Abstract
Background
Several Trypanosoma species transmitted by leeches infect marine and freshwater fish worldwide. To date, all South American fish trypanosome species identified have been based on unreliable morphological parameters. We recently isolated and cultured trypanosomes from the Brazilian armoured catfishes Hypostomus luetkeni and H. affinis. Here, we report the first phylogenetic analyses of South American (Brazilian) trypanosomes isolated from fish, and from leeches removed from these fish. We also analysed morphologically and morphometrically the different forms of fish, leech and cultured trypanosomes.
Methods
V7V8 SSU rRNA and gGAPDH sequences were used for phylogenetic analysis of Brazilian fish and leech trypanosomes. Trypanosomes from cultures, fish blood and leech samples were also characterized morphologically and morphometrically by light and electron microscopy.
Results
In blood smears from fish high trypanosome prevalence (90–100 %) and parasitemia (0.9-1.0x102) were observed. Phylogenetic relationships using SSU rRNA and gGAPDH showed that, despite relevant sequence divergence, all Brazilian fish (and derived cultures) and leech trypanosomes clustered together into a single clade. The Brazilian clade clustered with European, North American and African fish trypanosomes. Based on sequence analysis, we uncovered a new species of Brazilian fish trypanosome, Trypanosoma abeli n. sp. Trypanosoma abeli cultures contained pleomorphic epimastigotes, small trypomastigotes and rare sphaeromastigotes. Ultrastructural features of T. abeli included a cytostome-cytopharynx complex in epi- and trypomastigotes, a compact rod-like kinetoplast, lysosome-related organelles (LROs) and multivesicular bodies. Trypanosomes found in fish blood smears and leech samples were highly pleomorphic, in agreement with sequence data suggesting that catfishes and leeches often have mixed trypanosome infections.
Conclusions
Trypanosoma abeli n. sp. is the first trypanosome from South American fishes isolated in culture, positioned in phylogenetic trees and characterized at the ultrastructural level. Trypanosoma abeli n. sp. is highly prevalent in H. luetkeni and H. affinis armoured catfish from the Atlantic Forest biome, and in other catfish species from the Amazon and the Pantanal. Sequencing data suggested that Brazilian catfish often have mixed trypanosome infections, highlighting the importance of molecular characterization to identify trypanosome species in fishes and leeches.
Similar content being viewed by others
Background
Trypanosomes are widespread and highly prevalent in freshwater and marine fishes, including teleost and elasmobranch species. Since the initial report of trypanosomes in the blood of trouts from the species Salmo fario [1], more than 200 species of fish trypanosomes have been identified under the taxonomic criteria of morphology, and geographical and host origin [2, 3]. Although most infected fish are asymptomatic, fish trypanosomiasis can be severe at high parasitemias, and symptoms include anaemia, leukocytosis, hypoglycemia and splenomegaly [4, 5]. Aquatic leeches are both hosts and vectors of fish trypanosomes [6, 7]. Despite the reports of trypanosomes infecting leech-parasitizing fishes [7–10], host-vector relationships were only recently demonstrated by molecular comparison of trypanosomes from fish and leeches removed from the same fish [11].
In Brazil, more than 60 species of trypanosomes were recorded in marine and freshwater fishes [3] including loricariid (catfish) species such as Trypanosoma hypostomi [12], Trypanosoma chagasi and Trypanosoma guaibensis [13] and Trypanosoma lopesi [14]. Catfishes belong to the order Siluriformes, which comprises more than ~3,093 species of freshwater and salt water fishes inhabiting every continent except Antarctica, although more than 50 % of all catfish species, including all members of Loricariidae (>680 species), live in the tropical Americas. Loricariidae species vary largely in size, inhabit almost all freshwater habitats and are mostly bottom feeders. The genus Hypostomus is native to South America and comprises of large armoured catfish species used extensively as ornamental fish and food [15–17].
After the initial description of fish trypanosome cultivation by Thomson [18], several authors reported culturing of a variety of trypanosomes from the blood of freshwater and marine fish species from Europe, North America and Africa [7, 18–22]. However, only recently trypanosomes from South American (Brazilian) fish – the catfish H. luetkeni and H. affinis - were established in culture [23], despite the numerous reports of fish trypanosomes in Brazil. While there are many species descriptions based on Giemsa-stained bloodstream trypomastigote forms of fish trypanosomes [24–29], comparatively less is known about their ultrastructure, since only a few species were analysed at this level, either in culture [19, 30], or in vivo, in fish [5, 31, 32] and leech samples [33, 34].
Nevertheless, all descriptions of Brazilian fish trypanosome species available to date were based exclusively on morphology and host taxonomy criteria [3, 12, 14, 35–37]. These criteria are insufficient for species identification, because some species are morphologically indistinguishable (aside from intra-specific pleomorphism), and not host-specific, with a high frequency of mixed infections detected in molecular studies [11, 38–41]. Barcoding by variable SSU rRNA sequences is capable of distinguishing new trypanosome species/genotypes from previously known ones [11, 41–47].
The number of phylogenetic studies of fish trypanosomes has increased in recent years, and these studies have focused on trypanosomes removed directly from the blood of European [38, 41, 48, 49], African [11, 39] and Asian [5, 40, 50] marine and freshwater fishes. These trypanosomes segregate, in general, in two groups within a main clade [5, 11, 38, 40, 41, 49, 50]. However, molecular phylogenetic analysis currently adopted for the description of trypanosome species is still lacking for Brazilian fish trypanosomes. These limitations render necessary a molecular revision of the profusion of fish trypanosome species, including all of those described in Brazil before this study.
In this study, specimens of the ornamental armoured catfishes H. luetkeni and H. affinis were captured in The Atlantic Forest biome of Southeast Brazil and examined to assess trypanosome prevalence and parasitemia. We used phylogenetic analysis and both light and electron microscopy to characterize trypanosomes from blood samples, cultures and leeches removed from catfish. Also, the phylogenetic analysis included novel data on additional trypanosome isolates from other loricariid fishes captured in northeast (Amazonia) and central (Pantanal) Brazil, aiming to assess the genetic diversity. By integrating the phylogeny of fish trypanosomes with the morphological and ultrastructural features of cultured parasites, our findings enabled the description of Trypanosoma abeli n. sp. of Brazilian armoured catfishes.
Methods
Fish and leech collection, studied area, parasitemia and trypanosome prevalence
Armoured catfish, identified as Hypostomus affinis and Hypostomus luetkeni, were captured in the city of Guarani, state of Minas Gerais, Brazil (21°21’S, 43°02’W), in the banks of Pomba River, a 300-km long affluent of the Paraiba do Sul River basin that extends through the States of São Paulo, Minas Gerais and Rio de Janeiro, in the Atlantic Forest biome. Fish capture was performed according to procedures D-075 of the National Forests Institute (IEF) and N° 24402–1 of the Brazilian Institute of Environment and Renewable Natural Resources (IBAMA). If present, leeches were removed manually from the body surface and the oral and branchial cavities of captured fish. Leeches were macerated and sectioned in longitudinal and transversal directions for trypanosome detection. For blood sample collection, fish were anaesthetized and blood was collected by cardiac puncture, and blood from the heart, liver and kidney were used for smears. Only blood from cardiac puncture was used for trypanosome hemoculture, performed as described previously [23].
Trypanosome prevalence was determined by light microscopy examination of stained blood smears, and parasitaemia was determined by direct counting of trypanosome cells in fish blood using a hemocytometer. Samples of leeches macerated in sterile PBS were examined by light microscopy for the presence of trypanosomes. Fish blood and leech samples were processed immediately for electron microscopy (see SEM and TEM).
Hemocultures of fish trypanosomes
Here, we used 16 cultured fish trypanosome isolates whose in vitro cultivation was first described by Lemos & Souto-Padrón [23]. Isolates were cultured in three different biphasic media: a) Ponsele medium overlaid with Eagle’s Basal Medium (BME) (CULTILAB, Brazil) diluted to different concentrations; b) blood agar base (BAB) (HIMEDIA, Brazil) overlaid with BME diluted to 50 %, and c) NNN blood agar base overlaid with Fish ringer’s medium. All media were supplemented with 10 % heat-inactivated fetal calf serum (FCS) and cultures were maintained at 22 °C.
DNA extraction, PCR amplification, sequencing and phylogenetic analysis
DNA was extracted from trypanosome cultures in the exponential phase of growth using phenol-chloroform, and from fish blood and leech samples using the method described by Fermino et al. [44]. PCR amplification of the V7V8 region of SSU rRNA genes was performed using the primers 609 F (5’CACCCGCGGTAATTCCAGC3’) and 706R (5’TCTGAGACTGTAACCTCAA3’), and the primers GAP3F (5’GTGAAGGCGCAGCGCAAC3’) and GAP5R (5’CCGAGGATGYCCTTCATG3’) were used for the amplification of gGAPDH sequences. The PCR reaction conditions employed for the two sequences were detailed previously [45]. Amplified DNA fragments were cloned in pGEM-T-easy, and 3–5 clones were sequenced for each sample, except for culture L4100, for which an additional 15 clones were sequenced, aiming at detecting mixed trypanosome infections. Sequences were deposited in GenBank under the accession numbers shown in Table 1. For comparative purposes, DNA from blood trypanosomes of loricariid fishes captured in the Amazonia and the Pantanal biomes (Fermino et al., in preparation) were included in this study (Table 1).
Trypanosome SSU rRNA and gGAPDH gene sequences determined in this study were aligned with those from several fish trypanosomes available in GenBank. Two alignments were created: One comprising SSU rRNA sequences (800 ~ bp of variable V7V8 region of SSU rRNA) from Brazilian fish trypanosomes in cultures and trypanosomes from leeches, aligned with sequences from freshwater and marine fish trypanosomes from Europe, Africa and Asia; trypanosomes from crocodilians, lizards and snakes were used as outgroup. The second alignment was composed of gGAPDH sequences (~600 bp) of fish trypanosomes obtained from cultures, blood samples and leeches taken from captured fishes. This alignment includes GenBank sequences from a variety of ‘Aquatic clade’ trypanosomes (found in fishes and in other aquatic and semi-aquatic vertebrates), as well as those from trypanosomes outside the Aquatic clade, and other non-trypanosome trypanosomatids, which were used as outgroups. The complete list of samples, host and geographic origin, and Genbank accession numbers of all sequences determined in this work are shown in Table 1. Phylogenetic analysis of the new sequences (barcodes) were done by maximum parsimony using V7V8 SSU rRNA sequences (dendrogram of Aquatic clade trypanosomes), and maximum likelihood (ML), maximum parsimony (MP) and Bayesian inference (BI) for inferences of phylogenetic trees based on gGAPDH sequences, as described previously [43, 44, 46, 47].
Light microscopy and morphometry
Smears of fish blood samples, leech gut contents and cultures were fixed with methanol, stained with Giemsa, and examined using a Zeiss Axioplan II light microscope equipped with a Color View XS digital camera. Morphometry analysis of the different developmental forms was performed using the AnalySIS® soft, using images of 80 bloodstream trypomastigotes (20 images per developmental form). Data were analysed by descriptive statistics and Kruskal-Wallis variance analysis (p < 0.05 was considered statistically significant, using the BioEstat software, version 5.0).
Fluorescence microscopy
Trypanosomes in the logarithmic phase of growth were washed in BME at 4 °C and adhered to glass coverslips coated with 0.1 % of poly-l-lysine in PBS (pH 7.2), for 20 min. Then, cells were washed, fixed in methanol at 20 °C, and incubated at 22 °C with 1 μg/mL Hoechst H33342 (Molecular Probes) in BME, for 15 min. Coverslips were mounted using 0.2 M N-propyl-gallate (Sigma, USA) in glycerol and 0.01 M PBS (pH 7.2), and analyzed by epifluorescence microscopy using a Zeiss Axioplan II light microscope equipped with a Color View XS digital camera.
Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM)
Blood and axenic trypanosome cultures were fixed in 2.5 % glutaraldehyde in 0.1-M cacodylate buffer (pH 7.2) containing 5 mM calcium chloride and 3.7 % sucrose. For SEM, cells were washed and adhered to glass coverslips previously coated with 0.1 % of poly-l-lysine and processed (dehydrated, critical point dried and coated), according Lemos et al. [51]. Leeches were sectioned in the longitudinal and transverse directions, fixed and critical point dried for SEM (as described above), adhered to glass coverslips using adhesive tape, and coated with a 20-nm-thick gold layer, using a Bal-Tec CPD030 sputtering device. SEM samples were examined in a QUANTA 250 (Fei Company) Scanning Electron Microscope operated at 15 kV.
For TEM, flagellates from blood samples, cultures and leeches were fixed as described above and post-fixed for 30 min (in the dark) in 1 % osmium tetroxide (OsO4), 0.8 % potassium ferrocyanide and 5 mM calcium chloride, in 0.1 M cacodylate buffer (pH 7.3). Then, samples were washed, dehydrated in a series of acetone solutions of ascending concentrations, and embedded in Polybed 812 resin. Ultrathin sections were stained with uranyl acetate and lead citrate, and observed in a FEI Morgagni F268 Transmission Electron Microscope (Eindhoven, The Netherlands), operating at 80 kV.
Results and discussion
Prevalence of trypanosomes in armoured catfishes and in leeches removed from these fishes
In this study, Giemsa-stained blood smears from 40 specimens of H. affinis were all positive for trypanosomes by microscopy, yielding 100 % prevalence, with an average of 0.9 × 102 parasites/ml. The prevalence of trypanosomes in 10 specimens of H. luetkeni was 90 %, and the average parasitaemia was 1 × 102 parasites/ml. Similar levels of parasitaemia were detected in blood samples collected from the heart, liver and kidneys, for both fish species (Additional file 1). The fish trypanosome T. cobitis concentrates in the visceral circulation, mainly of the kidney of the hosts Cottus gobio, Phoxinus phoxinus, Nemacheilus barbatulus and Gobio gobio [9], while T. mioxocephali concentrates in the heart of Myoxocephalus octodecimspinosus [52]. These studies suggested that blood samples obtained from the heart and kidneys may allow improved sensitivity for the detection of fish trypanosomes, since these may be a preferred infection site for some trypanosomes.
The first reports of fish trypanosomes in Brazil date from the early 20th century, and surveys performed in several regions described diverse trypanosomes in catfish [35, 53, 54]. Splendore [12] was the first to provide data on the prevalence (9.5 %) of Trypanosoma hypostomi in blood smears from Hypostomus aurogutatus caught in the Tietê River (SP). Bara and Serra-Freire [13] reported high prevalences of Trypanosoma chagasi (95 %) and low of Trypanosoma guaibensis (7 %) in Hypostomus punctatus. Several armoured fish species captured in the hydrological basin of the Guamá River (in the state of Pará, in Northern Brazil) showed variable prevalence of trypanosome infection; the prevalence in Hypostomus sp. was of 20 %, and these fish showed the highest parasitaemia and haematological changes, but no evidence of disease [55]. Here, despite relevant parasitaemia, no macro pathological signs were observed in H. affinis and H. luetkeni infected with trypanosomes.
The leeches removed from H. affinis and H. luetkeni were identified morphologically as Haementeria sp. and molecularly as Haementeria brasiliensis [56]. On average, two and nine leeches per fish (for H. affinis and H. luetkeni, respectively) were found attached to the skin, mouth and, occasionally, to the pectoral fins of armoured catfish, at all seasons. The prevalence of trypanosomes in leeches was 81 % and 75 %, for leeches taken from H. affinis and H. luetkeni, respectively. The number of trypanosomes infecting leeches was high, ranging from 1.6 to 1.1 × 104 parasites/ml of leech macerate. Our data showing high trypanosome prevalence in leeches infecting fish is in line with that reported in previous studies: 61 % of Actinobdella inequiannulata leeches were infected by T. catostomi [57] and 60–100 % of Johanssonia arctica leeches were infected with T. murmanensis [58]. Importantly, these leech species were capable of transmitting trypanosomes to fish, by feeding [10]. High prevalence was also reported in the leech Z. arugamensis, considered the vector of Trypanosoma nudigobii [11]. In Brazil, a single report related the presence of trypanosomes in freshwater leeches (from the species Batracobdella gemmata) removed from H. punctatus catfish [59]; however, no prevalence data was reported in that study.
Culture of fish trypanosomes
The isolation and in vitro maintenance of fish trypanosomes has been a challenge since the initial description of T. danilewskyi isolation [18]. Recently, we isolated trypanosomes from Brazilian fish for the first time, successfully culturing these parasites in haemocultures [23]. In the present study we analysed 16 of these culture isolates obtained from 6 H. affinis and 4 H. luetkeni specimens.
Barcoding of Brazilian fish and leech trypanosomes and phylogenetic relationships
Most molecular and phylogenetic studies of fish trypanosomes relied on SSU rRNA sequences, and show a major clade harbouring all fish trypanosomes, divided in two clades of species from marine or freshwater fishes. The positioning of trypanosomes from turtles and platypuses was unresolved, but they always clustered closely related to fish trypanosomes in the phylogenetic trees [5, 11, 38–41, 50].
In the present study, PCR-amplified trypanosome SSU rRNA and gGAPDH sequences obtained directly from fish blood samples had to be cloned before sequencing due to the presence of more than one sequence per sample, indicating that the Brazilian armoured catfishes examined in the present study had mixed trypanosome infections (Table 1). Although other trypanosome species were identified by sequence analyses, the lack at this moment of appropriate type-materials prevented their description as additional species. According to the V7V8 SSU rRNA analysis, sequences from Brazilian fish and leech trypanosomes examined in this study clustered together in a clade with small heterogeneity (average of ~0.8 %); however, relevant sequence divergences (up to 3.8 %) separated this clade from available sequences from European, Asian and African fish trypanosomes (Fig. 1). The closest relatives of the Brazilian fish trypanosome was T. sp. K&A [60] from the European freshwater leech Piscicola geometra, and a trypanosome from European percae (Perca fluviatilis) [41] (Fig. 1). Sequences of trypanosomes from diverse fish orders and families, including catfishes from other parts of the world, did not cluster with those from Brazilian loricariid catfish, or from leeches that were attached to these fish (Fig. 1). Other catfish trypanosomes included in the phylogenetic analysis were isolated from widespread species of Clariidae (Africa), Siluridae (Europa) and Bagridae (Asia) [38, 39, 41].
Barcoding and phylogenetic relationships of fish trypanosomes. Phylogenetic analysis inferred by maximum parsimony based on V7V8 SSU rRNA sequences (barcodes) of fish trypanosomes from Brazilian armoured catfish (Loricariidae), or from leeches taken from these fishes. Note the inclusion of a new species, Trypanosoma abeli n. sp. established in culture. Freshwater and marine fish trypanosomes from different continents were included in the analysis, as well as other trypanosome species of the Aquatic clade. Numbers at nodes are bootstrap values derived from 100 replicates. GenBank accession numbers of SSU rRNA sequences determined in this study are listed in Table 1. Codes in parenthesis are GenBank accession numbers available for previously described species
Aside from yielding highly consistent alignments, gGAPDH sequences are much more polymorphic than SSU rRNA sequences for most trypanosome clades, resulting in phylogenies with considerably improved resolution [43, 44, 46, 47, 61]. Indeed, our analysis showed an average of 3.8 % gGAPDH sequence divergence within the clade formed by the Brazilian trypanosomes of fishes and leeches, compared with ~0.8 % of divergence for V7V8 SSU rRNA sequences. The phylogenetic inferences based on gGAPDH strongly supported the positioning of the trypanosomes found in Brazilian Hypostomus catfishes, as well as of the whole assemblage of fish trypanosomes (Fig. 2).
Phylogenetic tree of fish trypanosomes with the positioning of Trypanosoma abeli n. sp.: Maximum likelihood phylogenetic tree inferred from gGAPDH sequences of Trypanosoma abeli (cultures) and other trypanosomes from Brazilian loricariids and leeches, including sequences from European, African and Asian fish trypanosomes, as well as turtle and platypus trypanosomes placed within the Aquatic clade. Sequences from other trypanosomes and non-trypanosome trypanosomatids were used as outgroups in the phylogenetic tree (− Ln = − 7078.905197). Numbers on branches represent bootstrap support (>50) estimated with 500 pseudoreplicates in RAxML, using GTRGAMMA. GenBank accession numbers of gGAPDH sequences determined in this study are listed in Table 1. Codes in parenthesis are GenBank accession numbers available for previously described species
To better assess the genetic diversity, we compared gGAPDH sequences obtained from blood samples and cultures of trypanosomes from Brazilian catfishes and leeches removed from these fishes. We detected sequence polymorphisms among different blood samples from the same fish specimen (not shown), but also between samples from fishes and their respective leeches, and between samples from different specimens of the same fish species. Importantly, blood samples frequently harboured different sequences, indicating mixed trypanosome infections, in contrast to cultured isolates, from which, in general, only one sequence was recovered, likely due to selection by culturing (Table 1). It appears likely that more mixed samples would be identified by sequencing a larger number of cloned PCR-amplified DNA sequences obtained from blood samples of each fish specimen.
All trypanosome gGAPDH sequences from Brazilian armoured catfish obtained in this study clustered together in phylogenetic inferences (Fig. 2), supporting the existence of a separate clade exclusive for trypanosomes from Brazilian catfishes and from leeches taken from these fishes. The clade of Brazilian catfish trypanosomes was nested into the “Aquatic clade” composed by trypanosomes from anurans, fishes, turtles, platypuses and aquatic leeches [38, 61–63]. In agreement with previous phylogenetic studies [38, 39, 41], all fish trypanosomes nested into a single clade that also harboured trypanosomes from turtles and platypuses; this complex branching pattern needs the inclusion of more trypanosomes for improved resolution.
In our study, no sequence from trypanosomes found in leeches completely matched those found in the corresponding fish blood samples. In contrast, a recent study reported identical SSU rRNA sequences for trypanosomes in marine fishes and leeches taken from the same fishes, confirming host-vector relationships [11]. Our findings support the existence of a large repertoire of trypanosome species and genotypes infecting Brazilian catfishes and leeches, including relevant divergences distinguishing trypanosomes found in sympatric Hypostomus fishes and within the same fish specimens. Therefore, results from the present study reinforce the need for molecular characterization using sensitive approaches in order to evaluate species repertoire before ascertaining any parasite-vector association between trypanosome species infecting these hosts.
Taken together, our phylogenetic analyses using V7V8 SSU rRNA and gGAPDH sequences support the identification of a clade of new trypanosome species from Brazilian loricariid fishes differing from previously molecularly characterized fish trypanosomes, including those from other catfish species (Figs. 1 and 2). This clade may be associated with the evolutionary history of the Loricariidae restricted to Central and South America [15, 16]. In contrast, phylogenetic studies of fish trypanosomes from Europe, Africa and Asia did not support geographical structure or host-restrictions at species, genus or family levels [11, 38, 41]. Further studies are required to verify whether trypanosomes from other Neotropical fish families also cluster within this clade.
Description of Trypanosoma abeli n. sp. in Brazilian armoured catfishes
Fish trypanosomes characterized in this study shared similar sequences regardless of host species (H. affinis and H. luetkeni). Within the group of sequences obtained, we identified unique SSU rRNA and gGAPDH sequences (as determined by BLAST searches and positioning in phylogenetic trees) that supported the description of a new trypanosome species – hereby named Trypanosoma abeli n. sp. (Figs. 1 and 2). Here, we characterized this species using as type material the cultured isolate L4100, which showed a single trypanosome sequence in the analysis of sequences from 15 clones of the PCR-amplified DNA. In addition to H. affinis and H. luetkeni captured in Southeast Brazil, in the Atlantic Forest biome, isolates of T. abeli n. sp. sharing identical or very similar sequences were found in loricariids from other genera captured in Amazonia (Ancistrus sp.) and The Pantanal (Liposarcus anisistis) biomes (Figs. 1 and 2).
In the absence of cultures, new fish trypanosome species have been described based on the combination of morphological and molecular phylogenetic data from blood flagellates [5, 11, 40, 50]. Our finding that fish trypanosomes often harbour mixed infections, combined with similar data from previous studies [40, 41], suggests that this approach requires a careful assessment to exclude the possibility of mixed infections, which preclude the unambiguous association between trypanosome sequences and parasite morphotypes found in fish blood and leech samples.
Morphological and developmental analyses of Trypanosoma abeli n. sp. in cultures
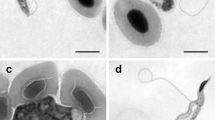
In primary hemocultures, bloodstream trypomastigotes (Fig. 3, a and b) transformed into short trypomastigotes (Fig. 3c) and, within two days, into epimastigotes (Fig. 3d). Established cultures in the logarithmic phase of growth consisted mostly of epimastigotes that ranged in shape and size from small and often “pear-shaped” (Fig. 3 d and e) to elongated and slender (Fig. 3f), with a long flagellum, a rod-like kinetoplast and an oval nucleus. Small (Fig. 3c) and elongated (Fig. 3g) trypomastigotes, as well as spheromastigotes with a round kinetoplast and a short free flagellum (Fig. 3, h and i), appeared in stationary-phase cultures. We also observed dividing epimastigotes (Fig. 3, j-m) and trypomastigotes (Fig. 3, n-t). In dividing trypomastigotes that had not reached the stage of cytokinesis, the two kinetoplasts located considerably posterior to the nuclei (Fig. 3, p and q). In cells that seemed to be at later stages in cell division, the kinetoplasts and nuclei appeared closer to each other (Fig. 3 r and s), but probably separated once again later in cytokinesis (Fig. 3t).
Light microscopy analysis of Trypanosoma abeli n. sp. cultured in vitro. Parasites were visualized by Giemsa staining (a-g, j, k, n, o, r-t), differential interference contrast (h, l, and p), and fluorescence microscopy using Hoechst H33342 (i, m, and q). Elongated (a) and short and broad (b) bloodstream trypomastigotes. Short trypomastigotes (c) and ‘pyriform’ (i.e. “pear shaped”) epimastigotes (d and e) and were the first developmental forms observed in primary cultures. Although trypomastigotes (g) were again observed in stationary phase, as well as spheromastigotes (h and i), with a round kinetoplast (arrowhead in i) and short flagellum (arrow in h). Cultures contained dividing epimastigotes (j-m) and trypomastigotes (n-t) at different stages of cell division, with duplicated single-copy structures, including the flagellum (F), the nucleus (N) and the kinetoplast (k). G, granules; V, vacuoles. Scale bar, 10 μm
Measurements of culture forms (type material) are still important for species description and must be included in the taxonomic section. Thus, we performed a detailed morphometric analysis of the different cell types found in cultures of isolate L4100 of T. abeli n. sp. (Additional file 2). Slender and pyriform epimastigotes had 19.8 ± 6.3 and 13.3 ± 5.4 μm in body length along cell midline, respectively, while trypomastigotes were longer, with body lengths along cell midline of 34.4 ± 1.6 and 22.8 ± 5 μm, for long and short forms, respectively.
Scanning electron microscopy (SEM) of fish and leech trypanosomes
The SEM analysis of T. abeli n. sp. highlighted the morphological diversity of cultured epimastigotes (Fig. 4, a and b) and trypomastigotes (Fig. 4, c and d), corroborating the light microscopy findings. The pleomorphism of bloodstream trypanosomes from H. affinis and H. luetkeni was clear in SEM images (Fig. 4 e-g). Trypanosomes from fish blood and leeches were, in fact, too pleomorphic (possibly due, in part, to mixed infections) to allow identification of the forms that corresponded to the bloodstream trypomastigotes of T. abeli. Flagellates found in the stomach caeca of leeches (Fig. 4h-j) were similar to those observed in cultures of T. abeli, and comprised of epimastigote (Fig. 4h), elongated trypomastigote (Fig. 4i) and spheromastigote (Fig. 4j) forms.
Morphological analysis of Brazilian catfish trypanosomes by scanning electron microscopy (SEM). a-d Trypanosoma abeli n. sp. e-g Bloodstream trypomastigotes of Trypanosoma sp. from fish blood samples. h-j Trypanosoma sp. from leech samples. Cultured epimastigotes with pyriform (a) or elongated (b) bodies. Short (c) and elongated (d) cultured trypomastigotes of T. abeli. Bloodstream trypomastigotes had elongated (e) or short (f and g) bodies, with a long section of ‘free’ flagellum portion (arrows). Samples from leech stomach caecum contained pyriform epimastigotes (h), short trypomastigotes (i) and short and wide forms (j) similar to spheromastigotes. Scale bars, 5 μm
Ultrastructural analysis of Trypanosoma abeli n. sp. by transmission electron microscopy (TEM)
To examine in detail the ultrastructure of T. abeli n. sp., we analysed log-phase cultures of this parasite by transmission electron microscopy (TEM). When observed by TEM, T. abeli n. sp. trypomastigotes and epimastigotes had a rod-like kinetoplast containing a compact array of kDNA fibrils (Fig. 5, a and c) similar to that observed in the kinetoplast of bloodstream trypomastigotes found in H. affinis and H. luetkeni (Fig. 5 b). The kinetoplast pattern observed in T. abeli was similar to that of freshwater and marine fish trypanosomes, such as T. pseudobagre and T. triglae [31, 32], but differed from that of T. cruzi, which exhibits a round kinetoplast with a relatively ‘loose’ kDNA network [64].
Morphological analysis of culture forms Trypanosoma abeli n. sp. by transmission electron microscopy (TEM). Cultured trypomastigotes (a) had a rod-like kinetoplast (k) similar to that of bloodstream trypomastigotes of Trypanosoma sp. from H. affinis and H. luetkeni (b), and epimastigotes of T. abeli (c). Lysosome related organelles (LROs; in c and e) had an electron-lucent matrix containing small vesicles (arrowheads in e) and rod-shaped electron-lucent bodies (arrows in e). Epimastigotes (c) and trypomastigotes (d) had numerous lipid bodies (L) of different sizes located near the kinetoplast (k) and the nucleus (N). Adjacent lipid bodies appeared to fuse, generating larger ones (f). A multivesicular body (MVB) containing small electron lucent (white arrow) and electron dense (black arrows) vesicles was located near the flagellar pocket (g), and a microtubule quartet (arrowheads in g) subtended the flagellar pocket membrane. Trypomastigotes had a cytostome (Ct)-cytopharynx (Cy) complex (d and h) that extended deep into the cytoplasm (h), reaching the perinuclear region (d). Nucleus, N; Nu, nucleolus; k, kinetoplast; FP, flagellar pocket; F, flagellum, Ac, acidocalcisomes; GC, Golgi Complex; L, lipid body; LRO, lysosome-related organelle; MVB, multivesicular body; Cy, cytostome; Ct, cytopharynx (Ct). Scale bars: 1 μm (c, f); 500 nm (a, b, h); 250 nm (d); 200 nm (e); 400 nm (g)
Numerous lipid bodies with variable sizes (from 50 nm to 1.5 μm) were observed in the cytoplasm of T. abeli n. sp. (Fig. 5c, d, f), and we also observed smaller lipid bodies that appeared to fuse with (or bud from) larger ones (Fig. 5f). At the posterior region, T. abeli n. sp. had large membrane-bound structures reminiscent of lysosomes-related organelles (LROs), containing linear membrane profiles and small vesicles (Fig. 5e) immersed in an electron-dense matrix (Fig. 5c). These putative LROs were morphologically similar to those of T. cruzi (referred to as ‘reservosomes’) [65, 66], which contain rod-shaped electron-lucent bodies [67] also observed in T. abeli n. sp. (Fig. 5e).
Epimastigotes of T. abeli n. sp. had multivesicular bodies near the flagellar pocket containing numerous vesicles of regular size (~80 nm) and with an electron-dense matrix (Fig. 5g). Multivesicular bodies were also observed in Trypanosoma terena from cayman [44], Trypanosoma serpentis and Trypanosoma cascaveli from snakes [43] and Trypanosoma cobitis from fish [34]; however, they had different localization and internal vesicles of different size compared with the T. abeli n. sp. multivesicular bodies. The nature of these structures in trypanosomatids remains unknown, and hypotheses suggest that they are formed by viral particles, or may be associated with the endocytic or exocytic pathways [34, 68].
Both epimastigotes and bloodstream trypomastigotes of T. abeli n. sp. displayed structures morphologically similar to acidocalcisomes, an acidic organelle rich calcium and polyphosphates and involved in osmoregulation, in some protozoa [69]. These structures have been described in other fish trypanosomes (Trypanosoma pseudobagri and T. danilewskyi) [31, 32], and in Trypanosoma fallisi from anurans [70], T. serpentis and T. cascavelli from snakes [43], T. terena and T. ralphi from caymans [44], as well as in several trypanosomes of mammals, including T. cruzi, Trypanosoma erneyi and Trypanosoma brucei [46, 71].
The cytostome-cytopharynx complex - an invagination of plasma membrane penetrating deep into the cytoplasm and sometimes reaching the perinuclear region - was observed in trypomastigotes (Fig. 5d, h) and epimastigotes (not shown) of T. abeli. In some images, the cytostome appears to ‘open’ into the flagellar pocket (not shown), differently from that observed in other trypanosomes [30, 31, 34, 43, 44, 47, 64, 70, 72]. The cytopharynx was first observed in epimastigotes and trypomastigotes of the fish trypanosomes T. raiae, T. danilewskyi and T. cobitis [30, 31, 34], and a cytostome-cytopharynx complex was also observed in epimastigotes of trypanosomes from anurans, snakes and crocodilians [43, 44, 70]. The cytostome-cytopharynx complex has also been observed in epimastigotes of T. cruzi and in other species of the subgenus Schyzotrypanum; however, trypomastigotes of these species lacked this complex [46, 72, 73]. Thus far, the presence of the cytostome-cytopharynx complex in blood and culture trypomastigotes appears to be a common and exclusive characteristic of fish trypanosomes [31], as confirmed here by our TEM analysis of T. abeli n. sp.
Morphology features of blood trypomastigotes found in H. affinis and H. luetkeni
Bloodstream trypomastigotes from both H. affinis and H. luetkeni were highly polymorphic (Fig. 6), and the number and proportion of morphotypes varied between blood samples. The morphological variation observed here, if used as the sole taxonomic criterion, would support the description of more than one species in these samples. In fact, the different morphotypes may represent distinct developmental stages from the same species, instead of different species. In fish blood samples, we could not distinguish between intra- and inter-specific polymorphism and, thus, could not assign a given morphotype to a given species.
Morphological analysis of different developmental forms of Trypanosoma sp. observed in the blood of H. affinis and H. luetkeni (a-g), or in the gut contents of Haementeria brasiliensis leeches removed from these fish (h-n). Bloodstream trypomastigotes were divided into four morphotypes as follows: a morphotype 1 has a long and broad body; b morphotype 2 has a long and slender body, and several vacuoles (arrowheads); c morphotype 3 has a broader body with striations (arrowheads); and d-g morphotype 4 has a short and slender body. The developmental forms isolated from leeches were trypomastigotes (h), slender epimastigotes (i, k, and l), and pyriform epimastigotes (j). Dividing forms included epimastigotes undergoing cytokinesis (m), with two nuclei (arrows) and 2 kinetoplasts (arrowheads), and trypomastigotes at a post-mitotic stage of the cell duplication cycle (N; arrows indicate duplicated nuclei). N, nuclei; F, flagellum; k, kinetoplast; S, striae; V, vacuoles; G, granules. Scale bar = 10 μm
To facilitate morphological description of blood flagellates, morphometrical data were used to divide bloodstream trypomastigotes into four morphotypes, according to their body length along cell midline (short vs. elongated, with p < 0.001 by Kruskal-Wallis) and width (slender vs. broad, p < 0.00001 by Kruskal-Wallis) (Fig. 6, a-g; Additional file 3). Morphotype 1 trypomastigotes (Fig. 6a; Additional file 3) have a long and broad body (67.3 ± 1.2 μm in length and 4.08 ± 0.1 μm in width), with a tapering anterior end, a broad posterior end (often C-shaped), and a long section of free flagellum (20.8 ± 0.6 μm). The nucleus is elongated and positioned in the anterior end, and the kinetoplast is close to the posterior extremity of the cell. Morphotype 2 (Fig. 6b; Additional file 3) has a long and slender body (64.57 ± 2.3 μm length and 2.73 ± 0.06 μm width) and a long free flagellum (21.07 ± 1.4 μm). The rod-like kinetoplast is close to the posterior end, the nucleus is oval and eccentrically placed in an anterior position, and the cytoplasm may contain several vacuoles. Morphotype 3 (Fig. 6c; Additional file 3) has a long, broad and S-shaped body (length 61 ± 3 μm, width 5.5 ± 0.4 μm), with striations along the main axis, a tapered anterior end, a wide posterior end, and a long (41.6 ± 8.8 μm) free flagellum. A round kinetoplast is found at the posterior end and the nucleus is almost oval, large and anterior in position (Fig. 6c). Morphotype 4 (Fig. 6, d-g; Additional file 3) has a short and slender body (length 28.01 ± 1.4 μm, width 1.58 ± 0.08 μm), a short free flagellum (13.49 ± 1.4 μm), a rod-like kinetoplast located at the posterior end, and an anterior and oval-shaped nucleus (Fig. 6 d-g), similar to that observed in morphotypes 2 and 3.The four morphotypes described here may represent a mixture of trypanosome species/genotypes as indicated by our sequence analyses (Figs. 1 and 2; Table 1).
There are more than 30 trypanosome species reported in Brazilian catfishes and at least 25 of armoured catfishes [3], including ~15 species described in Hypostomus spp. However, no trypanosome species was described in H. affinis or H. luetkeni before this study. The morphotypes identified in the blood samples of catfishes examined in this study broadly resembled those previously reported in other catfishes. However, the trypomastigotes observed in blood smears from fishes infected with T. abeli n. sp. were in general longer and larger than those found in other species of armoured catfishes, although trypomastigotes of different trypanosome species share morphological similarities with at least one of the four morphotypes identified in fishes infected with T. abeli. For instance, the trypomastigotes of Trypanosoma chagasi are quite similar to morphotype 1 [55], whereas Trypanosoma loricariae [35] and Trypanosoma hypostomi [12] share blood forms more similar to morphotype 2 of T. abeli. Altogether, our findings corroborated that morphology and morphometry are not sufficient to distinguish species of fish trypanosomes, although these data are relevant as a complementary data for the description of any new trypanosome species.
Morphological features of trypanosomes found in leeches removed from H. affinis and H. luetkeni
Developmental forms of trypanosomes were observed in the digestive tract of H. brasiliensis leeches taken from H. affinis and H. luetkeni. Trypomastigotes (Fig. 6h) were observed in the proboscis and stomach caeca, and epimastigotes (Fig. 6, i-l) in the stomach caeca and intestines. Slender (Fig. 6 i, k and l) and pyriform (Fig. 6j) epimastigotes had an oval nucleus and a rod-like kinetoplast. Trypomastigotes had slender bodies, with an oval nucleus located in the anterior region of the cell body, and a rod-like kinetoplast close to the nucleus (Fig. 6h). Leech samples also contained dividing epimastigote (Fig. 6, m) and trypomastigote (Fig. 6n) forms, including trypomastigotes with two nuclei (Fig. 6n). Morphometrical data of trypomastigotes and epimastigotes observed in leech samples are shown in Additional file 4.
In leeches examined in a previous study, spheromastigotes occurred in the crop, and this and a variety of epimastigote forms were observed in the intestine, while metacyclic trypomastigotes were generally found in the proboscis following complete digestion of the blood meal. However, migration to the leech proboscis sheath by fish trypanosomes has not been elucidated [7, 8, 11, 57].
Variable forms of trypanosomes in leeches can represent mixtures of trypanosome species, different stages of one trypanosome species, or flagellates from old and new infections [11]. Our results show that leeches recovered from fish could be infected with trypanosomes different from those infecting their host fish. Unfortunately, we did not recover T. abeli n. sp. sequences from DNA preparations from Haementeria brasiliensis leeches. Therefore, trypanosomes found in Giemsa-stained leeches samples could be not identified as T. abeli n. sp., and further surveys are necessary to evaluate the role of H. brasiliensis as vector of this trypanosome species.
Taxonomical summary
Phylum Euglenozoa Cavalier-Smith, 1981. Class Kinetoplastea Honigberg, 1963, emend. Vickerman, 1976. Order Trypanosomatida Hollande 1952. Family Trypanosomatidae Doflein, 1951 Trypanosoma abeli n. sp. Lemos and Souto-Padrón 2015.
Type material
Hapantotype, culture of the isolate L4100 from Hypostomus luetkeni deposited in the FIOCRUZ Protozoan Collection (code COLPROT - 700) of the Instituto Oswaldo Cruz, Fundação Oswaldo Cruz, Rio de Janeiro, RJ, Brazil. Details of T. abeli has been registered in ZooBank with the following Life Science Identifier (LSID): zoobank.org:pub:AAD60F84-2E99-46A4-929B-BBA1AEDE437C.
Type host
Hypostomus luetkeni (Siluriformes, Loricariidae). Additional host: Hypostomus affinis (Siluriformes, Loricariidae).
Type locality
Pomba River (21°21’S, 43°02’W), a small tributary of the Paraíba do Sul River, Minas Gerais, Southeast Brazil.
Morphological characteristics
In logarithmic cultures, pyriform (length, 13.3 μm; width, 1.6 μm) and slender (length, 19.8 μm; width, 1.7 μm) epimastigotes (Fig. 3 e-f). In stationary cultures, elongated (length, 34.4 μm; width, 1.6 μm; Fig. 3 c) and short (length, 22.8 μm; width, 2.1 μm) trypomastigotes (Fig. 3g). Ultrastructural features include a rod-shaped and compact kinetoplast, LRO structures, multivesicular bodies and a cytostome-cytopharynx complex (Fig. 5a, c, e, g and h).
Species diagnosis
Unique sequences of SSU rRNA and gGAPDH of T. abeli n. sp., deposited in GenBank under accession numbers KR048310 (SSU rRNA gene) and KR048292 (gGAPDH gene).
Etymology
The species was named in honour of José Abel, for his constant encouragement and genuine interest in science.
Conclusions
Trypanosoma abeli n. sp. is the first trypanosome from South American fishes isolated in culture, positioned in phylogenetic trees, and characterized at morphological and ultrastructural levels. Phylogenetic relationships inferred using SSU rRNA and gGAPDH gene sequences positioned the trypanosomes of armoured catfishes within an assemblage of closely related trypanosomes from Brazilian loricarids and leeches removed from these fishes, but separated from the other fish trypanosomes, all from other continents. Our studies revealed the presence of T. abeli n. sp. in H. luetkeni and H. affinis caught in a small river of the Atlantic Forest, and in other loricariid fishes from rivers of Amazonia and the Pantanal biomes.
Ultrastructural analyses confirmed that T. abeli n. sp. have a cytostome-cytopharynx complex in both epimastigotes and trypomastigotes, as shown in other fish trypanosomes, large lysosome-related organelles and multivesicular bodies. In spite of considerable resemblance between blood trypomastigotes morphotypes detected in blood smears of H. luetkeni and H. affinis, we obtained molecular evidence for mixed trypanosome infections in these fishes, which prevented the establishment of morphotype-species associations. In addition, we obtained different sequences from fish blood samples and from leeches taken from these fishes; thus, further and more detailed molecular characterization is required to assess the repertoire of H. luetkeni and H. affinis trypanosomes and any link between these trypanosomes and H. brasiliensis leeches.
References
Valentin P. Ueber ein entozoon im blute von Salmo fario. Arch Anal Phys Wiss Med. 1841; 435–436.
Lom J, Dykova I. Flagellates (Phylum Mastigophora). In: Protozoan parasites of fishes. Amsterdam: Elsevier; 1992. p. 32–8.
Eiras JC, Takemoto RM, Pavanelli GC, Luque JL. Checklist of Protozoan parasites of fishes from Brazil. Zootaxa. 2012;3221:1–25.
Islam NAKM, Woo PTK. Anorexia in goldfish Carassius auratus infected with Trypanosoma danilewskyi. Dis Aquatic Org. 1991;11:45–8.
Su Y, Feng J, Jiang J, Guo Z, Liu G, Xu L. Trypanosoma epinepheli n. sp. (Kinetoplastida) from a farmed marine fish in China, the brown-marbled grouper (Epinephelus fuscoguttatus). Parasitol Res. 2014;113:11–8.
Khan RA. The life cycle of Trypanosoma murmanensis Nikitin. Can J Zool. 1976;54:1840–9.
Jones SRM, Woo PTK. Development and infectivity of Trypanosoma phaleri in leech and fish hosts. Can J Zool. 1991;68:1951–5.
Khan RA. A redescription of Trypanosoma cotti Brumpt and Lebailly,1904 and its development in the leech, Calliobdella punctata. Ann Parasitol Hum Comp. 1978;53:461–6.
Letch CA. The life-cycle of Trypanosoma cobitis Mitrophanow 1883. Parasitol. 1980;80:163–9.
Karlsbakk E, Haugen E, Nylund A. Morphology and aspects of growth of a trypanosome transmitted by the marine leech Johanssonia arctica (Piscicolidae) from Northern Norway. Fol Parasitol. 2005;52:209–15.
Hayes PM, Lawton SP, Smit NJ, Gibson WC, Davies AJ. Morphological and molecular characterization of a marine fish trypanosome from South Africa, including its development in a leech vector. Parasit Vectors. 2014; doi:10.1186/1756-3305-7-50.
Splendore A. Trypanosomes de poissons brésiliens. Bull Soc Pathol Exotique. 1910;3:521–4.
Bara MDA, Serra-Freire NM. Aspectos epidemiológicos da infecção por tripanossomas em Hypostomus punctatus Valenciennes, 1840 (Osteichthyes, Loricariidae) no lago Açu da UFRRJ, Brasil. Rev Bras Med Vet. 1985;7:46–9.
Ribeiro RD, Satake T, Nutisobrinho AIG, Brentegani HA, Britski HA, Lopes RA. Trypanosomes of brazilian fishes. 4. Trypanosoma lopesi sp. n. from armoured catfish Rhinelepis aspera Agassiz, 1829 (Teleostei, Loricariidae). Zool Anz. 1989;222:244–8.
Da Silva M, Ribeiro ED, Matoso DA, Sousa LM, Hrbek T, Py-Daniel LR, et al. Chromosomal polymorphism in two species of Hypancistrus (Siluriformes: Loricariidae): an integrative approach for understanding their biodiversity. Genetica. 2014;142:127–39.
Roxo FF, Albert JS, Silva GS, Zawadzki CH, Foresti F, Oliveira C. Molecular phylogeny and biogeographic history of the armoured neotropical catfish subfamilies hypoptopomatinae, neoplecostominae and otothyrinae (siluriformes: loricariidae). PLoS One. 2014;9 Suppl 8:e105564.
Silva GdeS, Roxo FF, Britzke R, Oliveira C. New species of the Pseudancistrus barbatus group (Siluriformes, Loricariidae) with comments on its biogeography and dispersal routes. Zookeys. 2014; doi:10.3897/zookeys.406.7011.
Thomson JD. Cultivation of the trypanosome found in the blood of the goldfish. J Trop Med Hyg. 1908;8:75–82.
Lewis JW, Ball SJ. Micro-organisms in Trypanosoma cobitis. Int J Parasitol. 1981;11:121–5.
Rouskova H, Kulda J. Growth and transformation of Trypanosoma danilewskyi in a liquid medium. Fol Parasitol. 1983;30:295–301.
Islam NAKM, Woo PTK. Effects of temperature on the in vivo and in vitro multiplication of Trypanosoma danilewskyii Laveran & Mesnil. Fol Parasitol. 1992;39:1–12.
Davies AJ, Thomson RJ, Sappal GK. Behaviour of two fish trypanosomes over cultured fish cells. Bull European Assoc Fish Pathol. 1996;16:172–5.
Lemos M, Souto-Padrón T. Isolation and In Vitro Maintenance of Trypanosomes from Naturally Infected and Commercially Important Brazilian Fish. J Parasitol. 2014;100:687–91.
Hasan R, Qasim SZ. Trypanosoma punctati n. sp. from the fish Ophiocephalus punctatus (Bloch), common fresh water murrel of India. Zeit fur Parasitenkd. 1962;22:118–22.
Daly JJ, DeGiusti DL. Trypanosoma catostomi n. sp. from the White Sucker Catostomus c. commersoni (Lacepede). J Protozool. 1971;18:414–7.
Narasimhamurti CC, Saratchandra B. Two new species of trypanosomes, Trypanosoma channai n. sp. From channai punctata and Trypanosoma quadrii n. sp. from Clarias batrachus. Prec Indian Acad Sci. 1980;89:371–6.
Morilas J, George-Nascimento M, Valeria H, Khan RA. Trypanosoma humboldti n. sp. from the Chilean catshark, Schroederichthys chilensis (Guichenot, 1848). J Protozool. 1987;34 Suppl 3:342–4.
Gupta DK, Gupta N, Gangwar R. Two new species of Trypanosoma from freshwater fish (Heteropneustes fossilis and Channa punctatus) from Bareilly. India J Parasitic Dis. 2006;30:58–63.
Yeld EM, Smit NJ. A new species of Trypanosoma (Kinetoplastida: Trypanosomatidae) infecting catsharks from South Africa. J Mar Biol Ass. 2006;86:829–33.
Preston TM. The forms of the cytostome-cytopharynx of the culture forms of the elasmobranch haemoflagellate Trypanosoma raiae Laveran & Mesnil. J Protozool. 1969;16:320–33.
Lom J, Paulin JJ, Nohýnková E. Ultrastructure of the fish trypanosome, Trypanosoma danilewskyi: I. Presence of a cytopharyngeal complex in bloodstream trypomastigotes. Protistol. 1980;16:365–73.
Gu Z, Wang J, Zhang J, Li M, Ke X, Gong X. Ultrastructure of the bloodstream forms of the fish trypanosomeTrypanosoma pseudobagri Dogiel et Achmerov, 1959. Acta Protozool. 2007;46:139–45.
Lewis JW, Ball SJ. Prevalence of Trypanosoma cobitis Mitrophanow, 1883 in fishes from River Lee. Parasitol. 1979;79:119–24.
Lewis JW, Ball SJ. Ultrastructure of the epimastigotes of the fish trypanosome Trypanosoma cobitis Mitrophanow, 1883, in the crop oh the leech vector, Hemiclepsis marginata. J Parasitol. 1980;66:948–53.
Fonseca F, Vaz Z. Novos tripanossomas de peixes brasileiros. Ann Fac Med São Paulo. 1928;3:69–94.
Fróes OM, Fortes E, Lima DF, Leite VRV. Tripanosomas (Protozoa, Kinetoplastida) de peixes de água doce do Brasil. II Novos tripanossomas de cascudos (Pisces, Loricariidae). Rev Bras Biol. 1979;39:425–9.
Eiras JC, Rego AA, Pavanelli GC. Trypanosoma scrofae SP. N. (Protozoa, Kinetoplastida) parasitizing Prochilodus scrofa (Osteichtyes, Prochilodontidae) from Paraná River. Rev Ibérica Parasitol. 1989;49:111–5.
Gibson WC, Lom J, Pecková H, Ferris VR, Hamilton PB. Phylogenetic analysis of freshwater fish trypanosomes from Europe using ssu rRNA gene sequences and random amplification of polymorphic DNA. Parasitol. 2005;130:405–12.
Davies A, Gibson W, Ferris V, Basson L, Smit N. Two genotypic groups of morphologically similar fish trypanosomes from the Okavango Delta, Botswana. Dis Aquat Org. 2005;66:215–20.
Gu Z, Wang J, Li M, Zhang J, Ke X, Gong X. Morphological and genetic differences of Trypanosoma in some Chinese freshwater fishes: difficulties of species identification. Parasitol Res. 2007;101:723–30.
Grybchuk-Ieremenko A, Losev A, Kostygov AY, Lukeš J, Yurchenko V. High prevalence of trypanosome co-infections in freshwater fishes. Folia Parasitol. 2014;61:495–504.
Ferreira RC, Campaner M, Viola LB, Takata CSA, Takeda GF, Teixeira MMG. Morphological and molecular diversity and phylogenetic relationships among anuran trypanosomes from the Amazonia, Atlantic Forest and Pantanal biomes in Brazil. Parasitol. 2007;137:1623–38.
Viola LB, Attias M, Takata CSA, Campaner M, De Souza W, Camargo EP, et al. Phylogenetic analyses based on small subunit rRNA and glycosomal glyceraldehyde-3-phosphate dehydrogenase genes and ultrastructural characterization of two snake Trypanosomes: Trypanosoma serpentis n. sp. from Pseudoboa nigra and Trypanosoma cascavelli from Crotalus durissus terrificus. J Eukaryot Microbiol. 2009;56:594–602.
Fermino BR, Viola LB, Paiva F, Garcia HA, de Paula CD, Botero-Arias R et al.. The phylogeography of trypanosomes from South American alligatorids and African crocodilids is consistent with the geological history of South American river basins and the transoceanic dispersal of Crocodylus at the Miocene. Parasit Vectors. 2013; 6: doi:10.1186/1756-3305-6-313.
Borghesan TC, Ferreira RC, Takata CS, Campaner M, Borda CC, Paiva F, et al. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013;164:129–52.
Lima L, Silva FM, Neves L, Attias M, Takata CS, Campaner M, et al. Evolutionary Insights from Bat Trypanosomes: Morphological, Developmental and Phylogenetic Evidence of a New Species, Trypanosoma (Schizotrypanum) erneyi sp. nov., in African Bats Closely Related to Trypanosoma (Schizotrypanum) cruzi and Allied Species. Protist. 2012;163:856–72.
Lima L, Espinosa-Alvarez O, Hamilton PB, Neves L, Takata CS, Campaner M et al.. Trypanosoma livingstonei: a new species from African bats supports the bat seeding hypothesis for the Trypanosoma cruzi clade. Parasit Vectors. 2013; doi:10.1186/1756-3305-6-221.
Figueroa F, Mayer WE, Lom J, Dykova I, Weller M, Peckova H, et al. Fish trypanosomes: their position in kinetoplastid phylogeny and variability as determined from 12S rRNA kinetoplast sequences. J Euk Microbiol. 1999;46:473–81.
Karlsbakk E, Nylund A. Trypanosomes infecting cod Gadus morhua L. in the North Atlantic: a resurrection of Trypanosoma pleuronectidium Robertson, 1906 and delimitation of T. murmanense Nikitin, 1927 (emend.), with a review of other trypanosomes from North Atlantic and Mediterranean teleosts. Syst Parasitol. 2006;65:175–203.
Gu Z, Wang J, Ke X, Liu Y, Liu X, Gong X, et al. Phylogenetic position of the freshwater fish trypanosome, Trypanosma ophiocephali (Kinetoplastida) inferred from the complete small subunit ribosomal RNA gene sequence. Parasitol Res. 2010;106:1039–42.
Lemos M, Souza CS, da Costa SC G, Souto-Padrón T, D’ Agosto M. Isolation and In Vitro Culture of Trypanosomes From Leptodactylus ocellatus From the Atlantic Forest in a New Experimental Culture Medium. J Parasitol. 2013;99:164–7.
Fantham HB, Porter A, Richardson LR. Some haematozoa observed in vertebrates in eastern Canada. Parasitol. 1942;34:194–205.
Botelho JC. Sur deux nouveaux trypanosomes de poissons. C R Acad Sci Biol. 1907;59:128–9.
Horta PP. Trypanosoma chagasi n. sp. Parasito de Plecostomus punctatus. Brasil Médico. 1910;24:273.
Fujimoto RY, Neves MS, Santos RFB, Souza NC, Do Couto MVS, Lopes JNS, et al. Morphological and hematological studies of Trypanosoma spp. infecting ornamental armoured catfish from Guamá River-PA, Brazil. An Acad Bras Cien. 2013;85:1149–56.
Oceguera-Figueroa A. Molecular phylogeny of the New World bloodfeeding leeches of the genus Haementeria and reconsideration of the biannulate genus Oligobdella. Mol Phylogenet Evol. 2012;62:508–14.
Jones SR, Woo PT. Vector specificity of Trypanosoma catostomi and its infectivity to freshwater fishes. J Parasitol. 1992;78:87–92.
Khan RA. Trypanosome occurrence and prevalence in the marine leech Johanssonia arctica and its host preferences in the northwestern Atlantic Ocean. Can J Zool. 1991;69:2374–80.
Bara MDA, Serra-Freire NM. Infecção natural de sanguessugas do gênero Diplobdella por tripanosomas de Hypostomus punctatus Valencienne (Osteichthyes, Loricariidae). Rev Fac Vet. 1984;1:55–7.
Stevens JR, Noyes HA, Gibson WC. The Evolution of Trypanosomes Infecting Humans and Primates. Mem Inst Oswaldo Cruz. 1998;93 Suppl 5:669–76.
Hamilton P, Stevens J, Gaunt M, Gidley J, Gibson W. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol. 2004;12:1393–404.
Stevens JR, Noyes HA, Schofield CJ, Gibson W. The molecular evolution of Trypanosomatidae. Adv Parasitol. 2001;48:1–56.
Hamilton PB, Gibson WC, Stevens JR. Patterns of co-evolution between trypanosomes and their hosts. Deduced from ribosomal RNA and protein-coding gene phylogenies. Mol Philogenet Evol. 2007;44:15–25.
De Souza W. A Short Review on the Morphology of Trypanosoma cruzi: from 1909 to 1999. Mem Inst Oswaldo Cruz. 1999;94:17–36.
Soares MJ, De Souza W. Cytoplasmic organelles of trypanosomatids: a cytochemical and stereological study. J Submicrosc Cytol Pathol. 1988;20:349–61.
Sant’ Anna C, Parussini F, Lourenço D, De Souza W, Cazzulo JJ. All Trypanosoma cruzi developmental forms present lysosome-related organelles. Histochem Cell Biol. 2008;130:1187–98.
Pereira MG, Nakayasu ES, Sant’anna C, De Cicco NN, Atella GC, De Souza W, et al. Trypanosoma cruzi epimastigotes are able to store and mobilize high amounts of cholesterol in reservosome lipid inclusions. PLoS One. 2011;8 Suppl 6:e22359.
Souto-Padrón T, Lima VMQ, Roitman I, De Souza W. Fine structure of Leptomonas samueli by freeze-fracture technique. Z Parasitenk. 1980;62:145–57.
Docampo R, Moreno SNJ. Acidocalcisomes. Cell Calcium. 2011;50:113–9.
Martin DS, Desser SS. Development of Trypanosoma fallisi in the leech, Desserobdella picta, in toads (Bufo americanus), and in vitro. A light and electron microscopic study. Parasitol Res. 1991;77:18–26.
Docampo R, De Souza W, Miranda K, Rohloff P, Moreno SNJ. Acidocalcisomes - conserved from bacteria to man. Nature reviews Microbiol. 2005;3:251–61.
Nakamura CV, Ueda-Nakamura T, De Souza W. Visualization of the cytostome in Trypanosoma cruzi by high resolution field emission scanning electron microscopy using secondary and backscattered electron imaging. FEMS Microb Lett. 2005;242:227–30.
De Souza W. Structural organization of Trypanosoma cruzi. Mem Inst Oswaldo Cruz. 2009;104:89–100.
Acknowledgments
We are grateful to Mr. Alcir Pereira de Souza, Ms. Alessandra Simões de Toledo Pereira and Mrs. Tereza da Silva Lemos for their valuable support with the fieldwork, and to Dr. José Carlos Oliveira and Valter M. Azevedo-Santos for fish identification. DNA sequencing and phylogenetic analyses done in USP were supported by grants from CNPq and CAPES to MMGT. ML is a postdoctoral fellow funded by CNPq, BRF is a PhD student funded by CAPES, CSR is PhD student funded by CNPq, and LH is postdoctoral fellow funded by FAPERJ.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ML performed the isolation and culture of trypanosomes, morphological, morphometrical and ultrastructural analyses. BRF was responsible for phylogenetic analyses and participated in DNA sequencing and manuscript preparation. ML, CSR, LH and RS contributed to sequencing and phylogenetic inferences. BRF, CSR, LH, RS and EPC provided critical comments to improve the manuscript. ML, MMTG and TSP wrote the manuscript. TSP is the senior author. All authors read, revised and approved the final version of the manuscript.
Additional files
Additional file 1:
Trypanosome parasitemia in blood removed from organs of Hypostomus affinis and H. luetkeni armored catfish. (XLSX 8 kb)
Additional file 2:
Morphometric analysis of the different in vitro cultured forms of Trypanosoma abeli n. sp. (XLSX 9 kb)
Additional file 3:
Morphometric analysis of bloodstream trypomastigotes. (XLSX 10 kb)
Additional file 4:
Morphometric analysis of the different developmental forms of trypanosomes found in leeches of the species Haementeria brasiliensis . (XLSX 9 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lemos, M., Fermino, B.R., Simas-Rodrigues, C. et al. Phylogenetic and morphological characterization of trypanosomes from Brazilian armoured catfishes and leeches reveal high species diversity, mixed infections and a new fish trypanosome species. Parasites Vectors 8, 573 (2015). https://doi.org/10.1186/s13071-015-1193-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-1193-7