Abstract
Background
Animal African Trypanosomiasis (AAT) is caused by several species of trypanosomes including Trypanosoma congolense, T. vivax, T. godfreyi, T. simiae and T. brucei. Two of the subspecies of T. brucei also cause Human African Trypanosomiasis. Although some of them can be mechanically transmitted by biting flies; these trypanosomes are all transmitted by tsetse flies which are the cyclical vectors of Trypanosoma congolense, T. godfreyi, T. simiae and T. brucei. We present here the first report assessing the prevalence of trypanosomes in tsetse flies in Nigeria using molecular tools.
Methods
488 tsetse flies of three species, Glossina palpalis palpalis, G. tachinoides and G. morsitans submorsitans were collected from Wuya, Niger State and Yankari National Park, Bauchi State in 2012. Trypanosomes were detected and identified using an ITS1 PCR assay on DNA purified from the ‘head plus proboscis’ (H + P) and abdomen (ABD) parts of each fly.
Results
T. vivax and T. congolense Savannah were the major parasites detected. Trypanosomes prevalence was 7.1 % in G. p. palpalis, 11.9 % in G. tachinoides and 13.5 % in G. m. submorsitans. Prevalences of T. congolense Savannah ranged from 2.5 to 6.7 % and of T. vivax were approximately 4.5 %. Trypanosoma congolense Forest, T. godfreyi and T. simiae were also detected in the site of Yankari. The main biological and ecological determinants of trypanosome prevalence were the fly sex, with more trypanosomes found in females than males, and the site, with T. congolense subspp. being more abundant in Yankari than in Wuya. As expected, the trypanosome species diversity was higher in Yankari National Park than in the more agricultural site of Wuya where vertebrate host species diversity is lower.
Conclusions
Our results show that T. congolense Savannah and T. vivax are the main species of parasite potentially causing AAT in the two study sites and that Yankari National Park is a potential reservoir of trypanosomes both in terms of parasite abundance and species diversity.
Similar content being viewed by others
Background
Tsetse flies are the cyclical vectors of African trypanosomes. Many of these trypanosomes are pathogens of humans and livestock, causing human African trypanosomiasis (HAT) and animal African trypanosomiasis (AAT), diseases with major medical and economic impact. The human disease is caused by two subspecies of Trypanosoma brucei while AAT is caused by at least seven species/subspecies of trypanosomes [1]. Trypanosoma vivax and Trypanosoma congolense Savannah are the most important pathogens causing AAT, because of their predominance throughout sub-Saharan Africa and of their economic impact on animal production [1]. AAT is recognized as an important constraint to agricultural development, causing losses to livestock producers and consumers exceeding $1300 million per year [2]. The number of disease control methods within the vertebrate host is limited. Moreover, the evolution of resistances to trypanocidal drugs makes chemotherapy difficult to sustain for the control of AAT [3]. For these reasons, vector control remains a very important part of an integrated management of AAT [4]. Detecting and identifying trypanosome species in tsetse flies is fundamental to understanding the epidemiology of the associated diseases and to inform the establishment of efficient tsetse control programs aiming at reducing the prevalence of AAT in a particular area. That way, tsetse control programs can, for example, focus on localities where a high prevalence of the most virulent pathogens is found and use appropriate technologies depending on the vector species.
Little information exists about trypanosome biodiversity and distribution in Nigerian tsetse populations. Eleven species of tsetse were reported in Nigeria [5] infesting approximately 70 % of the country’s land mass [6]. Some of those species are major vectors of T.vivax, T. brucei subspp. and T. congolense subspp. [7, 8]. However, most of the available data rely on microscopy which has been shown to be far less sensitive and accurate than DNA-based detection and identification methods [9]. DNA-based methods have the advantage of being more sensitive and able to identify trypanosomes to the subspecies level and to detect mixed infections. Detailed information is required to better identify and characterize trypanosome-infested areas within the context of the eradication of AAT by the Pan African Tsetse and Trypanosomiasis Eradication Campaign (PATTEC).
Although all tsetse species are susceptible to trypanosomes infection, differences in susceptibility exist between species. In general, species from the morsitans group are considered more susceptible to trypanosome infection than species from the palpalis group. Within each species, a variety of factors intrinsic to the insect host can influence the competence of each individual fly to trypanosomes. For example, it has been shown that the susceptibility to trypanosomes is influenced by the sex or the age at the time of the first infected meal [10]. Studies describing the distribution of trypanosome species diversity among different tsetse species can bring insights about the vector competence of the different tsetse species highlighting non-random associations between tsetse and trypanosome species.
The present study is focused on the three main tsetse vector of AAT in Nigeria, namely Glossina palpalis palpalis, Glossina tachinoides and Glossina morsitans submorsitans. Individuals from these three species were collected in two localities (two species in one locality and the third species in the second locality) in central Nigeria. We then used PCR-based methods to detect, identify and determine the prevalence of trypanosomes in the tsetse collected. The main objectives of the study were to (i) evaluate the species diversity of trypanosomes causing AAT in northern Nigeria; and (ii) attempt to identify some ecological and biological factors responsible for potential differences in the trypanosome prevalence.
Methods
Sample collection and DNA purification
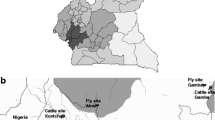
Tsetse fly samples were collected from Yankari National Park and Wuya areas between March and August 2012 using biconical traps. Yankari National Park is in the south-central part of Bauchi State (9.749°N, 10.499°E), in northeastern Nigeria. It is composed of savannah grassland with well-developed patches of woodland. Wuya is in Niger State (10.000°N, 6.167°E) located in north-western Nigeria. It is a mountainous area with wooded savannah vegetation. Glossina palpalis palpalis flies were caught in Wuya while G. morsitans submorsitans and G. tachinoides were captured in Yankari. Flies were killed by desiccation and stored as dry carcasses until further analysis. Samples are available upon request.
The sex of each fly was recorded and then ‘head plus proboscis’ (H + P) and abdomen (ABD) were cut from the thorax. Before tissue lysis, each ABD sample was cut into four pieces using a single-use scalpel blade. For each H + P, the sample was frozen in liquid N2 and then ground using a pestle (only the external part of the tube containing the sample was in contact with liquid N2, not the sample itself). DNA extraction and purification was carried out on all samples using the DNeasy blood and tissue kit (Qiagen, Manchester, UK) according to the manufacturer’s instruction for animal tissues except for the final elution volumes: one elution of 100 μl for ABD samples and two elutions of 35 and 30 μl for H + P (the two elutions were done in the same tube and a final volume of ~60 μl was obtained).
As trypanosome positive controls, we used purified DNA samples from parasites cryopreserved in mouse blood: T. b. brucei (strain STIB 247), T. vivax (ILRAD V-34), T. congolense Savannah (IL3000), T. congolense Forest (ANR3). DNA was purified from 100–300 μl cryostabilates with > 6.3 × 107 trypanosomes/ml using the DNeasy blood and tissue kit (Qiagen, Manchester, UK) according to the manufacturer’s instructions for cultured cells.
Trypanosome detection and identification by PCR
Detection and identification of trypanosomes were undertaken using the ITS1 PCR assay developed by Njiru et al. [9]. This PCR assay allows the detection and identification of Trypanosoma vivax, T. simiae Tsavo, T. godfreyi, T. simiae, T. congolense Kilifi and the subgenus Trypanosoon (T. brucei ssp. and T. evansi). Using this PCR assay, a single band at ~700 bp is obtained for both T. congolense Savannah and T. congolense Forest and species-specific PCRs are thus subsequently required to identify these trypanosomes at the subspecies level.
ITS1 PCR amplifications were carried out in 10 μl reaction mixture containing 5.9 μl distilled water, 1 μl of 10× Custom PCR Master Mix (45 mM Tris-HCl (pH 8.8), 11 mM (NH4)2SO4, 4.5 mM MgCl2, 0.113 mg/ml BSA, 4.4 μM EDTA, 1 mM each of dATP, dCTP, dGTP and dTTP) (Thermo Scientific, ABgene®UK), 1 μl of each primer solution at 10 μM (see Table 1 for primer sequences), 0.1 μl of Taq polymerase at 5 U/μl (Thermo Scientific, ABgene®UK) and 1 μl DNA template. The cycling conditions were as follows: (95 °C, 2 min); 30 cycles of (95 °C, 50 s), (60 °C, 50 s) and (72 °C, 1 min); (72 °C, 5 min). For each ITS1 PCR performed a negative control (distilled water) as well as positive controls for T. brucei brucei, T. congolense Savannah and T. vivax (see the previous section for details) were included. Gel electrophoreses were carried out using 5 μl of PCR product in 1.5 % agarose gels stained with ethidium bromide (5 μg/ml). PCR products were then visualized using UV and gels were photographed.
Samples positive for T. congolense (ITS1 band at ~700 bp) were further screened using subspecies-specific PCR to determine the subspecies of T. congolense (Savannah or Forest) the fly was infected with. Subspecies-specific PCR primer pairs were used (see Table 1). For both subspecies, PCR amplifications were carried out in 10 μl reactions containing 1 μl of template DNA, 3 μl of distilled water, 1 μl of a primer mix (F + R) at 10 μM each and 5 μl of Dream Taq green Mater Mix (Thermo Scientific, UK). The cycling conditions were as follows: (95 °C, 3 min); 30 cycles of (95 °C, 30 s), (58 °C, 30 s) and (72 °C, 30 s); (72 °C, 5 min). For each T. congolense subspecies-specific PCR performed, a negative control (distilled water) as well as a positive control, for T. congolense Savannah or Forest (see the previous section for details), were included. Gel electrophoreses were carried out using 4.5 μl of PCR product in 1.5 % agarose gels stained with ethidium bromide (0.8 mg/ml). PCR products were then visualized using UV and gels were photographed.
Statistics
The data for H + P and ABD samples were compared using McNemar’s test. We tested the effect of the following explanatory variables on trypanosome prevalence using multiple logistic regression analyses: ‘tsetse species’, ‘collection site’ and ‘fly sex’. For all multiple regressions analyses performed on trypanosome prevalence a fly was considered infected if either the ABD or the H + P sample was infected. Multiple logistic regressions were performed for “all trypanosomes”, T. congolense subspp. (which refers to the prevalence of both T. congolense subspecies combined), T. congolense Savannah, T. congolense Forest and T. vivax. The prevalence of other types of infection present only in one tsetse species (T. simiae and T. godfreyi) was considered as part of the category “all trypanosomes” rather than individually.
Because one tsetse species is only present in one of the sites (G. p. palpalis in Wuya) and the other two species are only present in the second site (G. tachinoides and G. m. submorsitans in Yankari) it was not possible to include ‘tsetse species’ and ‘collection site’ as explanatory variables in a multiple logistic regression applied to the complete dataset.
Thus, ‘collection site’, ‘fly sex’ and their interaction were included as explanatory variables in a multiple logistic regression applied to the complete dataset [prevalence ~ ‘collection site’ + ‘fly sex’ + (‘collection site’ × ‘fly sex’)]. The association between the trypanosome prevalence and the tsetse species could only be investigated within the site of Yankari. ‘tsetse species’, ‘fly sex’ and their interaction were thus included as explanatory variables in a multiple logistic regression applied to the Yankari dataset [prevalence ~ ‘tsetse species’ + ‘fly sex’ + (‘tsetse species’ × ‘fly sex’)].
All statistical analyses were performed using R [11]. Multiple logistic regressions were performed using the glm function from the package stats. A binomial probability distribution was used and a logit link function was applied to the dependent variable, i.e. trypanosome prevalence. Backward model selection was performed using the stepAIC function from the MASS package [12] to identify the most relevant explanatory variables among the ones considered. The complete trypanosome prevalence dataset is provided in Additional file 1 and the R code used for the multiple logistic regression analyses is provided as Additional file 2.
Results
The average apparent tsetse densities recorded during the trapping period (March-August 2012) were 27.31 flies/trap/day for G. p. palpalis in Wuya and 128.03 and 101.21 flies/trap/day for G. tachinoides and G. m. submorsitans in Yankari, respectively. We screened 488 tsetse flies of three species for the presence of trypanosomes using the ITS1 PCR approach developed by Njiru et al. [9]. All samples that were positive for T. congolense subspp. (PCR product at ~700 bp, n = 26) were then screened using subspecies-specific PCR to determine whether the fly was infected by T. congolense Savannah and/or by T. congolense Forest.
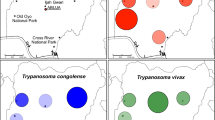
Trypanosome species diversity seemed higher in Yankari than in Wuya (five and two species, respectively; Table 2 and Fig. 1) with T. simiae, T. congolense Forest and T. godfreyi detected in Yankari at low prevalence but not detected in Wuya. It must be noted that the upper limits of the prevalence confidence intervals indicate that T. simiae, T. congolense Forest and T. godfreyi could be present in Wuya. The most prevalent trypanosomes species in both sites were T. congolense Savannah and T. vivax (Table 2 and Fig. 1). Only one infection of T. simiae and T. congolense Forest were found in G. m. submorsitans. Similarly, only one infection of T. godfreyi and four infections of T. congolense Forest were detected in G. tachinoides. In G. p. palpalis, the tsetse species collected in Wuya, only T. congolense Savannah and T. vivax were detected (Table 2 and Fig. 1).
To collate these data we screened DNA preparations from both ABD and H + P samples from more than 95 % of the flies (this could not be done for all the flies because some fly heads detached from their body during storage). Comparisons of the results obtained for each of these body parts are shown in Tables 3 and 4 for T. congolense subspp. and T. vivax respectively. For T. congolense subspp., there were very few flies that were positive in both ABD and H + P and most infections were found only in the abdomen samples (χ 2 = 8.47, df = 1, P < 0.01). In contrast, T. vivax was detected only in the H + P samples (χ 2 = 20.05, df = 1, P < 0.001). In G. p. palpalis, T. congolense Savannah was detected in some ABD and some H + P samples. In G. tachinoides, T. congolense Savannah and Forest were detected in some ABD and some H + P samples while T. godfreyi was detected in a single H + P sample. In G. m. submorsitans, T. congolense Savannah was detected in some ABD and some H + P samples while T. congolense Forest and T. simiae were only detected in ABD samples.
The differences in prevalence between male and female flies of each of the three species of tsetse are shown in Table 2 for each trypanosome species detected as well as the categories “All trypanosomes” and T. congolense subspp. To investigate the differences in prevalence between ‘tsetse species’, ‘collection site’ and ‘fly sex’ we undertook multiple logistic regression analyses, the outputs of which are summarised in Table 5. Infection prevalence of ‘All trypanosomes’ was higher in female than in male flies (Tables 2 and 5, Z = 2.165, P = 0.030). The prevalence of T. congolense subspp. was significantly higher in Yankari (considering both tsetse species together in this locality) than in Wuya (Fig. 1b, Table 5; Z = 2.185, P = 0.029). There were no statistically significant differences in trypanosome prevalence between G. tachinoides and G. m. submorsitans in Yankari (Fig. 1a, Table 5).
Discussion
Identification of major pathogens causing AAT
Our results show that tsetse flies in the two study sites harbour mature infections of T. vivax, T. congolense Savannah, T. congolense Forest and T. godfreyi. This implies that there is an active transmission of these trypanosomes in the two sites studied. Among the trypanosome species associated with mature infections in tsetse, T. vivax and T. congolense Savannah are likely to be the main parasites responsible for African Animal Trypanosomiasis (AAT) in livestock. T. vivax was only detected in the H + P samples. This is expected since this parasite only develops in the mouthparts [13] although it is possible to detect T. vivax in the abdomen if the fly has recently taken an infected meal. These results are in agreement with a recent survey carried out in cattle in northern Nigeria to discriminate T. congolense subspecies using molecular tools [14]. The presence of both subspecies of T. congolense is of importance for the management of AAT in Nigeria because T. congolense Savannah is more virulent than T. congolense Forest [15]. Indeed, the management of the disease caused by each T. congolense subspecies would likely be more effective if it could be subspecies-specific.
Trypanosome diversity and prevalence are different between sites
Although the upper limits of the prevalence confidence intervals suggest that T. simiae, T. congolense Forest and T. godfreyi could be present in Wuya, trypanosome species diversity seemed higher in Yankari (five species) than in Wuya (two species). In Wuya, only the two main pathogens responsible for AAT (T. congolense Savannah and T. vivax) were detected. This probably reflects the fact that tsetse in the Yankari National Park have a more diverse host range, feeding on wild animals, than the tsetse from Wuya that are likely feeding on a restricted number of livestock species. Indeed, the availability of hosts can influence trypanosome infection rates in tsetse [16]. Yankari game reserve is home to both wild and domestic animals while only domestic animals are seen in Wuya.
Combining the data for both species in Yankari, we were able to identify a difference between sites with the prevalence of T. congolense subspp. being higher in Yankari than in Wuya. The fact that the prevalence of T. congolense subspp. is higher in flies from Yankari than in flies from Wuya is probably a consequence of the higher prevalence of T. congolense subspp. in wild tsetse hosts in Yankari than in domesticated animals in Wuya. A complementary or alternative hypothesis would be that tsetse flies from Yankari are more susceptible to T. congolense subspp. than flies from Wuya. Our sampling design does not allow testing of that hypothesis because we could not sample the same species in both sites.
In Yankari, there were no statistically significant differences in trypanosome prevalence between G. tachinoides and G. m. submorsitans. This is a particularly interesting result because it goes against the traditional view that tsetse from the morsitans group are considered to be more susceptible to trypanosomes than tsetse from the palpalis group. The data generally gathered to support this concept have often been collected using approaches where the ‘species effect’ is confounded with a potential geographical effect. Indeed, experimental approaches have generally used tsetse laboratory colonies originating from different regions e.g. [17] and field studies often compared trypanosome prevalence of allopatric species. This is not the case in the present study as G. m. submorsitans and G. tachinoides are sympatric in Yankari. However, one cannot make conclusions on tsetse susceptibility based on data collected at a single field site. It must be noted that some experimental data have also shown similar vectorial capacity between tsetse from the morsitans and the palpalis group e.g. [11].
Trypanosome prevalence is higher in females than males
Our results clearly show that the trypanosome prevalence is higher in the female than the male flies analysed. Although this effect is likely mainly driven by a stronger effect for T. vivax infections, it must be noted that it was also the case for other trypanosome species (T. congolense Savannah in G. m. submorsitans and T. congolense Savannah and Forest in G. tachinoides). Although experimental data do not support significant differences in trypanosome infections between male and female tsetse [18, 19], higher trypanosome prevalence in females than in males have been previously reported in the field e.g. [20]. Such a result could be explained by the fact that female tsetse flies live longer than males. Females are thus more likely than males to be exposed to trypanosome infection during their lifetime. In line with that assertion, a study has shown that the prevalence of T. vivax rises approximately linearly with fly age [21]. The higher trypanosome prevalence observed in females would thus imply that the collected females were on average older than the collected males. Such a hypothesis could thus be tested in the future by estimating the age of the flies collected.
Conclusions
Our results show that T. congolense Savannah and T. vivax are the main species of parasite potentially causing AAT in the two study sites and that Yankari National Park is a potential reservoir of trypanosomes both in terms of parasite abundance and species diversity. This study constitutes the first report of T. congolense Savannah and T. congolense Forest in tsetse in Nigeria; T. congolense Savannah being more prevalent than T. congolense Forest. The presence of both subspecies and the difference in their prevalences are of importance for the management of AAT in Nigeria because T. congolense Savannah is more virulent than T. congolense Forest [15].
References
CFSPH, IICAB. African animal trypanosomiasis. USA: Center for Food Security & Public Health and Institute for International Cooperation in Animal Biologics; 2009. http://www.cfsph.iastate.edu/Factsheets/pdfs/trypanosomiasis_african.pdf. Accessed May 2016.
Kristjanson PM, Swallow BM, Rowlands GJ, Kruska RL, de Leeuw PN. Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agr Syst. 1999;59(1):79–98.
Delespaux V, Geysen D, Van den Bossche P, Geerts S. Molecular tools for the rapid detection of drug resistance in animal trypanosomes. Trends Parasitol. 2008;24(5):236–42.
Holmes P. Tsetse-transmitted trypanosomes - Their biology, disease impact and control. J Invertebr Pathol. 2013;112:S11–4.
Baldry DAT, Riordan K. A review of 50 years entomology of insect-borne diseases of veterinary importance in Nigeria, with special reference to tsetse flies and trypanosomiasis. In Gerard, B. M. (Ed.), Fifty years applied entomology in Nigeria. Paper presented at a conference of the Entomological Society of Nigeria, University of Ife, Ibadan, 5-7 Aprilia. 1967. pp.43–55.
Dede PM, Halid I, Omoogun GA, Uzoigwe NR, Njoku CI, Daniel AD, et al. Current tsetse and trypanosomiasis situation on Jos Plateau, Nigeria. Epizootiological factors that may enhance transmission and spread. Rev Elev Med Vet Pay. 2005;58(1–2):31–5.
Ahmed AB. High trypanosome infections in Glossina palpalis palpalis Robineau-Desvoidy 1830 in southern Kaduna State, Nigeria. Sci World J. 2007;2(2):1–7.
Madubunyi LC. Trypanosome infections in Glossina spp. inhabiting peridomestic agroecosystems in Nsukka area, Anambra State, Nigeria. Nlm. 1987;81(3):319–29.
Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RCA, et al. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95(3):186–92.
Peacock L, Ferris V, Bailey M, Gibson W. The influence of sex and fly species on the development of trypanosomes in tsetse flies. Plos Neglect Trop Dis. 2012;6(2):e1515.
R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2015. https://www.R-project.org/.
Venables WN, Ripley BD. Modern Applied Statistics with S, Fourth edition edn: Springer-Verlag New York; 2002
Gardiner PR. Recent studies of the biology of Trypanosoma vivax. Adv Parasitol. 1989;28:229–317.
Takeet MI, Fagbemi BO, De Donato M, Yakubu A, Rodulfo HE, Peters SO, et al. Molecular survey of pathogenic trypanosomes in naturally infected Nigerian cattle. Res Vet Sci. 2013;94(3):555–61.
Bengaly Z, Sidibe I, Ganaba R, Desquesnes M, Boly H, Sawadogo L. Comparative pathogenicity of three genetically distinct types of Trypanosoma congolense in cattle: clinical observations and haematological changes. Vet Parasitol. 2002;108(1):1–19.
Tarimo SRA, Snow WF, Butler L. Trypanosome infections in wild tsetse, Glossina pallidipes Austen on the Kenya coast. Int J Trop Insect Sci. 1984;5(Special Issue 05):415–8.
Reifenberg JM, Cuisance D, Frezil JL, Cuny G, Duvallet G. Comparison of the susceptibility of different Glossina species to simple and mixed infections with Trypanosoma (Nannomonas) congolense savannah and riverine forest types. Med Vet Entomol. 1997;11(3):246–52.
Welburn SC, Maudlin I. The nature of the teneral state in Glossina and its role in the acquisition of trypanosome infection in tsetse. Nlm. 1992;86(5):529–36.
Moloo SK, Sabwa CL, Kabata JM. Vector competence of Glossina pallidipes and G. morsitans centralis for Trypanosoma vivax, T. congolense and T. b. brucei. Acta Trop. 1992;51(3–4):271–80.
Desta M, Menkir S, Kebede A. The study on tsetse fly (Glossina species) and their role in the trypanosome infection rate in Birbir valley, Baro Akobo River system, western Ethiopia. J Vet Med Anim Health. 2013;5(7):186–94.
Kubi C, Billiouw M, Van Den Bossche P. Age prevalence of trypanosomal infections in female Glossina morsitans morsitans (Diptera: Glossinidae) on the plateau area of eastern Zambia. Onderstepoort J Vet. 2007;74(3):223–9.
Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;22(7):909–18.
Isaac C, Hamilton A, Scullion K, Ciosi M, Dede PM, Igbinosa IBI et al. ISID Fellowship Report, Distribution of trypanosomes within tsetse in northern Nigeria. ISID NEWS. 2013; December 2013:12–14, available at http://www.isid.org/publications/isid_news/ISIDnewsletter201312.pdf. Accessed May 2016.
Acknowledgements
This project was funded by the International Society for Infectious Diseases (ISID), the European Society of Clinical Microbiology and Infectious Diseases (ESCMID), the Tertiary Education Trust Fund (TETFUND) of Nigeria and The Wellcome Trust (grant number: 093692). We thank Anne-Marie Donachie (University of Glasgow) for technical and administrative assistance, as well as for the flowers; and Laurie Baker (University of Glasgow) for her help with the statistical analyses.
Authors’ contributions
CI and CMRT conceived and designed the study. CI, AH, KS and MC conducted the laboratory work. IBI, OPGN, DM and CMRT helped obtain funding and provide oversight of the work. CMRT and MC analysed the data. CI, CMRT and MC wrote the manuscript. CI and MC contributed equally to this work. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Complete trypanosome prevalence dataset: file “Isaac_et_al_additional_file_1_complete_trypanosome_prevalence_dataset.txt”. (TXT 13 kb)
Additional file 2:
R code used for the multiple logistic regression analyses: file “Isaac_et_al_additional_file_2_R_code_for_multiple_logistic_regressions.txt”. (TXT 7 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Isaac, C., Ciosi, M., Hamilton, A. et al. Molecular identification of different trypanosome species and subspecies in tsetse flies of northern Nigeria. Parasites Vectors 9, 301 (2016). https://doi.org/10.1186/s13071-016-1585-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-016-1585-3