Abstract
Background
Amblyomma ticks parasitize a wide range of animals in tropical regions. This study describes the identification of Amblyomma ticks from wild snakes in Malaysia and the detection of potential human pathogens such as Rickettsia, Anaplasma, Ehrlichia and bartonellae in the ticks.
Findings
Twenty one adult ticks (twelve A. varanense and nine Amblyomma helvolum ticks) identified from seven Python molurus snakes in Sepang and a pool of six A. helvolum ticks from a Naja sumatrana snake in Johore, Malaysia were investigated in this study. Amplification of the citrate synthase (gltA), 190-kDa surface antigen gene (ompA), 135-kDa surface antigen (ompB) and surface cell antigen (sca4) genes followed by sequence analysis confirmed the presence of two potential novel spotted fever group rickettsiae in the ticks. Candidatus Rickettsia sepangensis from an engorged A. varanense tick demonstrated high sequence similarity to Rickettsia tamurae; while Candidatus Rickettsia johorensis from two samples (individual and pooled) of A. helvolum and two A. varanense ticks were closely related to Rickettsia raoultii. Anaplasma and Ehrlichia DNA were detected from seven and two ticks, respectively. No bartonellae was detected from any of the ticks.
Conclusion
The finding in this study suggests that Amblyomma ticks parasitizing wild snakes may serve as reservoir hosts and carriers for rickettsioses, anaplasmosis and ehrlichiosis in this region.
Similar content being viewed by others
Background
Ticks are the vector for numerous emerging zoonotic diseases which can be severe and life-threatening to humans. In nature, ticks and a wide range of animals may act as reservoirs or amplifiers for human pathogens such as spotted fever group rickettsiae, anaplasma, ehrlichiae and bartonellae. Humans can be accidentally infected with these organisms through tick bites. The ticks belonging to the genus Amblyomma have been implicated as a carrier for several pathogenic rickettsiae including Rickettsia rickettsii, R. aeschlimannii, R. raoultii, and R. tamurae [1], Anaplasma phagocytophilum, Ehrlichia chaffeensis and E. ewingii [2,3]. Additionally, Bartonella DNA has also been detected in A. americanum ticks [4].
Amblyomma ticks parasitize a wide range of animals and are often seen on mammalian hosts, reptiles and amphibians [5,6]. However, information is lacking on tick carriage of emerging human pathogens in the tropical region. In this study, we assessed the occurrence of these microorganisms in Amblyomma ticks parasitizing wild snakes in Malaysia by using molecular approach.
Methods
Twenty-one adult ticks (12 A. varanense and nine A. helvolum) from seven Python molurus snakes from Sepang (2°49′10.862″N, 101°44′1.262″E) and a pool of six A. helvolum ticks from a Spitting cobra (Naja sumatrana) in Johore, Malaysia (1°43′58.321″N, 103°54′5.082″E) collected from August-October 2012 were investigated in this study. The ticks were identified based on the taxonomic keys of Burridge [5] and Kohls [7].
Tick DNA was extracted using QIAamp DNA mini kit (Qiagen, Hilden, Germany) in accordance to the manufacturer’s instruction. Four rickettsial-specific genes were targeted for amplification from the tick samples, i.e., citrate synthase gene (gltA), 190-kDa outer membrane protein gene (ompA), 135-kDa outer membrane protein gene (ompB) and surface cell antigen (sca4) [8-11]. Identification of Anaplasma and Ehrlichia DNA in the samples was performed using a PCR assay targeting 16S rRNA gene of the organisms [12] followed by sequence analysis. For further differentiation of Anaplasma spp., amplification of the full length sequences of 16S rDNA and msp4 genes were performed [13]. A PCR assay targeting citrate synthase (gltA) gene was performed for detection of bartonellae DNA [14]. Cloned PCR2.1-TOPO T/A plasmids (Invitrogen, USA) with amplified gltA fragment from R. honei (strain TT118), ompA and ompB fragments from rickettsial endosymbionts (98% similarity to R. heilongjiangensis and R. raoultii, respectively) of tick samples were used as positive controls. BLAST analysis was performed to search for homologous sequences in the GenBank database. To determine the phylogenetic position of the rickettsiae identified in this study, dendrogram was constructed based on concatenated sequences of gltA (1040–1046 nucleotides) and ompA (407–431 nucleotides) genes using neighbour-joining method of MEGA software [15].
Findings
Table 1 shows the amplification of rickettsial gltA gene from three A. varanense (S5, S4-2 and S7-2) and two A. helvolum tick samples (S6-1, P1). The gltA and ompA sequences from the S5 tick was almost similar (99.0% and 97.7%, respectively) with R. tamurae strain AT-1 from A. testudinarium tick in Japan [16]. However, the ompB gene of the rickettsia was unable to be amplified and no significant similarity was obtained for the amplified sca4 fragment.
BLAST analysis of the rickettsial gltA sequence from two samples (individual and pooled) of A. helvolum (S6-1, P1) and two A. varanense (S4-2 and S7-2) ticks demonstrated the closest match (99.7%) to R. raoultii strain Khabarovsk (Table 1), which was cultivated from Dermacentor ticks in Russia and France [17]. The sequence similarity of the ompA, ompB and sca4 sequences of these ticks with those of R. raoultii strain Khabarovsk was 97.4%, 98.3% and 97.4%, respectively.
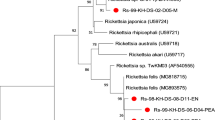
According to the current criteria for speciation of rickettsial species, uncultured rickettsia exhibiting sequence similarity of ≤99.9% for gltA, ≤ 98.8% for ompA, ≤99.2% ompB and ≤99.3% for sca4 genes with a validated Rickettsia species may be given Candidatus status [18]. Hence, the rickettsiae are thus named as Candidatus Rickettsia sepangensis and Candidatus Rickettsia johorensis, respectively, in accordance to the location of their first sample collection. The dendrogram constructed using concatenated sequence of gltA and ompA gene fragments (Table 2 and Figure 1) confirmed the clustering of Candidatus Rickettsia sepangensis with the type strain of R. tamurae, and Candidatus Rickettsia johorensis with R. raoultii type strains.
Phylogenetic placement of concatenated sequences ( gltA and ompA ) of known rickettsial species in Table 2 . Bootstraps analysis was performed with 1000 replications. Numbers in brackets are GenBank accession numbers. Scale bar indicates the nucleotide substitutions per sites.
Several spotted fever group rickettsiae with unknown or potentially pathogenicity for humans have been reported in the Southeast Asia region, mainly in Thailand. R. honei (strain TT-118) and R. thailandii sp. nov. have been identified from Ixodes and Rhipicephalus ticks [19,20]. Closely related species of R. raoultii have also been detected from A. helvolum from a lizard (Varanus salvator) in Thailand [21]. Exposure to infected snake ticks may pose risks to human health as R. tamurae and R. raoultii have been implicated in human infections [22,23]. High antibody prevalence to R. honei (TT118 strain) has been reported in febrile patients in rural areas in Malaysia [24]. However, information on the type of spotted fever group rickettsiae is still lacking.
Anaplasma DNA was amplified from seven ticks (Table 1). Based on the 256 nucleotides of the amplified 16S rDNA partial gene fragments, sequences from three A. varanense and two A. helvolum ticks showed the closest similarity to those of A. phagocytophilum [Genbank accession no.: AY551442, 99%, 253/256] or A. platys [Genbank accession no.: JX261979, 99%, 253/256]. A. bovis DNA [Genbank accession no.:AB983438, 99%, 253/256] was amplified from two A. varanense ticks, whereas DNA of Ehrlichia spp. [Genbank accession no.: KJ410257, 99%, 249/256] was amplified from two A. varanense ticks. Attempts to determine the full length sequence of 16S rRNA and msp4 genes were not successful as the sequences obtained were not satisfactory for analysis. No bartonellae was detected from any of the ticks understudied.
There is no report on the human infections caused by tickborne pathogens with reptile as a host in Southeast Asia. The presence of SFG rickettsiae (Rickettsia species closely related to R. raoultii, R. tamurae and R. bellii) has been recently shown in A. varanense and A. helvolum in Thailand [25]. Detection of R. honei in a reptilian tick, Bothriocroton hydrosauri (formerly Aponomma hydrosauri) has been reported in Australia [26]. Rickettsia spp. closely related to R. tamurae has also been detected in A. fimbriatum ticks collected from reptiles (yellow-spotted monitor, water python and green-tree snake) in the Northern Territory of Australia [27], and A. exornatum tick from a lizard (Varanus olivaceus) in United States of America [28]. In the South America, Rickettsia sp. strain Colombianensi has been identified from A. dissimile ticks parasitizing iguanas in Colombia [29]. All these findings suggest the existing of a natural cycle of spotted fever group rickettsial infection in ticks and snakes in different geographical regions. A. phagocytophilum has been detected in A. flavomaculatum tick collected from a Varanus exanthematicus lizard imported into Poland [30]. Meanwhile, the detection of Ehrlichia spp. from ticks collected from snakes has not been reported previously and thus, merits further investigation.
A. helvolum ticks have been identified from different snakes including Python sp., Ptyas (Zamensis) korros and Naja naja (Kohls) [7] in Malaysia. A. varanense is also one of the most widespread Amblyomma ticks in large snakes in Southeast Asia [5]. As P. molurus and N. sumatrana snakes are native to Southeast Asia [31,32], ticks parasitizing the snakes could be endemic where the animal hosts are available. Although there is no data about the affinity of the ticks to bite humans yet, the detection of rickettsial agents in the snake ticks poses a risk to both wildlife and human. Further work is required to assess the prevalence of these potential tick-borne pathogens on a larger scale.
Conclusions
This study presented the molecular evidence of the presence of potential novel spotted fever group rickettsiae closely related to R. tamurae and R. raoultii, Anaplasma and Ehrlichia spp. in two species of Amblyomma ticks parasitizing P. molurus and N. sumatrana snakes. The finding in this study suggests the potential role of Amblyomma ticks as a reservoir host and carrier for rickettsioses, anaplasmosis and ehrlichiosis in this region.
References
Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702.
Bekker CP, De Vos S, Taoufik SA, Sparagano OA, Jongejan F. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridization. Vet Microbiol. 2002;89:223–38.
Cohen SB, Yabsley MJ, Freye JD, Dunlap BG, Rowland ME, Huang J, et al. Prevalence of Ehrlichia chaffeensis and Ehrlichia ewingii in ticks from Tennessee. Vector Borne Zoonotic Dis. 2010;10:435–40.
Billeter SA, Miller MK, Breitschwerd EB, Levy MG. Detection of two Bartonella tamiae-like sequences in Amblyomma americanum (Acari: Ixodidae) using 16S-23S intergenic spacer region-specific primers. J Med Entomol. 2008;45:176–9.
Burridge MJ. Ticks (Acari: Ixodidae) spread by the international trade in reptiles and their potential roles in dissemination of diseases. Bull Entomol Res. 2001;91:3–23.
Pandit P, Bandivdekar R, Geevarghese G, Pande S, Mandke O. Tick infestation on wild snakes in northern part of Western Ghats of India. J Med Entomol. 2011;48:504–7.
Kohls GM. Malaysian parasites XVII ticks (Ixodoidea) of Borneo and Malaya. Stud Inst Med Res Malaya. 1957;28:65–94.
Labruna MB, Whitworth T, Horta MC, Bouyer DH, McBride JW, Pinter A, et al. Rickettsia species infecting Amblyomma cooperi ticks from an area in the state of Sao Paulo, Brazil, where Brazilian spotted fever is endemic. J Clin Microbiol. 2004;42:90–8.
Regnery RL, Spruill CL, Plikaytis BD. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. 1991;173:1576–89.
Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmp (ompB). Int J Syst Evol Microbiol. 2000;50:1449–55.
Sekeyova Z, Roux V, Raoult D. Phylogeny of Rickettsia spp. inferred by comparing sequences of ‘gene D’, which encodes an intracytoplasmic protein. Int J Syst Evol Microbiol. 2001;51:1353–60.
Parola F, Roux V, Camicas JL, Baradji I, Brouqui P, Raoult D. Detection of ehrlichiae in African ticks by polymerase chain reaction. Trans R Soc Trop Med Hyg. 2000;94:707–8.
Bown KJ, Lambin X, Ogden NH, Petrovec M, Shaw SE, Woldehiwet Z, et al. High-resolution genetic fingerprinting of European strains of Anaplasma phagocytophilum by use of multilocus variable-number tandem-repeat analysis. J Clin Microbiol. 2007;45:1771–6.
Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–803.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Fournier PE, Takada N, Fujita H, Raoult D. Rickettsia tamurae sp nov, isolated from Amblyomma testudinarium ticks. Int J Syst Evol Microbiol. 2006;56:1673–5.
Mediannikov O, Matsumoto K, Samoylenko I, Drancourt M, Roux V, Rydkina E, et al. Rickettsia raoultii sp. nov., a spotted fever group rickettsia associated with Dermacentor ticks in Europe and Russia. Int J Syst Evol Microbiol. 2008;58:1635–9.
Raoult D, Fournier PE, Eremeeva M, Graves S, Kelly PJ, Oteo JA, et al. Naming of Rickettsiae and Rickettsial diseases. Ann N Y Acad Sci. 2005;1063:1–12.
Graves S, Stenos J. Rickettsia honei: a spotted fever group Rickettsia on three continents. Ann N Y Acad Sci. 2003;990:62–6.
Kollars TMJ, Tippayachai B, Bodhidatta DL. Short report: Thai tick typhus, Rickettsia honei, and a unique Rickettsia detected in Ixodes granulates (Ixodidae: Acari) from Thailand. Am J Trop Med Hyg. 2001;65:535–7.
Doornbos K, Sumrandee C, Ruang-Areerate T, Baimai V, Trinachartvanit W, Ahantarig A. Rickettsia sp. closely related to Rickettsia raoultii (Rickettsiales: Rickettsiaceae) in an Amblyomma helvolum (Acarina: Ixodidae) tick from a Varanussalvator (Squamata: Varanidae) in Thailand. J Med Entomol. 2013;50:217–20.
Imaoka K, Kaneko S, Tabara K, Kusatake K, Morita E. The first human case of Rickettsia tamurae infection in Japan. Case Rep Dermatol. 2011;3:68–73.
Phongmany S, Rolain JM, Phetsouvanh R, Blacksell SD, Soukkhaseum V, Rasachack B, et al. Rickettsial infections and fever, Vientiane, Laos. Emerg Infect Dis. 2006;12:256–62.
Tay ST, Ho TM, Rohani MY, Shamala D. Antibody prevalence of Orientia tsutsugamushi, Rickettsia typhi and TT118 spotted fever group rickettsiae among febrile patients in rural areas of Malaysia. Trans R Soc Trop Med Hyg. 2000;94:280–4.
Sumrandee C, Hirunkanokpun S, Doornbos K, Kitthawee S, Baimai V, Grubhoffer L, et al. Molecular detection of Rickettsia species in Amblyomma ticks collected from snakes in Thailand. Ticks Tick Borne Dis. 2014;5:632–40.
Stenos J, Graves S, Vsevolod LP, Walker D. Aponomma hydrosauri, the reptile-associated tick reservoir of Rickettsia honei on Flinders Island, Australia. Am J Trop Med Hyg. 2003;69:314–7.
Vilcins IME, Fournier PE, Old JM, Deane E. Evidence for the presence of Francisella and Spotted Fever Group Rickettsia DNA in the tick Amblyomma fimbriatum (Acari:Ixodidae), Nothern Territory, Australia. J Med Entomol. 2009;46:926–33.
Reeves WK, Durden LA, Darch GA. A spotted fever group Rickettsia from an exotic tick species, Amblyomma exornatum (Acari: Ixodidae), in a reptile breeding facility in the United States. J Med Entomol. 2006;43:1099–101.
Miranda J, Portillo A, Oteo JA, Mattar S. Rickettsia sp. Strain colombianensi (Rickettsiales: Rickettsiaceae): a new proposed Rickettsia detected in Amblyomma dissimile (Acari: Ixodidae) from iguanas and free-living larvae ticks from vegetation. J Med Entomol. 2012;49:960–5.
Nowak M, Cieniuch S, Stanczak J, Siuda K. Detection of Anaplasma phagocytophilum in Amblyomma flavomaculatum ticks (Acari: Ixodidae) collected from lizard Varanus exanthematicus imported to Poland. Exp Appl Acarol. 2010;51:363–71.
Wuster W. Taxonomic changes and toxinology: systematic revisions of the Asiatic cobras (Naja naja species complex). Toxicon. 1996;34:399–406.
Barker DG, Barker TM. The distribution of the Burmese Python, Python bivittatus, in China. Bull Chicago Herp Soc. 2010;45:86–8.
Acknowledgments
The authors would like to thank Mr. Chai Koh Shin, Mr. Saidon, Mdm Asha Devi Amarajothi and the research group from TIDREC, UM who provided assistance in this project. This project was funded by High Impact Research-MOHE Grant [E000013-20001 (subprogramme-4)], University Malaya Research Grant (RP013-2012A) and Postgraduate Research Fund (PG006-2013B) from University of Malaya, Kuala Lumpur, Malaysia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KKL carried out the identification of ticks, PCR for detection of rickettsiae and bartonellae, data analysis and wrote the manuscript. KFX performed PCR for detection of ehrlichiae and anaplasma and data analysis. TST initiated and designed the study, supervised the laboratory work, data analysis and wrote the manuscript. All authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kho, K.L., Koh, F.X. & Tay, S.T. Molecular evidence of potential novel spotted fever group rickettsiae, Anaplasma and Ehrlichia species in Amblyomma ticks parasitizing wild snakes. Parasites Vectors 8, 112 (2015). https://doi.org/10.1186/s13071-015-0719-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-015-0719-3