Abstract
Background
Trypanosomiasis is a neglected tropical disease caused by the trypanosome parasite and transmitted by the tsetse fly vector. In Sub-saharan Africa, both the human and animal variants of the disease are a great obstacle towards agriculture, development, and health. In order to better understand and therefore combat Trypanosomiasis, characterizing disease hotspots across species is critical.
Methods
In this study, 193 samples from cattle, sheep, and goats were collected from eight sites. Samples were taken from animals belonging mostly to Maasai herdsmen in the Ngorongoro Crater Conservation Area (NCA) and analysed for the presence of trypanosomiasis infection using PCR techniques. Those that tested positive for T. brucei parasite were further tested using SRA LAMP technique to check for T. brucei rhodesiense, the human infective subspecies of parasite.
Results
Our study found a high incidence of Trypanosoma brucei infections across species. Of animals tested, 47 % of cattle, 91.7 % of sheep, and 60.8 % of goats were infected. Most of the infections were of the T. brucei species. We also identified sheep and goats as carriers of the T. brucei rhodesiense subspecies, which causes acute human trypanosomiasis.
Conclusions
Together, these results point toward the need for stricter control strategies in the area to prevent disease outbreak.
Similar content being viewed by others
Background
Tsetse flies (Glossina) are one of the insect vectors that have most greatly affected human and animal populations in Tanzania, as they transmit the blood-borne flagellate protozoa known as the Trypanosome parasite. The diseases caused by trypanosomes are Human African Trypanosomiasis (HAT or Sleeping Sickness) and Animal African Trypanosomiasis (AAT or Nagana). Trypanosomiasis and tsetse flies remain the greatest obstacles to livestock development in Africa [1]. The wide occurrence of trypanosomiasis in areas of tsetse habitation [2] has effectively prevented the establishment of sustainable development of viable agricultural systems in many areas of great agricultural potential [3].
In Tanzania, tsetse flies infest approximately 33 % of the 940,027 sq km of the mainland and have been a limitation to the country’s development [4, 5]. These parasites cause sub-Saharan Africa about $4.75 billion USD of loss in agricultural potential due to livestock losses to AAT. It is estimated that productivity in livestock is heavily reduced by this disease: 70 % reduction of cattle density, 50 % in sale of meat and milk, 20 % in calving rates, and increases in calf mortality by 20 % [1].
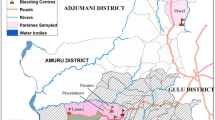
Due to recent changes in habitat because of deforestation, climate change, and loss of native blood meal species, domestic animals, particularly livestock, are becoming an important source of food for tsetse flies [6]. In areas where there is a high density of human, cattle, and fly cohabitation, the risk of disease transmission across species is high, as it is estimated that sleeping sickness is more likely to be transmitted by a cattle-fly-human transmission cycle than a human-fly-human cycle [7]. In order to prevent large outbreaks of HAT, it is critical to identify reservoirs of HAT within cattle populations and to implement appropriate control strategies, whether at the parasite or vector level [8]. In Tanzania, the Ngorongoro Conservation Area (NCA) is one of the most popular tourist attractions and is bordered by Serengeti National Park, Maswa Game Reserve and Loliondo Game Reserve, all areas which are tsetse infested and at risk of HAT. NCA practices multiple land use whereby wildlife, livestock and people co-exist. The large pastoral Maasai population which lives and herds cattle in this area is therefore at great risk.
The ability to identify different species of trypanosomes has evolved from pure microscope observation [9] and unreliable field diagnostic techniques with low sensitivity to faster and more reliable gene sequencing techniques. These included both species-specific DNA probes [10–12], as well as species-specific Polymerase Chain Reaction (PCR) tests [13–15]. The newest methods use generic primers to sequence highly conserved regions across trypanosome species in between which are regions of varying length. Of these methods, one takes advantage of the interspecies size variation of the PCR product of the ITS-1 ribosomal RNA region [16–18]. The other technique uses fluorescent primers to detect length variations in multiple fragments in 18S and 28S ribosomal RNA regions [19, 20]. This allows for quick and specific parasite recognition without the use of various PCR tests, as was the case with the species-specific primers. It also allows for the quick detection of multiple infections within the same host. Using this last technique, our laboratory has previously detected a new species of the subgenus Trypanozoon [20]. In this study, samples from the NCA were analysed for trypanosome infection and further tested for presence of human infective T. brucei rhodesiense.
Methods
Ethical approval
Approval for this research project was granted by the Tanzania Commission for Science and Technology (COSTECH).
Data collection
Samples were collected twice, once in November 2013 and again in February 2014, both times before the onset of the rainy season from eight sites (Table 1; Fig. 1). Briefly, herdsmen were alerted one day earlier. We requested for their consent to sample their animals. Because most pastoralists in the area are familiar with the disease, they easily agreed to have their animals sampled. A small puncture was done on the peripheral ear vein and blood collected in duplicate heparinised capillary tubes for further processing back in the field station. Most samples were taken in the morning, before cattle were taken out to graze.
Sample storage
Capillary tubes were spun centrifuged at 12,000 rpm for about 5 min in Hawksley® centrifuge machine (Hawksley Ltd, Sussex, UK). Once proper separation of blood had taken place, the buffy coat only was spotted onto Whatman FTA cards for sample storage. Previous studies have shown this to be the most convenient and efficient method for collecting parasite DNA [21–25]. All samples were then transported to the lab and stored until all samples had been collected.
DNA extraction
DNA extraction from Whatman FTA cards and filter paper have been previously described [21, 22]. Briefly, 10 punches of 0.1 mm were taken from each card and pooled. DNA was then eluted using Chelex®100 20 % solution at 55 °C for 60 min and then briefly vortexed before boiling for 10 min.
qPCR and LAMP
DNA was then tested for the presence of Trypanosome DNA. Primers for the conserved ITS region (purchased from Bionner ISO 9001:2000) were used as previously described [17, 26]. PCR reactions were carried out using Phusion PCR kit (Thermo Fischer F-530S).
Those samples that tested positive for T.brucei were tested using the SRA LAMP technique as previously described [27] to determine whether they have the human resistance associated (SRA) gene uniquely expressed by T. b. rhodesiense (SRA-LAMP) [28, 29]. In this technique samples were considered positive if they turned green when Sybergreen was added. Those which never changed colour were assumed to contain samples that were SRA negative and were thus considered to contain T. brucei only. In this technique distilled water was used as a negative control and T. b. rhodesiense DNA, kindly donated by Dr. Enock Matovu (Makerere University, Kampala), was used as a positive control.
Statistical analysis
95 % confidence intervals for infection prevalence within populations sampled were calculated using the Wilson method for calculating 95 % confidence intervals of binomial populations [30]. Briefly, the populations sampled were assumed to follow a binomial distribution in which two outcomes were possible: either the samples tested positive (success) or negative (failure). The Wilson method was then applied: the total number of samples within a given subpopulation represented the total number of trials and the number of positive samples represented the number of successes. For example, in Table 1, the site of Kakesio was considered to be an independent set of 40 trials (n = 40) with 9 successes (x = 9). In Table 2, as another example, the adult bovine population was considered an independent set of 70 trials (n = 70) with 38 successes (x = 38). Note that for these calculations, each subpopulation was treated as an independent set of trials. Because of this, the “tightest” confidence intervals come from those calculated for all species in the bovine, ovine, or caprine populations, and these represent the prevalence of infections in those species throughout the area. All calculations were carried out using R statistics software with the “binom” package.
Results
As can be seen in Tables 1 and 2, the majority of the samples were taken from Bovine specimens. This can be attributed to the fact that most of the herdsmen visited owned more cattle than goats and sheep, but were also more readily available and willing for us to sample their cows.
Of those samples testing positive for parasite DNA, the majority, 85 out of 108, were infected by a single trypanosome, T. brucei (Table 3, Fig. 2). Double infections were less rare (14). They were strictly limited to bovine hosts and all but one were T. brucei and T. congolense. Only 9 samples were single infections due to parasites other than T. brucei. For the T. brucei infections tested for T. brucei rhodesiense, only 5 out of the 85 tested positively (Table 3).
Conclusion
This is the first study that reports on the role of ovine and caprine species in the transmission of T.brucei rhodesiense in Tanzania. While both cattle and more recently, pigs have been reported as potential reservoirs of acute human sleeping sickness [31, 32] this is the first study to address this in small ruminants. These findings, along with our discovery of a high incidence of animals infected with trypanosomiasis in the area, call for a shift in the care of livestock among the Maasai herdsmen. Normally, cattle are given the required attention and treated accordingly once animals become symptomatic. Small ruminants, even when symptomatic, are not given the required treatments. In areas of tsetse infestation, vector control and treatment of diseased animals should be common practice. For the prevention of outbreaks of the human version of the disease, these should be coupled with periodic screening of patients in areas where humans, domestic animals, wild animals, and tsetse flies cohabitate.
References
Swallow BM: Impacts of trypanosomiasis on African agriculture. Rome: FAO; 1999.
Steverding D: The history of African trypanosomiasis. Parasites and Vectors 2008;1(3):1–8.
Ilemobade AA. Tsetse and trypanosomosis in africa: the challenges, the opportunities. Onderstepoort J Vet Res. 2009;76:35–40.
Daffa J, Byamungu M, Nsengwa G, Mwambembe E, Mleche W. Tsetse distribution in Tanzania: 2012 status. Tanzania Veterinary Journal. 2013;28(Special Issue):1–11.
Malele I, Nyingilili H, Msangi A. Factors defining the distribution limit of tsetse infestation and the implication for livestock sector in Tanzania. Afr J Agric Res. 2011;6(10):2341–7.
Van den Bossche P, Stephane dLR, Hendrickx G, Bouyer J. A changing environment and the epidemiology of tsetse-transmitted livestock trypanosomiasis. Trends Parasitol. 2010;26(5):236–42.
Hide G. History of Sleeping Sickness in East Africa. Clin Microbiol Rev. 1999;12(1):112–25.
Rogers DJ, Hendrickx G, Slingenbergh JHW. Tsetse flies and their control. Rev sci tech Off int Epiz. 1994;13(4):1075–124.
Lloyd L, Johnson W. The Trypanosome infections of the tsetse flies in Northern Nigeria and a new method of estimation. Bull Entomol Res. 1924;14:265–88.
Gibson WC. Species-specific DNA probes for the identification of trypanosomes in tsetse. Parasitology. 1988;97:63–73.
Kukla B, Majiwa P, Young C. Use of species-specific DNA probes for the detection and identification of trypanosome infections in tsetse flies. Parasitology. 1987;95:1–26.
Majiwa PAO, Otieno LH. Recombinant DNA probes reveal simultaneous infection of tsetse flies with different trypanosome species. Mol Biochem Parasitol. 1990;40:245–54.
Majiwa P, Thatthi R, Moloo S. al. e: Detection of trypanosome infections in the saliva of tsetse flies and buffy coat samples from antigenaemic but aparitaemic cattle. Parasitology. 1994;108:313–22.
Masiga DK, Smyth AJ, Hayes P, Bromidgge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol. 1992;7:909–18.
DR M, GA C, DE O, Al E. Detection of Trypanosoma congolense and T. brucei subspecies by DNA amplification using the polymerase chain reaction. Parasitology. 1989;99:57–66.
Adams E, Malele I, Msangi A, Gibson W. Trypanosome Identification in wild tsetse populations in Tanzania using generic primers to amplify the ribosomal RNA ITS-1 Region. Acta Trop. 2006;100:103–9.
Malele I, Craske L, Knight C, Ferris V, Njiru Z, Hamilton P, et al. The use of specific and generic primers to identify trypanosome infections of wild tsetse flies in Tanzania by PCR. Infect Genet Evol. 2003;3:271–9.
Cox A, Tilley A, McOdimba F, Fyfe J, Eisler M, Hide G, et al. A PCR based assay for detection and differentiation of African trypanosome species in blood. Exp Parasitol. 2005;111:24–9.
Adams ER, Hamilton PB, Malele II, Gibson WC. The identification, diversity and prevalence of trypanosomes in field caught tsetse in Tanzania using ITS-1 primers and fluorescent fragment length barcoding. Infect Genet Evol. 2008;8:439–44.
Hamilton P, Adams E, Malele I, Gibson W. A novel, high-throughput technique for species identification reveals a new species of tsetste-transmitted trypanosome related to the Trypanosoma brucei subgenus, Trypanozoon. Infect Genet Evol. 2008;8:26–33.
Cox A, Tosas O, Tilley A, Picozzi K, Coleman P, Hide G, Welburn S: Constraints to estimating the prevalence of trypansome infections in East African zebu cattle. Parasites and Vectors 2010;3(1):82.
Katakura K, Lubinga C, Chitambo H, Tada Y. Detection of Trypanosoma congolense and T. brucei subspecies in cattle in Zambia by polymerase chain reaction from blood collected on a filter paper. Parasitol Res. 1997;83:241–5.
Ahmed HA, MacLeod ET, Hide G, Welburn SC, Picozzi K: The best practice for preparation of samples from FTA cards for diagnosis of blood borne infections using African trypanosomes as a model system. Parasites and Vectors 2011;4(68):1–7.
Reifenberg J-M, Solano P, Duvallet G, Cuisance D, Simpore J, Cuny G. Molecular characterization of trypanosome isolates from naturally infected domestic animals in Burkina Faso. Vet Parasitol. 1997;71:251–62.
Solano P, Michel JF, Lefrancois T, De la Rocque S, Sidibe I, Zoungrana A, et al. Polymerase chain reaction as a diagnosis tool for detecting trypanosomes in naturally infected cattle in Burkina Faso. Vet Parasitol. 1999;86:95–103.
Malele II, Magwisha HB, Nyingilili HS, Mamiro KA, Rukambile EJ, Daffa JW, Lyaruu EA, Kapange LA, Kasilagila G, Lwitiko N et al.: Multiple Trypanosoma infections are common amongst Glossina species in the new farming areas of Rufiji district, Tanzania. Parasites and Vectors 2011;4:217.
Njiru ZK, Mikosza ASJ, Armstrong T, Enyaru JC, Ndung’u JM, Thompson ARC. Loop-Mediated Isothermal Amplification (LAMP) Method for Rapid Detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis. 2008;2(2):e147.
Namangala B, Hachaambwa L, Kajino K, Mweene AS, Hayashida K, Simuunza M, et al. The use of Loop-mediated Isothermal Amplification (LAMP) to detect the re-emerging Human African Trypanosomiasis (HAT) in the Luangwa and Zambezi Valleys. Parasites and Vectors. 2012;5:282.
Namangala B, Juma Haji I, Sugimoto C, Kajino K, Malele I, Simukoko H, Chitambo H: Determination of the prevalence of trypanosome species in cattle from Monduli district, northern Tanzania, by loop mediated isothermal amplification. Tropical Health and Animal Production 2015;1–5
Brown LD, Cai TT, DasGupta A. Interval estimation for a binomial proportion (with discussion). Stat Sci. 2001;16:101–33.
Hamill LC, Kaare MT, Welburn SC, Picozzi K: Domestic pigs as potential reservoirs of human and animal trypanosomiasis in Northern Tanzania. Parasites and Vectors 2013;6(1):322.
Kaare MT, Picozzi K, Mlengeva T, Fevre EM, Mellau LS, Mtambo M, et al. Sleeping sickness- A re-emerging disease in the Serengeti? Travel Med Infect Dis. 2007;5(2):117–24.
Acknowledgments
The opportunity to work in Tanzania in conjunction with the Vector and Vector Borne Diseases Research Institute, formerly Tsetse and Trypanosomiasis Research Institute (TTRI), would not have been possible without the aid of a Fulbright Student Research Grant, from which this study is funded. The authors also thank NCAA for logistic and funding support during sample collections and Dr. Enock Matovu (Makerere University, Kampala) for his kind donation of T.b.rhodesiense DNA to be used as a positive control at the Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiment: JPR, HSN, GHM, and IIM. Performed the experiment: JPR, HSN, and GHM. Analysed the data: JPR, HSN, GHM, and IIM. Wrote the paper: JPR, HSN, GHM, and IIM. All authors read and approved the final version of the MS.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ruiz, J.P., Nyingilili, H.S., Mbata, G.H. et al. The role of domestic animals in the epidemiology of human african trypanosomiasis in Ngorongoro conservation area, Tanzania. Parasites Vectors 8, 510 (2015). https://doi.org/10.1186/s13071-015-1125-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-1125-6