological profile of the disease must be studied from a novel perspective because the control measures that are used for the elimination of T. infestans (domiciliary vector) are not well suited for the current threat. Moreover, all rural locations in Guarapari municipality are now at risk of experiencing CD because residents continue to observe triatomine invasions in their residences and, further, are not aware of the oral route of transmission of T. cruzi.
In this study, valuable information was collected regarding ACD due to the unfortunate death of a young patient. Indeed, based on the direct evaluation of infected tissue, a mixed infection by four T. cruzi DTUs (TcI, TcII, TcIII and TcIV) was detected in a concomitant infection with T. dionisii, a bat trypanosomatid. Fixed biological material, such as tissue embedded in paraffin, for diagnoses is an important source to investigate and understand epidemiology [64]. DNA recovered from this type of material is well maintained and does not result in non-specific bands [65–67]. Moreover, T. cruzi has already been diagnosed from mummies [68–70], whose tissue is highly degraded.
This is the first report of a mixed T. cruzi infection by four DTUs, identified using DNA extracted directly from human cardiac tissue. Mixed T. cruzi DTU infections have been described in several different mammal and triatomine species. Cura et al. [71] reported mixed infections by TcI-IV, TcI-III/IV and TcIII-TcIV in different triatomine species on the American continent. In the Amazon region, Lima et al. [72] reported that R. pictipes exhibited a mixed infection of TcI and TcII. Concomitant infections (TcI-TcII and TcII-TcIV) were detected in tissue samples of rodents from the USA [73]. Mixed infections by two or three DTUs in free-living wild mammals have also been described [74]. In humans, mixed infections by two or three DTUs in chronically infected patients have been described in Colombia, Argentina, Chile and in Bolivian patients’ residing in Spain [75–79].
T. vitticeps specimens exhibiting mixed infections with TcI-IV, TcII-III-IV and TcI-TcII in ES have previously been observed by our group (Dario, unpublished data). In the present study, observations were conducted in Guarapari, a municipality of ES, where we observed mixed infections of T. vitticeps by TcI and TcIV. The finding of simultaneous infection by four T. cruzi DTUs in cardiac tissue is consistent with the genotype diversity observed in the state. A high diversity of T. cruzi DTUs is not usually observed in other regions in Brazil: in Poço das Antas (Rio de Janeiro state), where TcII is the main DTU infecting monkeys, TcI infection is rare; in Piauí state, TcI and a few cases of TcII infection have been reported; in Santa Catarina state, both TcI and TcII were reported; and in Pará state, TcI infection has been reported [21, 37, 80–82]. In addition, mixed DTU infections in T. vitticeps may be attributable to differentially infected blood meal sources or mixed DTU infections in mammals.
The DTUs that we detected infecting the patient have already been described in human infections in addition to presenting a large host range. TcI is the most widespread DTU in nature and is primarily responsible for human infections in the Amazon basin in Brazil, Colombia and Venezuela [11, 16, 83]. TcII, which was classically associated with the Southern cone of South America [10, 84], has already been found in the Brazilian Amazon basin, Colombia, Mexico and USA [15, 72, 73, 85, 86]. In nature, TcIII, which was classically associated with the terrestrial transmission cycle and the armadillo from Dasypus novemcinctus, has previously been identified infecting dogs, rodents and marsupials [5, 74, 87–89]. TcIV has also demonstrated a much larger host range as this DTU has been isolated from primates, coati, marsupial, bats, rodent species and Rhodnius brethesi triatomines [73, 74, 90]. This study has shown for the first time that TcIII and TcIV are related to human infection in ES. Until now, these DTUs have only been reported in the Amazon—TcIII and TcIV [12, 13], in Bolivian patients in Spain-TcIV [79], in the Southern and Northeast parts of Brazil—TcIII [91, 92], in Minas Gerais state-TcIII [93] and in Argentina-TcIII [94]. These findings show that the distribution of TcIII and in particular TcIV is higher than has been assumed up to now and confirm that these two DTUs are involved in human infection. Moreover, this finding warns of the danger of establishing associations between a parasite species or a parasite genotype and pathogenicity, course of infection or epidemiology. Indeed, a disease is the result of the interaction of several variables, including the peculiarities of a host specimen.
For the first time, the presence of T. dionisii has been observed in a human sample. This species, which is closely related to T. cruzi, is able to invade mammalian cells as previously demonstrated experimentally [95, 96] and to form cysts in cardiac tissue [97]. We detected T. dionisii directly from cardiac tissue more than two weeks after the infection of the patient. This finding indicates that we demonstrated that T. dionisii is able to invade and differentiate in human cells. Bat trypanosomatid infections are likely self-resolving and we hypothesize that we would not have been able to detect the parasite at later stages of infection. T. dionisii is widely distributed in ES and has been reported in the northern part of the state (Pinheiros municipality) in the bat species Sturnira lillium, Carollia perspicillata, Desmodus rotundus, Myotis nigricans and Lophostoma brasiliensis [98], and particularly in Guarapari, in bats of Carollia species (Dario, unpublished data). The vector of T. dionisii is still unknown. There has only been one report on Cimicid insects that maintained an experimental infection by T. dionisii [99].
Monogenetic and non-human digenetic trypanosomatid species have already been described to infect humans. Trypanosomes from the subgenus Herpetosoma (T. lewisi, T. lewisi-like), subgenus Dutonella (T. vivax), subgenus Trypanozoon (T. b. brucei and T. evansi), subgenus Nannomonas (T. congolense) and the genus Leptomonas (Leptomonas seymouri) were identified as infecting humans in Africa and Asia [100–103]. Leishmania tarentolae, a species that typically occurs in lizards, has been identified in mummies [104]. Additionally, TcBat, a T. cruzi DTU reported in bats from different Latin American countries [8, 56, 105], has been described in mummies [71] and in a child from Colombia [106]. These findings show that trypanosomatids are biologically plastic and may be host generalist parasites.
Parasite maintenance via cultivation in axenic media or by passaging in experimentally infected animals results in selective pressure [17, 79, 107], making it difficult to detect the assemblage of clonal components of a given T. cruzi isolate. Additionally, during the course of infection, the infected host exerts selective pressures on the parasite population. As a result, during the course of the infection or due to differences in the growth rates of specific populations [108, 109], some populations may be preferentially selected over others [107]. Our study reinforces the importance of the direct characterization of biological samples. In this case, which was a case of CD acquired by the oral route; it is possible that the identification of all of the T. cruzi DTUs and T. dionisii would not have been possible if we had analyzed the hemoculture of the patient or the same cardiac tissue in the chronic phase of the disease.
In nature, the detection of several different parasite species in the same host is common. Furthermore, the impact of mixed infections on a host is still not well understood. Numerous models and experimental studies have been carried out and in general, they have concluded that mixed infections can affect the host immune response and result in increases in virulence [110, 111]. Female tamarins infected with T. cruzi and Acanthocephala (intestinal helminths) may experience increases in the rates of T. cruzi infection [112, 113]. Araujo et al. [114] observed that isolated TcI grew faster under culture conditions than Tcl in mixed infections with TcII. Single infections, if they occur in nature, are rare [115, 116]. However, the complexity of this phenomenon means that there are several aspects that still require clarification. Mixed infections may occur after serial exposure to different genotypes and species of parasites at different time intervals and by distinct routes. Here, we know for sure that the patient was infected by the oral route with the four T. cruzi genotypes and T. dionisii on the same occasion. The fact that T. cruzi and T. dionisii are within the subgenus Schyzotrypanum and were found to occupy the same habitat leads to the hypothesis that an increase in virulence and pathogenicity may occur through competition for resources or alterations in doubling time or impairment in immune clearance, or through a combination of all of these factors.
We observed that the Atlantic rainforest remnants in the study area are suffering from degradation, as demonstrated by the low capture success (1.3 %) during the fieldwork. However, despite degradation, six different mammal species were captured, indicating that the area contains a moderate diversity of small wild mammal species. It has been reported that T. vitticeps presents high T. cruzi infection rates (more than 60 % of the triatomines are infected) [30, 117, 118]. The absence of positive hemocultures and borderline serological titers showed that the animals examined presented low force of infection and strongly suggests that triatomines are not being infected in the peridomicile area but in distant forest fragments. In this case, the capacity for flight in T. vitticeps may be much higher than reported for other insects of this genus. Nothing is known concerning the flight capacity of T. vitticeps. T. infestans is capable of flying 200 m or more [119, 120] because it is capable of flying with wind assistance [121]. Another explanation for the high T. cruzi infection rates in T. vitticeps is that other non-sampled mammals, such as armadillos and bats, can be responsible for parasite maintenance [37, 87, 89, 122–124].
Conclusion
In conclusion, our results indicate that (i) mixed infections in humans may be more common than has been recognized up to now and should be taken into consideration in future studies; (ii) the distribution of T. cruzi TcIII and TcIV in Brazilian biomes is broader than has been assumed until now, and the putative associations between T. cruzi DTUs and host species, geographical distribution and pathogenicity still pose epidemiological challenges; and (iii) T. dionisii is able, at least, to colonize human heart muscle cells.
Abbreviations
- ACD:
-
Acute chagas disease
- BLAST:
-
Basic local alignment search tool
- CD:
-
Chagas disease
- COLTRYP:
-
Coleção de trypanosoma de mamíferos silvestres, domésticos e vetores
- DTU:
-
Discrete typing unit
- ELISA:
-
Enzyme-Linked Immunosorbent Assay
- ES:
-
Espírito Santo state
- GPI:
-
Glucose-phosphate isomerase
- IFAT:
-
Indirect Immunofluorescent Antibody Test
- LIT:
-
Liver Infusion Tryptose
- ML:
-
Maximum likelihood
- NNN:
-
Novy Mc Neal Nicole
- PCR:
-
Polymerase chain reaction
- RFLP:
-
Restriction fragment length polymorphism
- Sesa/ES:
-
Espírito Santo state Health Department
- ZCC:
-
Zoonosis Control Center
References
Marinkelle CJ. Biology of the trypanosomes of bats. In: Lumsden WHR, Evans DA, editors. Biology of the Kinetoplastida. London: Academic Press; 1976. p. 175–216.
Molyneux DH. Trypanosomes of bats. 2nd ed. London: Academic Press; 1991.
Hoare CA. The trypanosomes of mammals: a zoological monograph. Oxford: Blackwell Scientific Publications; 1972.
Baker JR. Bat trypanosome models for Trypanosoma cruzi. Parasitol Today. 1985;4:111–3.
Llewellyn MS, Lewis MD, Acosta N, Yeo M, Carrasco HJ, Segovia M, et al. Trypanosoma cruzi IIc: phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Negl Trop Dis. 2009; doi:10.1371/journal.pntd.0000510.
Jansen AM, Roque ALR. Domestic and wild mammalian reservoir. In: Telleria J, Tibayrenc M, editors. American trypanosomiasis Chagas Disease—100 years of research. London: Elsevier; 2010. p. 249–76.
Brisse S, Verhoef J, Tibayrenc M. Characterization of large and small subunit rRNA and mini-exon genes futher supports the distinction of six Trypanosoma cruzi lineages. Int J Parasitol. 2001;31:1218–26.
Marcili A, Lima L, Cavazzana M, Junqueira AC, Veludo HH, Da Silva FM, et al. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009;136:641–55.
Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernades O, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051–4.
Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, et al. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol. 2012;12:240–53.
Coura JR, Junqueira AC, Fernandes O, Valente SA, Miles MA. Emerging Chagas disease in Amazonian Brazil. Trends Parasitol. 2002;18:171–6.
Monteiro WM, Magalhães LK, Santana Filho FS, Borborema M, Silveira H. Barbosa Md. Trypanosoma cruzi TcIII/Z3 genotype as agent of an outbreak of Chagas disease in the Brazilian Western Amazonia. Trop Med Int Health. 2010;15(9):1049–51.
Monteiro WM, Magalhaes LKC, de Sá ARN, Gomes ML, Toledo MJO, Borges L, et al. Trypanosoma cruzi IV causing outbreaks of acute Chagas disease and infections by different haplotypes in the Western Brazilian Amazonia. PLoS One. 2010;7(7):e41284.
Carrasco HJ, Segovia M, Llewellyn MS, Morocoima A, Urdaneta-Morales S, Martínez C, et al. Geographical Distribution of Trypanosoma cruzi Genotypes in Venezuela. PLoS Negl Trop Dis. 2012; doi:10.1371/journal.pntd.0001707.
Guhl F, Ramírez JD. Retrospective molecular integrated epidemiology of Chagas disease in Colombia. Infect Genet Evol. 2013;20:148–54.
Ramírez JD, Montilla M, Cucunubá ZM, Floréz AC, Zambrano P, Guhl F. Molecular Epidemiology of Human Oral Chagas Disease Outbreaks in Colombia. PLoS Negl Trop Dis. 2013; doi:10.1371/journal.pntd.0002041.
Deane MP, Mangia RH, Pereira NM, Momen H, Goncalves AM, Morel CM. Trypanosoma cruzi: strain selection by different schedules of mouse passage of an initially mixed infection. Mem Inst Oswaldo Cruz. 1984;79:495–7.
Morel CM, Deane MP, Goncalves AM. The complexity of Trypanosoma cruzi populations revealed by schizodeme analysis. Parasitol Today. 1986;2:97–100.
Nobrega AA, Garcia MH, Tatto E, Obara MT, Costa E, Sobel J, et al. Oral transmission of Chagas disease by consumption of acai palm fruit. Br Emerg Infect Dis. 2009;15:653–5.
Valente SA, Valente VC, Neves PAY, de Jesus Barbosa CM, dos Santos MP, Miranda CO, et al. Analysis of an acute Chagas disease outbreak in the Brazilian Amazon: human cases, triatomines, reservoir mammals and parasites. Trans R Soc Trop Med Hyg. 2009;103:291–7.
Xavier S, Roque ALR, Bilac D, de Araujo VAL, Neto SFC, Lorosa ES, et al. Distantiae transmission of Trypanosoma cruzi: a new epidemiological feature of acute Chagas disease in Brazil. PLoS Negl Trop Dis. 2014;8(5):e2878.
Yoshida N. Molecular mechanisms of Trypanosoma cruzi infection by oral route. Mem Inst Oswaldo Cruz. 2009;104:101–7.
Yoshida N, Tyler KM, Llewellyn MS. Invasion mechanisms among emerging food-borne protozoan parasites. Trends Parasitol. 2011;27(10):459–66.
de Noya BA, González ON. An ecological overview on the factors that drives to Trypanosoma cruzi oral transmission. Acta Trop. 2015;51:94–102.
Rocha FL, Roque ALR, de Lima JS, Cheida CC, Lemos FG, de Azevedo FC, et al. Trypanosoma cruzi infection in neotropical wild carnivores (mammalia: carnivora): at the top of the Trypanosoma cruzi transmission chain. Plos one. 2013;8(7):e67463.
Leite GR, dos Santos CB, Falqueto A. Influence of the landscape on dispersal of sylvatic triatomines to anthropic habitats in the Atlantic Forest. J Biogeogr. 2011;38(4):651–63.
Silveira AC, Feitosa VR, Borges R. Distribuição de triatomíneos capturados no ambiente domiciliar no período de 1975–1983, no Brasil. Rev Bras Malariol Doencas Trop. 1984;36:15–312.
Galvão C, Carcavallo R, Rocha DS, Jurberg J. A checklist of the current valid species of the subfamily Triatominae Jeannel, 1909 (Hemiptera Reduviidae) and their geographical distribution, with nomenclatural and taxonomic notes. Zootaxa. 2003;202:1–36.
Santos CB, Ferreira AL, Leite GR, Ferreira GE, Rodrigues AA, Falqueto A. Peridomiciliary colonies of Triatoma vitticeps (Stal, 1859) (Hemiptera, Reduviidae, Triatominae) infected with Trypanosoma cruzi in rural areas of the state of Espírito Santo, Brazil. Mem Inst Oswaldo Cruz. 2005;100(5):471–3.
Santos CB, Leite GR, Ferreira GEM, Ferreira AL. Infecção natural de Triatoma vitticeps (Stal, 1859) por flagelados semelhantes a Trypanosoma cruzi (Chagas, 1909) no estado do Espírito Santo. Rev Soc Bras Med Trop. 2006;39(1):89–91.
Santos CB, Leite GR, Sessa PA, Ferreira AL, Ferreira GEM, Falqueto A. Potencial vetorial de Triatoma vitticeps (Stal 1859) (Hemiptera, Reduviidae, Triatominae) na transmissão do Trypanosoma cruzi (Chagas, 1909) (Kinetoplastidae, Trypanosomatidae) no estado do Espírito Santo, Brazil. Rev Soc Bras Med Trop. 2004;37(1):186.
Sikes RS, Gannon WL. Guidelines of the American Society of Mammalogist for the use of wild mammals in research. J Mammal. 2011;92:235–53.
Bonvicino CR, Lemos B, Weksler M. Small mammals of Chapada dos Veadeiros National Park (Cerrado of Central Brazil): ecology, karyologic and taxonomic considerations. Braz J Biol. 2005;65:395–406.
Camargo ME. Fluorescent antibody test for the serodiagnosis of American trypanosomiasis. Technical modification employing preserved culture forms of Trypanosoma cruzi in a slide test. Rev Inst Med Trop Sao Paulo. 1966;8:227–35.
Vaz VC, D’Andrea PS, Jansen AM. Effects of habitats fragmentation on wild mammal infection by Trypanosoma cruzi. Parasitology. 2007;134:1785–93.
Lent H, Wygodzinsky P. Triatominae. Bull Amer Mus Nat Hist. 1979;163:496–7.
Roque ALR, Xavier SCC, Rocha MR, Duarte ACM, D’Andrea PS, Jansen AM. Trypanosoma cruzi transmission cycle among wild and domestic mammals in three areas of orally transmitted Chagas disease outbreaks. Am J Trop Med Hyg. 2008;79:742–9.
Roque AL, Xavier SC, Gerhardt M, Silva MF, Lima VS, D’Andrea PS, et al. Trypanosoma cruzi among wild and domestic mammals in different areas of the Abaetetuba municipality (Pará State, Brazil), an endemic Chagas disease transmission area. Vet Parasitol. 2013;193(1–3):71–7.
Vallejo GA, Guhl F, Chiari E, Macedo AM. Species specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammal hosts by polymerase chain reaction amplification of kinetoplast minicircle DNA. Acta Trop. 1999;72:203–12.
Goelz SE, Hamilton SR, Vogelstein B. Purification of DNA from formaldehyde fixed and paraffin-embedded human tissue. Biochem Biophys Res Commun. 1985;130:118–26.
Coura R, Prolla JC, Meurer L, Ashton-Prolla P. An alternative protocol for DNA extraction from formalin-fixed and paraffin wax embedded tissue. J Clin Pathol. 2005;58:894–5.
Fernandes O, Santos SS, Cupolillo E, Mendonca B, Derre R, Junqueira ACV, et al. A mini-exon multiplex polymerase chain reaction to distinguish the major groups of Trypanosoma cruzi and T. rangeli in the Brazilian Amazon. Trans R Soc Trop Med Hyg. 2001;95:97–9.
Aliaga C, Breniere SF, Barnabe C. Further interest of miniexon multiplex PCR for a rapid typing of Trypanosoma cruzi DTU groups. Infect Genet Evol. 2011;11:1155–8.
Westenberger SJ, Barnabe C, Campbell DA, Sturm NR. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics. 2005;171:527–43.
Lima L, Espinosa-Álvarez O, Ortiz PA, Trejo-Varón JA, Carranza JC, Pinto CM, et al. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit). Acta Trop. 2015;151:166–77.
Rozas M, De Doncker S, Adaui V, Coronado X, Barnabé C, Tibyarenc M, et al. Multilocus polymerase chain reaction restriction fragment-length polymorphism genotyping of Trypanosoma cruzi (Chagas disease): taxonomic and clinical applications. J Infect Dis. 2007;195(9):1381–8.
Brenière SF, Aliaga C, Waleckx E, Buitrago R, Salas R, Barnabé C, et al. Genetic Characterization of Trypanosoma cruzi DTUs in wild Triatoma infestans from Bolivia: Predominance of TcI. Plos Neg Trop Dis. 2012;6(5):e1650.
Noyes HA, Stevens JR, Teixeria M, Phelan J, Holz P. A nested PCR for the sssrRNA gene detects Trypanosoma binney in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int J Parasitol. 1999;29:331–9.
Borghesan TC, Ferreira RC, Takata CS, Campaner M, Borda CC, Paiva F, et al. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypansomatidae) a genus of insect parasites associated with flies. Protist. 2013;164:129–52.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Machado CA, Ayala FJ. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc Natl Acad Sci U S A. 2001;98:7396–401.
Broutin H, Tarrieu F, Tibayrenc M, Oury B, Barnabe C. Phylogenetic analysis of the glucose-6-phosphate isomerase gene in Trypanosoma cruzi. Exp Parasitol. 2006;113:1–7.
Rozas M, De Doncker S, Coronado X, Barnabe C, Tibyarenc M, Solari A, et al. Evolutionary history of Trypanosoma cruzi according to antigen genes. Parasitology. 2008;135:1157–64.
Hamilton PB, Stevens RJ. Resolving relationships between Australian trypanosomes using DNA barcoding data. Trends Parasitol. 2010;11:99.
Lima L, Maia da Silva F, Neves L, Attias M, Takata CSA, Campaner M, et al. Evolutionary insights from bats trypanosomes: morphological, developmental and phylogenetic evidence of a new species, Trypanosoma (Schizotrypanum) erney sp. nov., in African Bats closely related to Trypanosoma (Schizotrypanum) cruzi and allied species. Protist. 2012;163:856–72.
Pinto CM, Kalko EK, Cottontail I, Wellinghausen N, Cottontail VM. TcBat a bat-exclusive lineage of Trypanosoma cruzi in the Panama Canal Zone, with comments on its classification and the use of the 18S rRNA gene for lineage identification. Infect Genet Evol. 2012;12:1328–32.
Cottontail VM, Kalko EKV, Cottontail I, Wellinghausen N, Tschapka M, Perkins SL, et al. High local diversity of Trypanosoma in a common bat species, and implications for the biogeography and taxonomy of the T. cruzi clade. PLoS One. 2014; doi:10.1371/journal.pone.0108603.
Lewis MD, Llewellyn MS, Yeo M, Acosta N, Gaunt MW, Miles M. Recent, independent and anthropogenic origins of Trypanosoma cruzi hybrids. PLoS Negl Trop Dis. 2011; doi:10.1371/journal.pntd.0001363.
Perez E, Monje M, Chang B, Buitrago R, Parrado R, Barnabé C, et al. Predominance of hybrid discrete typing units of Trypanosoma cruzi in domestic Triatoma infestans from the Bolivian Gran Chaco region. Infect Genet Evol. 2013;13:116–23.
Xavier SCdC, Roque ALR, Lima VdS, Monteiro KJL, Otaviano JCR, Ferreira da Silva LFC, et al. Lower richness of small wild mammal species and Chagas disease risk. PLoS Negl Trop Dis. 2012; doi:10.1371/journal.pntd.0001647
Toso MA, Vial UF, Galanti N. Oral transmission of Chagas’ disease. Rev Med Chil. 2011;139:258–66.
Shikanai-Yasuda MA, Carvalho NB. Oral transmission of Chagas disease. Clin Infect Dis. 2012;54:845–52.
Rueda K, Trujillo JE, Carranza JC, Valllejo GA. Oral transmission of Trypanosoma cruzi: a new epidemiological scenario for Chagas’ disease in Colombia and other South American countries. Biomedica. 2014;34(4):631–41.
Gilbert MTP, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, et al. The Isolation of Nucleic Acids from Fixed, Paraffin-Embedded Tissues-Which Methods Are Useful When? Plos One. 2007; doi:10.1371/journal.pone.0000537
Feldman MY. Reactions of nucleic acids and nucleoproteins with formaldehyde. Prog Nucleic Acid Res Mol Biol. 1973;13:1–49.
Greer CE, Peterson SK, Kiviat NB, Manos MM. PCR amplification from paraffin-embedded tissues. Effects of fixative and fixation time. Am J Clin Pathol. 1991;95:117–24.
Romero R, Juston AC, Ballantyne J, Henry BE. The applicability of formalin-fixed and formalin-fixed paraffin embedded tissues in forensic DNA analysis. J Forensic Sci. 1997;42:708–14.
Fernandes A, Iñiguez AM, Lima VS, Mendonça de Souza SMF, Ferreira LF, Vicente ACP, et al. Pre-Columbian Chagas disease in Brazil: Trypanosoma cruzi I in the archaeological remains of a human in Peruaçu Valley, Minas Gerais, Brazil. Mem Inst Oswaldo Cruz. 2008;103(5):514–6.
Lima VS, Iniguez AM, Otsuki K, Ferreira LF, Araújo A, Vicente ACP, et al. Chagas Disease in Ancient Hunter-Gatherer Population, Brazil. Emerg Infect Dis. 2008;14(6):1001–2.
Guhl F, Auderheide A, Ramírez JD. From ancient to contemporary molecular eco-epidemiology of Chagas disease in the Americas. Int J Parasitol. 2014;44:605–12.
Cura CI, Duffy T, Lucero RH, Bisio M, Péneau J, Jimenez-Coello M, et al. Multiplex real-time PCR assay using TaqMan probes for the identification of Trypanosoma cruzi DTUs in biological and clinical samples. PLoS Negl Trop Dis. 2015;9(5):e0003765.
Lima VS, Xavier S, Maldonado IFR, Roque ALR, Vicente ACP, et al. Expanding the knowledge of the geographic distribution of Trypanosoma cruzi TcII and TcV/TcVI genotypes in the Brazilian Amazon. Plos One. 2014;9(12):e116137.
Herrera CP, Licon MH, Nation CS, Jameson SB, Wesson DM. Genotype diversity of Trypanosoma cruzi in small rodents and Triatoma sanguisuga from a rural area in New Orleans, Louisiana. Parasit Vectors. 2015;8:123.
Jansen AM, Xavier SCC, Roque ALR. The multiple and complex and changeable scenarios of the Trypanosoma cruzi transmission cycle in the sylvatic environment. Acta Trop. 2015;151:1–15.
Mantilla JC, Zafra GA, Macedo AM, González CI. Mixed infection of Trypanosoma cruzi I and II in a Colombian cardiomyopathic patient. Hum Pathol. 2010;41:610–3.
Ramírez JD, Guhl F, Rendón LM, Rosas F, Marin-Neto JA, Morillo CA. Chagas cardiomyopathy manifestations and Trypanosoma cruzi genotypes circulating in chronic chagasic patients. PLoS Negl Trop Dis. 2010;4(11):e899.
Monje-Rumi MM, Brandán CP, Ragone PG, Tomasini N, Lauthier JJ, D’Amato AMA, et al. Trypanosoma cruzi diversity in the Gran Chaco: mixed infections and differential host distribution of TcV and TcVI. Infect Genet Evol. 2014;29:53–9.
Apt W, Arribada A, Zulantay I, Saavedra M, Araya E, Solari A, et al. Trypanosoma cruzi burden, genotypes and clinical evaluation of Chilean patients with chronic Chagas cardiopathy. Parasitol Res. 2015;114(8):3007–18.
Martinez-Perez A, Poveda C, Ramírez JD, Normana F, Gironés N, Guhl F, et al. Prevalence of Trypanosoma cruzi’s Discrete Typing Units in a cohort of Latin American migrants in Spain. Acta Trop. 2016;157:145–50.
Lisboa CV, Mangia RH, Rubião E, Lima NRC, Xavier SCC, Picinatti A, et al. Trypanosoma cruzi transmission in a captive primate unit, Rio de Janeiro, Brazil. Acta Trop. 2004;90:97–106.
Lisboa CV, Luz SL, Kluczkovski AA, Ferreira LF, Ribeiro CT, Fernandes O, et al. Stable infection of primates with Trypanosoma cruzi I and II. Parasitology. 2006;133:603–11.
Xavier SCC, D’Andrea PS, Herrera LM, Emperaire L, Vaz VC, Fernandes O, et al. Mapping of the distribution of Trypanosoma cruzi infection among small wild mammals in a conservation unit and its surroundings (Northeast-Brazil). Parasitol Int. 2007;56:119–28.
Anez N, Crisante G, da Silva FM, Rojas A, Carrasco H, Umezawa ES, et al. Predominance of lineage I among Trypanosoma cruzi isolates from Venezuelan patients with different clinical profiles of acute Chagas’ disease. Trop Med Int Health. 2004;9:1319–26.
Toledo A, Vergara F, Campos R, Botto-Mahan C, Ortiz S, Coronado X, et al. Trypanosoma cruzi genotypes in Meprais gajardoi from wild ecotopes in northern Chile. Am J Trop Med Hyg. 2013;88(2):285–8.
Ramos-Ligonio A, Torres-Montero J, Lopez-Monteon A, Dumonteil E. Extensive diversity of Trypanosoma cruzi discrete typing units circulating in Triatoma dimidiata from central Veracruz, Mexico. Infect Genet Evol. 2012;12(7):1341–3.
Ibanez-Cervantes G, Martinez-Ibarra A, Nogueda-Torres B, Lopez-Orduna E, Alonso AL, Perea C, et al. Identification by Q-PCR of Trypanosoma cruzi lineage and determination of blood meal sources in triatomine gut samples in Mexico. Parasitol Int. 2013;62(1):36–43.
Yeo M, Acosta N, Llewellyn M, Sanchez H, Adamson S, Miles GA, et al. Origins of Chagas disease: Didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int J Parasitol. 2005;35:225–33.
Cardinal MV, Lauricella MA, Ceballos LA, Lanati L, Marcet PL, Levin MJ, et al. Molecular epidemiology of domestic and sylvatic Trypanosoma cruzi infection in rural northwestern Argentina. Int J Parasitol. 2008;38(13):1533–43.
Marcili A, Lima L, Valente VC, Valente SA, Batista JS, Junqueira AC, et al. Comparative phylogeography of Trypanosoma cruzi TCIIc: new hosts, association with terrestrial ecotopes, and spatial clustering. Infect Genet Evol. 2009;9(6):1265–74.
Marcili A, Valente VC, Valente SA, Junqueira AC, da Silva FM, Pinto AY, et al. Trypanosoma cruzi in Brazilian Amazonia: Lineages TCI and TCIIa in wild primates, Rhodnius spp. and in humans with Chagas disease associated with oral transmission. Int J Parasitol. 2009;39(5):615–23.
Abolis NG, de Araujo SM, Toledo MJO, Fernandez MA, Gomes ML. Trypanosoma cruzi I-III in Southern Brazil causing individual and mixed infections in humans, sylvatic reservoirs and triatomines. Acta Trop. 2011;120:167–72.
Martins K, Andrade CM, Barbosa-Silva NA, Nascimento GB, Chiari E, Galvão LM, et al. Trypanosoma cruzi III causing the indeterminate form of Chagas disease in a semi-arid region of Brazil. Int J Infect Dis. 2015;39:68–75.
D’Ávila DA, Macedo AM, Valadares HM, Gontijo ED, Castro AM, Machado CR, et al. Probing population dynamics of Trypanosoma cruzi during progression of the chronic phase in chagasic patients. J Clin Microbiol. 2009;47:1718–25.
Monje-Rumi MM, Brandán CP, Ragone PG, Tomasini N, Lauthier JJ, Alberti D’Amato AM, et al. Trypanosoma cruzi diversity in the Gran Chaco: mixed infections and differential host distribution of TcV and TcVI. Infect Genet Evol. 2015;29:53–9.
Oliveira MP, Cortez M, Maeda FY, Fernandes MC, Haapalainen EF, Yoshida N, et al. Unique behavior of Trypanosoma dionisii interacting with mammalian cells: invasion, intracellular growth and nuclear localization. Acta Trop. 2009;110(1):65–74.
Maeda FY, Alves RM, Cortez C, Lima FM, Yoshida N. Characterization of the infective properties of a new genetic group of Trypanosoma cruzi associated with bats. Acta Trop. 2011;120:231–7.
Gardner RA, Molyneux DH. Schizotrypanum in British bats. Parasitology. 1988;97:43–50.
Acosta ICL, da Cosra AP, Gennari SM, Marcili A. Survey of Trypanosoma and Leishmania in wild and domestic animals in an Atlantic rainforest fragment and surroundings in the state of Espírito Santo, Brazil. J Med Entomol. 2014;51(3):686–93.
Gardner RA, Molyneux DH. Trypanosoma (Megatrypanum) incertum from Pipistrellus pipistrellus: development and transmission by cimicid bugs. Parasitology. 1988;96:433–47.
Truc P, Büscher P, Cuny G, Gonzatti MI, Jannin J, et al. Atypical Human Infections by Animal Trypanosomes. PLoS Negl Trop Dis. 2013;7(9):e2256.
Verma A, Manchanda S, Kumar N, Sharma A, Goel M, Banerjee PS, et al. Trypanosoma lewisi or Trypanosoma lewisi-like infection in a 37-day-old infant. Am J Trop Med Hyg. 2011;85:221–4.
Joshi PP, Shegokar V, Powar S, Herder S, Katti R, Salkar HR, et al. Human trypanosomiasis caused by Trypanosoma evansi in India: the first case report. Am J Trop Med Hyg. 2005;73:491–5.
Gosh S, Banerjee P, Sarkar A, Datta S, Chatterjee M. Coinfection of Leptomonas seymouri and Leishmania donovani in Indian Leishmaniasis. J Clin Microbiol. 2012;50(8):2774–8.
Novo SPC, Leles D, Bianucci R, Araújo A. Leishmania tarentolae molecular signatures in a 300 hundred-years-old human Brazilian mummy. Parasit Vectors. 2015;8:72.
Ramírez JD, Tapia G, Muñoz G, Poveda C, Rendón LM, Hincapié E, et al. Trypanosome species in neotropical bats: biological, evolutionary and epidemiological implications. Infect Genet Evol. 2014;22:250–6.
Ramírez JD, Hernández C, Montilla M, Zambrano P, Flórez AC, Parra E, et al. First report of human Trypanosoma cruzi infection attributed to TcBat genotype. Zoonoses Public Health. 2014;61:477–9.
Bosseno MF, Yacsik N, Vargas F, Brenière SF. Selection of Trypanosoma cruzi clonal genotypes (Clonet 20 and 39) isolated from Bolivian triatomines following subculture in liquid medium. Mem Inst Oswaldo Cruz. 2000;95(5):601–7.
Dvorak JA, Hartman DL, Miles MA. Trypanosoma cruzi: Correlation of growth kineties to zymodeme type in clones derived from various sources. J Protozool. 1980;27:472–4.
Dvorak JA, Hall TE, Crane MSJ, Engel JC, McDaniel JP, Uriegas R. Trypanosoma cruzi: flow cytometric analysis. I. Analysis of total DNA/organism by means of mitramycin-induced fluorescence. J Protozool. 1982;29:430–7.
Davies CM, Fairbrother E, Webster JP. Mixed strain schistosome infections of snails and the evolution of parasite virulence. Parasitol. 2002;124:31–8.
Mouton L, Dedeine F, Henri H, Boulétreau M, Profizi N, Vavre F. Virulence, multiple infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics. 2004;168:181–9.
Monteiro RV, Dietz JM, Raboy B, Beck B, Vleeschower KD, Baker A, et al. Parasite community interactions: Trypanosoma cruzi and intestinal helminths infecting wild golden lion tamarins Leontopithecus rosalia and golden-headed lion tamarins L. chrysomelas (Callitrichidae, L., 1766). Parasitol Res. 2007;101:1689–98.
Monteiro RV, Dietz JM, Jansen AM. The impact of cocomitant infections by Trypanosoma cruzi and intestinal helminths on the health of wild golden and golden-headed lion tamarins. Res Vet Sci. 2010;89(1):27–35.
Araújo CAC, Waniek PJ, Jansen AM. TcI/TcII co-infection can enhance Trypanosoma cruzi growth in Rhodnius prolixus. Parasit Vectors. 2014;7:94.
Read AF, Taylor LH. The ecology of genetically diverse infections. Science. 2001;292:1099–102.
Choisy M, Roode JC. Mixed infections and the evolution of virulence: effects of resource competition, parasite plasticity and impaired host immunity. Am Nat. 2010;175(5):105–18.
Sessa PA, Carias VRD. Infecção natural de triatomíneos do Espírito Santo for flagelados morfologicamente semelhantes ao Trypanosoma cruzi. Rev Soc Bras Med Trop. 1986;19:99–100.
Dias JCP, Feitosa VR, Filho ANF, Rodrigues VLC, Alencar AS, Sessa PA. Fonte alimentar e potencial vetorial deTriatoma vitticeps (Stal, 1859) com relação à doença de Chagas humana no estado do Espírito Santo, Brasil. Mem Inst Oswaldo Cruz. 1989;84:165–73.
Schofield CJ, Lehane MJ, McEwen P, Catalá SS, Gorla DE. Dispersive flight by Triatoma infestans under natural climatic conditions in Argentina. Med Vet Entomol. 1992;6:51–6.
Schweigmann N, Vallvé S, Muscio O, Ghillini N, Alberti A, Wisnivesky-Colli C. Dispersal flight by Triatoma infestans in an arid area of Argentina. Med Vet Entomol. 1988;2:401–4.
Gonçalves TC, de Oliveira E, Dias LS, Almeida MD, Nogueira WO, Pires FD. An investigation on the ecology of Triatoma vitticeps (Stal, 1859) and its possible role in the transmission of Trypanosoma cruzi, in the locality of Triunfo, Santa Maria Madalena municipal district, state of Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 1998;93:711–7.
Cavazzana Jr M, Marcili A, Lima L, da Silva FM, Junqueira AC, Veludo HH, et al. Phylogeographical, ecological and biological patterns shown by nuclear (ssrRNA and gGAPDH) and mitochondrial (Cyt b) genes of trypanosomes of the subgenus Schizotrypanum parasitic in Brazilian bats. Int J Parasitol. 2010;40(3):345–55.
Lisboa CV, Pinho AP, Herrera HM, Gerhardt M, Cupolillo E, Jansen AM. Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae) genotypes in neotropical bats in Brazil. Vet Parasitol. 2008;156(3–4):314–8.
Lisboa CV, Xavier SC, Herrera HM, Jansen AM. The ecology of the Trypanosoma cruzi transmission cycle: dispersion of zymodeme 3 (Z3) in wild host from Brazilian biomes. Vet Parasitol. 2009;165(1–2):19–24.
Acknowledgment
We would like to thank Dr Janaina A. Shineider Casotti, Dr Celia Marcia Birchler, Claudiney Biral dos Santos and the state health department (Sesa/ES) who kindly donated the material for this work. We are also thankful to Alcidelio Lovatti, Altemar Rodrigues Marques, Edmilson Vieira da Hora and Helton Meriguete for accompanying us during the fieldwork, to Cristiane Varella Lisboa and Socrates Fraga da Costa Neto who participated in the excursion, to Carlos Ardé and Marcos Antônio dos Santos Limas for technical support in the hemocultures, to Danielle Bilac da Silva and Valdirene dos Santos Lima for technical support in the molecular characterization and to the PDTIS/Fiocruz sequencing platform for sequence our samples.
Funding
This study was funded by Oswaldo Cruz Institute - FIOCRUZ; PAPES VI; CNPq and FAPERJ. A doctoral grant is provided by CNPq to MAD and by Oswaldo Cruz Institute to MRS. AMJ is a “Cientista do Nosso Estado”, provided by FAPERJ and is financially supported by CNPq (“Bolsista de Produtividade, nível 1”, CNPq). ALRR is a “Jovem Cientista do Nosso Estado” provided by FAPERJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All sequences analyzed were deposited in GenBank under the accession numbers KR905432–KR905446 for the 18S rRNA gene, KT737478 for GPI and KT983981 for 1f8. The GenBank accession numbers can be viewed in Additional file 1.
Authors’ contribution
MAD, ALRR and AMJ conceived and designed the experiments. PSD, ALRR and AMJ performed the fieldwork. MAD, MSR and JHSB performed and analyzed the molecular characterization. SCCX and JHSB performed and analyzed the serological characterization. MAD, ALRR and AMJ wrote the manuscript. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The capture of small wild mammals was licensed by the Sistema de Autorização e Informação em Biodiversidade - SISBIO of the Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA)-permanent license number 3365-1. Blood sample collection and euthanasia were performed and supervised by the Federal Counsel of Medical Veterinary under resolution number 1.000 approved on May 11th, 2012, according to the Ethical Committee for Animal Use of the Oswaldo Cruz Foundation (license 0015-07).
The cardiac fragment was kindly donated by Dr Janaina A. Shineider Casotti from the ES state health department (Sesa/ES).
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Table S1. Trypanosoma cruzi and Trypanosoma dionisii GenBank accession numbers for the 1f8, GPI and 18S rRNA genes. (DOCX 13 kb)
Additional file 2:
Table S2. Alignment sequences from Trypanosoma cruzi, Trypanosoma cruzi marinkellei, Trypanosoma dionisii species isolates, and V7V8 SSU rRNA clones obtained from cardiac tissue. The dots are representing same base position for T. cruzi. The stars are representing same base position for T. dionisii. (DOCX 17 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Dario, M.A., Rodrigues, M.S., Barros, J.H.d. et al. Ecological scenario and Trypanosoma cruzi DTU characterization of a fatal acute Chagas disease case transmitted orally (Espírito Santo state, Brazil). Parasites Vectors 9, 477 (2016). https://doi.org/10.1186/s13071-016-1754-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1754-4