Abstract
Single-cell protein (SCP) production in the methylotrophic yeast Pichia pastoris has the potential to achieve a sustainable protein supply. However, improving the methanol fermentation efficiency and reducing carbon loss has been a long-standing challenge with far-reaching scientific and practical implications. Here, comparative transcriptomics revealed that PAS_0305, a gene directly associated with cell wall thickness under methanol stress, can be used as a target for unlocking cell wall sensors. Intracellular trehalose accumulation confirmed that cell wall sensors were activated after knocking out PAS_0305, which resulted in increased cell wall permeability. Genome-wide signal perturbations were transduced through the HOG module and the CWI pathway, which was confirmed to connected by Pbs2-Mkk. As a consequence of CWI pathway activation, ΔPAS_0305 elicited a rescue response of cell wall remodeling by increasing the β-1,3-glucan content and decreasing the chitin/mannose content. Remarkably, perturbations in global stress signals led to a fine-tuning of the metabolic network of ΔPAS_0305, resulting in a superior phenotype with highest crude protein and methanol conversion rate of 67.21% and 0.46 gDCW/g. Further genome-scale metabolic models were constructed to validate the experimental results, confirming that unlocking cell wall sensors resulted in maximized flux from methanol towards SCP and effectively addressing the issue of carbon loss in methanol fermentation. This work sheds new light on the potential of manipulating cellular signaling pathways to optimize metabolic networks and achieve exceptional phenotypic characteristics, providing new strategies for constructing versatile cell factories in P. pastoris.
Similar content being viewed by others
Introduction
The growing world population along with a shift toward diets rich in animal protein has led to an increased consumption of animal products and an ever-increasing pressure on the food supply. Currently, the world consumes approximately 1.25 billion tons of meat and dairy products per year, which falls short of the projected global demand for animal-derived protein by 2050 [1]. While plant-based protein sources like beans are nutritionally valuable, they also require arable land and water, resources that are becoming increasingly scarce as the demand for protein grows [2]. However, the industrialization of alternative protein sources is predicated on new cost-effective avenues, ensuring both economic and environmental sustainability. One such avenue is the production of single-cell proteins (SCP) using yeast and algal cells. This approach holds promise for resolving issues of both food and feed supplies. SCP can be produced on a commercial scale, offering a viable alternative to traditional protein sources.
SCP production can utilize a variety of non-food raw materials, thereby reducing costs and enhancing sustainability. In particular, methanol has gained attention as a non-food one-carbon (C1) feedstock and a promising fermentation substrate due to its abundance, low cost, and high reduction potential [3]. Considerable breakthroughs were achieved in methanol synthesis via CO2 hydrogenation [4], opening an avenue for methanol to become a renewable raw material in the future. As the highly flexible methanol supply is derived from both fossil and renewable resources, methanol could serve as an alternative to sugars as a raw material for bio-based manufacturing.
Pichia pastoris, a representative methylotrophic eukaryote, is capable of using methanol as the sole carbon source for the synthesis of SCP and other products [5]. However, a mass balance analysis by Vanz et al. indicated that up to 80% of the methanol is converted into CO2 during the methanol fed-batch fermentation of P. pastoris [6]. Thus reduction of carbon loss and enhancement of carbon metabolism is essential for further improving the conversion of methanol into SCP. Some efforts have been made to regulate methanol metabolism, including peroxisomal compartmentalization of the methanol utilization pathway [7], balancing assimilation and dissimilation pathways to enhance methanol metabolism [8], as well as utilizing synthetic biology and genetic engineering to improve the methanol fermentation efficiency [9, 10]. However, P. pastoris is adapted to methanol utilization under ecological conditions, and uses a complex network to control methanol metabolism [11, 12]. Currently, there is limited knowledge on genetic mechanisms that can be used to generate desirable phenotypes via global perturbations in P. pastoris, beyond adaptive laboratory evolution [13]. In spite of significant engineering efforts, it remains challenging to improve the performance of P. pastoris by only altering one or several metabolic pathways. Therefore, although P. pastoris has been used to produce SCP from methanol on a laboratory scale for decades, improving the methanol conversion efficiency and reducing carbon loss remains a long-standing challenge [14, 15]. Therefore, there is a pressing need to explore novel strategies that comprehensively target and optimize the intricate network of methanol metabolism in P. pastoris.
Here, activation of cell wall sensors was used as an effective breeding strategy for improving the methylotrophic performance of P. pastoris. Genome-scale signal perturbations were triggered by activating the cell wall sensors, resulting in maximal flux distribution from methanol towards biomass in P. pastoris. Extensive investigations have successfully unraveled the intricate underlying mechanism responsible for generating superior phenotypes in P. pastoris following activation of cell wall sensors. The global metabolism of P. pastoris was successfully reprogrammed for improved robustness during SCP overproduction, providing a new strategy for constructing versatile cell factories in P. pastoris.
Results
Screening of key target genes related to cell wall sensors by RNA-Seq
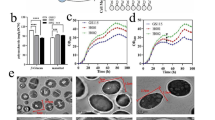
In this study, we found that the cell wall thickness of P. pastoris X33 increased with increasing methanol concentration (Fig. 1a, b). For example, the cell wall thickness of cells cultured in Delft basic salt medium containing 3% methanol was 22.8% greater than in the same medium with 0.5% methanol. To identify genes involved in cell wall synthesis in response to changes of methanol stress, RNA-Seq was conducted to compare gene expression changes between the cells cultured in media with different concentrations of methanol (Fig. 1c). The RNA-Seq data showed that 181 genes were significantly differentially expressed when the methanol concentration was increased from 0.5 to 3%, 115 of which were upregulated and 66 were downregulated. However, among these 181 differentially expressed genes, only 2 genes were related to cell wall synthesis, namely PAS_chr4_0305 (PAS_0305) encoding an O-glycosylated protein required for cell wall stability, and PAS_chr2-1_0454 (PAS_0454) encoding a major exo-1,3-beta-glucanase of the cell wall, involved in cell wall beta-glucan assembly (Additional file 1: Extended Data Table S1). The expression of PAS_0305 and PAS_0454 was upregulated by an average of 2.8- and 2-fold, respectively, with relative correlation with thickening of the cell wall in response to elevated methanol. Conversely, we hypothesized that deletion of PAS_0305 and PAS_0454 may alter the cell wall structure, thereby affecting the cell wall sensors. Therefore, we knocked out these two genes in wild-type P. pastoris. It is worth noting that there were no viable PAS_0454 mutant transformants on the selective plates, indicating that PAS_0454 is an essential gene for cell wall integrity of P. pastoris. However, the PAS_0305 gene may participate in maintaining P. pastoris cell wall properties, unveiling the potential impact of cell wall sensors.
Transmission electron micrographs of whole cells (a), with a zoomed-in view for measurement of cell wall thickness (b), and average analysis of cell wall thickness (c) of P. pastoris cells grown under 0.5% and 3% methanol. d Hierarchical clustering of significantly differentially expressed genes in P. pastoris X33 under 0.5% and 3% methanol
Knockout of PAS _0305 increased the cell wall permeability of P. pastoris
The cell wall serves as the robust outermost layer of yeast cells, acting as the primary defense mechanism against external stresses and the initial sensor to perceive the surrounding environment [16]. Cell wall permeability plays an important role in the sensing of external stress by P. pastoris. To investigate cell permeability changes after knock out of PAS_0305, we used fluorescence spectroscopy to measure the whole-well accumulation of propidium iodide and Nile red, representative hydrophilic and hydrophobic compounds, respectively. The results showed that the accumulation rate and final levels of the two compounds were higher in the ΔPAS_0305 strain than in the control strain, indicating an increase of cell wall permeability (Fig. 2a, b). This result was in agreement with a previous study, which found that the cell wall permeability of Mycolicibacterium smegmatis increased following alteration of SWU1gp39 [17].
Analysis of changes in P. pastoris cell wall permeability after knocking out ΔPAS-0305. Assessment of whole-cell permeability via a intracellular Nile red fluorescence measurement; b intracellular propidium iodide fluorescence measurement; and c measurement of CMCase activity of heterologously HgCel5A. d Comparison of P. pastoris extracellular protein. d1, SDS-PAGE. d2, extracellular protein concentration analysis
Compared with the fluorescence spectroscopy analysis, measurement of the extracellular levels of a heterologously expressed protein provides a more straightforward and intuitive assessment of cell wall permeability. Hence, identical HgCel5A expression cassette with the PAOX promoter and TGAP terminator was introduced into both the ΔPAS_0305 strain and the P. pastoris X33 strain at the same neutral PNSII-8 site using the Cas9 system. To further characterize HgCel5A secretion under identical growth conditions, fed-batch bioreactor cultivations were carried out. Throughout the entire induction period, both CMCase activity and protein secretion were superior in the ΔPAS_0305::HgCel5A strain compared to P. pastoris X33::HgCel5A. After 120 h, the extracellular protein levels and CMCase activity of ΔPAS_0305::HgCel5A reached a significantly higher level (Fig. 2c). During the fed-batch fermentation process, the extracellular protein concentration and CMCase activities of ΔPAS_0305::HgCel5A were 22.16% and 14.41% higher than those of P. pastoris X33::HgCel5A, at 5.50 vs. 4.28 and 7789 vs. 6808. IU/ml, respectively (Fig. 2c). The SDS-PAGE results further confirmed that knocking out PAS_0305 promoted protein secretion by increasing cell wall permeability (Fig. 2d). This finding was consistent with the results obtained from fluorescence spectroscopy analysis.
Further analysis confirmed that the ΔPAS_0305 strain exhibited significantly higher accumulation of intracellular osmolytes, particularly disaccharides, compared to P. pastoris X33 (Fig. 3a). LC–MSMS analysis further identified the accumulated disaccharide as trehalose (Fig. 3b–d). Previous studies have demonstrated that the expression of genes involved in trehalose metabolism is rapidly upregulated in response to various stressors, such as heat, resulting in trehalose accumulation in Saccharomyces cerevisiae [18, 19]. In comparison to P. pastoris X33, the ΔPAS_0305 strain exhibited a 32.71% increase of intracellular trehalose accumulation. This observation suggests that a cell wall sensor was activated by the increased cell wall permeability of P. pastoris after knockout of PAS_0305, triggering intracellular trehalose accumulation as a response to a perceived environmental stress (Fig. 3).
Enhanced environmental tolerance of P. pastoris after activating cell wall sensors
To investigate the impact of the PAS_0305 gene knockout on cell wall composition, we compared ΔPAS_0305 to its parental strain P. pastoris X33. A slight increase in lipid content was observed in the PAS_0305 strain (7.21%) compared to the control strain (6.50%) (P < 0.005) (Table 1). Notably, the cell wall of the ΔPAS_0305 strain exhibited significant decreases of 63.34 and 65.00% in chitin and mannose content, respectively. Conversely, the β-1,3-glucan content of the ΔPAS_0305 strain was 11.97% higher than that of the control strain(P < 0.005) (Fig. 4a). These findings suggest that the cell wall composition of P. pastoris undergoes remodeling following the disruption of the PAS_0305 gene. This observation is in agreement with previous research showing that overexpression of GPI-modifying cell wall proteins increased the β-1,3-glucan content, which enhances the resistance of P. pastoris X33 [20]. Additionally, the disruption of genes involved in chitin synthesis was found to increase cell wall resistance to CW, as chitin serves as the primary binding target for Congo red (CR) in yeast [21]. Ultimately, the increased β-1,3-glucan content and reduced chitin content in the P. pastoris cell wall contributed to its enhanced resistance to various environmental stresses.
Comparison of the methylotrophic performance and industrial robustness between P. pastoris X33 and ΔPAS-0305. a Analysis of cell wall composition. b Growth with different concentrations of methanol. c Sensitivity of strains to Congo red in YPD medium. d Sensitivity of strains to sodium dodecyl sulfate (SDS) in YPD medium
Based on these findings regarding cell wall composition, we further investigated the relationship between cell wall composition and environmental tolerance by comparing the growth of the ΔPAS_0305 strain and the parental strain P. pastoris X33 on plates containing cell wall-destabilizing reagents Congo red (CR) and sodium dodecyl sulfate (SDS). As shown in Fig. 4c, d, the ΔPAS_0305 strain exhibited faster growth on YPD + CR and YPD + SDS plates compared to P. pastoris X33, indicating that the ΔPAS_0305 strain had greater resistance to cell wall-destabilizing reagents. Thus, disruption of the PAS_0305 gene enhances the tolerance of P. pastoris cell walls to environmental stress.
The removal of the PAS_0305 gene activated the cell wall sensors of P. pastoris, activating adaptations to environmental changes. Remodeling of the cell wall composition and the accumulation of trehalose are both responses to multiple environmental stresses after cell wall sensor activation. This is similar to the evolutionary mechanism of adaptive laboratory evolution, where effective modifications to the cell wall can artificially create new environmental pressures as continuous signals.
Utilizing the superior phenotype of P. pastoris ΔPAS_0305 for SCP overproduction
According to the literature, methanol toxicity is a major challenge for improving the efficiency of methanol fermentation in P. pastoris [14]. Increased methanol tolerance has been shown to be crucial for enhancing methanol utilization in methylotrophs. Due to the enhanced robustness of strain ΔPAS_0305 after activating cell wall sensors, it exhibited significantly faster growth compared to P. pastoris X33 at various methanol concentrations (1.5%, 2%, 2.5%, and 3%). Notably, the strain ΔPAS_0305 achieved up to twofold higher OD600 values at a methanol concentration of 3.5% (Fig. 4b). These results demonstrate that both methanol tolerance and utilization in P. pastoris were enhanced after the PAS_0305 gene was knocked out. To confirm that the growth difference was indeed due to the deletion of the PAS_0305 gene, we constructed a PAS_0305-complementation strain, ΔPAS_0305-cp. The growth of the complemented strain was comparable to that of P. pastoris X33 at different methanol concentrations. At a methanol concentration of 1.5%, ΔPAS_0305 exhibited a higher methanol utilization rate of 0.482 mM/gDW/h, a significant improvement over P. pastoris X33, which had a rate of 0.333 mM/gDW/h. These findings indicate that the absence of the PAS-0305 gene promoted the growth of P. pastoris X33 on methanol. A previous study reported that the deletion of a glycosylphosphatidylinositol (GPI)-anchored protein increased the growth of P. pastoris GS115 when methanol was the sole carbon source [22], but there was no progress in understanding the underlying mechanisms involved in methanol utilization.
The utilization of methanol as a feedstock for the production of SCP in P. pastoris has been extensively studied [5]. One of the key nutritional aspects of SCP, either for human food or animal feed, is its high protein content. Here, high-density fermentation was employed to comprehensively examine the production process and evaluate the resulting SCP produced by P. pastoris. Compared with P. pastoris X33, the dry cell weight and crude protein content of the ΔPAS_0305 strain respectively increased by 18.59% (from 102.2 g to 121.2 g/L) and 18.70% (from 56.62 to 67.21%) (Table 2). In the fermentation process, strain ΔPAS_0305 achieved the highest methanol conversion ratio of 0.46 g DCW/g (Fig. 5a), which corresponds to 92% of the maximum theoretical yield (0.5 g DCW/g). This represents a significant improvement compared to the 0.37 g DCW/g achieved by P. pastoris X33. The absence of the gene PAS_0305 in strain ΔPAS_0305 led to a notable increase in the methanol utilization rate, resulting in enhanced dry cell weight and crude protein content. Conversely, the growth of the PAS-0305-complemented strain, ΔPAS_0305-cp, was similar to that of P. pastoris X33, resulting in comparable dry cell weight, crude protein content, and methanol conversion ratio. The ΔPAS_0305 strain exhibited a 15.78% higher methanol utilization rate than P. pastoris X33. These findings were in line with the results of shake-flask fermentation. The disruption of the PAS_0305 gene clearly contributed to the enhanced growth rate and biomass accumulation of P. pastoris when utilizing methanol. Increased crude protein content leads to improved nutritional value and quality of SCP in P. pastoris. Other yeast-based SCP sources such as Saccharomyces cerevisiae, Yarrowia lipolytica, and Candida tropicalis typically have crude protein contents ranging from 53 to 56% [23, 24], whereas Haematococcus pluvialis has a crude protein content of 64.9% [25]. However, SCP derived from ΔPAS_0305 is a high-quality alternative protein rich in 8 essential amino acids of 20.39%, specially methionine (0.82%), threonine (2.06%) and lysine (2.83%) (Fig. 5b), whereby the latter is deficient in cereals. Thus, P. pastoris demonstrated significantly higher SCP production efficiency compared to traditional protein sources. This was primarily attributed to several key factors, including a shorter production time, greatly reduced land use, and the independence from weather conditions.
Analysis of fed-batch fermentation, nutrient consumption and signal transduction pathways of ΔPAS_0305. a Fed-batch fermentation of strain ΔPAS_0305 in a 5-L bioreactor. b Amino acid content of ΔPAS_0305 biomass. c Sketch of DEGs related to methanol metabolism. Genes in red showed significant upregulation in expression. d Comparison of the transcription levels of genes related to the MAPK signaling pathways
Comparative transcriptomic analysis and validation using genome-scale metabolic models (GSMM)
To further understand the mechanisms underlying the improved performance, we performed a comprehensive whole-transcriptome analysis to identify differentially expressed genes (DEGs) between the P. pastoris X33 strain and the ΔPAS_0305 strain when cultivated in YPD medium. The strains exhibited significant differences in their transcriptomes, especially in pathways related to carbohydrate metabolism. Genes involved in methanol utilization, including both assimilation and dissimilation, were significantly upregulated in response to the rapid growth on methanol after knocking out the PAS_0305 gene. Thus, the results confirmed that upregulation of methanol metabolism played an important role in enhancing the cell growth of ΔPAS_0305 on methanol.
GSMMs can be used to predict the phenotype of a microorganism under a range of conditions, including those derived from genetic modification. A new version of GSMM (v3.0) for the growth of P. pastoris on glycerol and methanol was validated within the growth rate range. Based on the cell wall composition, GSMM could predict the metabolic phenotype of P. pastoris. The growth rates of P. pastoris X33 and the ΔPAS_0305 strain were calculated using the model iMT1026 v3.0 [26]. Firstly, the biomass composition in iMT1026 v3.0 was adjusted according to the experimentally determined ratios (Table 1). According to these calculations, Flux Balance Analysis (FBA) was performed with the methanol utilization rate set at the experimentally measured value [27], i.e., 1.238 mmol methanol/gDCW/h for P. pastoris X33 and 1.454 mmol methanol/gDCW/h for ΔPAS_0305. The calculated growth rates were 0.0315 h−1 for P. pastoris X33 and 0.0335 h−1 for the ΔPAS_0305 strain. Notably, these calculated results confirmed that deletion of PAS_0305 could enhance the growth rate on methanol, which was related to specific metabolic adaptations of the ΔPAS_0305 strain. The increase of the methanol utilization rate of ΔPAS_0305 was in agreement with experimental data.
As metabolic flux distributions are sensitive to changes of biomass composition [28], we adapted the biomass equation to the modified composition of P. pastorisΔPAS_0305, after which the GSMM could calculate the in vivo flux distribution accurately. The calculated flux distribution indicated that the carbon loss was 1.156 mmol CO2/mmol methanol for P. pastoris X33, compared to 1.056 mmol CO2/mmol methanol for ΔPAS_0305 (Table 1). Based on these GSMM calculation results, the absence of ΔPAS_0305 could reduce fluxes towards the dissimilatory pathway, leading to less carbon loss and higher methanol utilization efficiency compared with P. pastoris X33. These results confirmed that the internal metabolism of P. pastoris underwent significant changes, maximizing the flux toward biomass accumulation while minimizing the metabolic burden caused by the stress response to methanol.
Underlying mechanism of cell wall sensor activation
The cell wall is the first line of defense of yeast cells against external stresses [16]. The yeast cell wall is a highly dynamic structure whose properties change constantly in response to stress, including adjustment of cell wall composition, or incorporation of new synthesized polysaccharides into the preexisting cell wall core. Therefore, the changes of cell wall permeability were perceived as a new environmental stress by P. pastoris, triggering cell wall sensor activation after knockout of ΔPAS_0305. MAPK signaling pathways play a crucial role in transmitting signals from the cell wall to the nucleus, ultimately regulating gene expression and cellular responses. These pathways are highly conserved across yeast species, making them a promising area of exploration for understanding cellular adaptation and stress responses [29]. Accordingly, it is crucial to investigate whether the signal transduction pathway has been affected in the ΔPAS_0305 strain, as this could potentially explain the underlying mechanism, providing a new rational strategy for generating desirable phenotypes in P. pastoris.
As MAPK signaling pathways are highly conserved among yeast species, the key genes involved in the cell wall integrity (CWI) and high-osmolarity glycerol (HOG) pathways have been identified and annotated through comparative transcriptomics between P. pastoris X33 and ΔPAS_0305 comparing whole-transcriptome comparison of P. pastoris X33 and ΔPAS_0305 revealed that critical regulatory elements within CWI and HOG pathways exhibit variable transcriptional abundance (Fig. 5c, d). Firstly, the upregulation of Sho1 in response to hyperosmotic stress conditions in ΔPAS_0305 was consistent with the observed increase of intracellular osmolytes due to enhanced cell wall permeability. This suggests that Sho1 plays a role as a sensor in the stress response of ΔPAS_0305. Secondly, the Rho GTPase Ste50 was downregulated in the sho1 branch, which synchronously led to the downregulation of pbs2 transcription. This result suggested that the Hog signaling pathway was perturbed. However, there is a lack of detailed understanding regarding the connection between the HOG and CWI pathways [30]. Previous studies demonstrated that the MAP kinases Mkk1 and Pbs2 of the HOG pathway, as well as Slt2 and Hog1 of the CWI interact physically, forming a complex [31]. In ΔPAS_0305, a positive correlation between Pbs2 and Mkk1 transcription was observed (Fig. 6), indicating similar perturbations in response to cell wall sensor activation. These findings shed new light on the interplay between the HOG and CWI pathways, suggesting that alterations in one pathway can influence the other. However, further studies are needed to unravel the precise mechanisms underlying these connections, as well as their implications for cell wall dynamics and the stress response of P. pastoris.
Analysis of signal transduction pathways of ΔPAS_0305. a Sketch of DEGs related to the CWI and HOG MAPK signaling pathways. Genes in green showed significant downregulation, while those in red showed significant upregulation. b Amino acid residues engaged in binding between Pbs2 and Mkk1. c Three-dimensional structure of FKS1 and FKS2 from P. pastoris modeled by SWISS-MODEL using Saccharomyces cerevisiae FKS1 as the template
To investigate the potential interaction between Pbs2 and Mkk1, we utilized RosettaDock, a protein docking and simulation tool [32]. According to the docking results, the P value between Pbs2 and Mkk1 was less than 0.5, indicating that the results are credible. The ΔG of the composite system was − 9.7, which indicated that the binding between Pbs2 and Mkk1 is spontaneous. The protein docking complex revealed that approximately 15.4% of amino acid residues in Pbs2 and 13.6% of those in Mkk1 were involved in the binding interface. Additionally, the binding area of Pbs2 was found to be marginally larger than that of Mkk1, with the latter occupying 9.9% and Pbs2 occupying 10.7% of the binding interface (Fig. 6b). Notably, the interaction between Pbs2 and Mkk1 was stabilized by four specific amino acids (Mkk1: F198, R275; Pbs2: R323, G471), which formed stable hydrogen bonds (Fig. 6b). This indicates a strong interaction between Pbs2 and Mkk1. The consistency between the transcriptional changes and the molecular docking results further supports the hypothesis of a signaling interaction between Pbs2 and Mkk1, bridging the HOG and CWI pathways. These findings also provide valuable insights into the molecular mechanisms underlying the cross-talk between the HOG and CWI pathways in response to hyperosmotic stress.
The Pbs2-Mkk1 signaling pathway was found to transmit the signal from the HOG to the CWI MAPK module, confirming that activation of the CWI pathway depended on signal transduction via the HOG pathway. Finally, the downregulation of MAPKs Mkk1/Mkk2 led to the activation of the MAPK Slt2, thereby activating the CWI signaling pathway. In the ΔPAS_0305 strain, the transcription of Slt2 was indeed upregulated, while other phosphatases such as Ptp2/Ptp3 were downregulated. This result was in agreement with previous reports that Slt2 activity can be negatively regulated by dephosphorylation through the activity of different phosphatases such as Ptp2 and Ptp3 [33, 34]. It is possible that Slt2 is activated by either the CWI signal or the pbs2-Mkk1 mediated HOG signal. The knockout of the PAS_0305 gene in P. pastoris resulted in sequential signal transduction through two MAPK pathways, leading to the elicitation of a rescue response, including the remodeling of the cell wall.
Significantly, whole-transcriptome analysis showed that FKS1 and FKS2 were respectively upregulated by 2.8- and 1.7-fold. The FKS family genes are responsible for β-1,3-glucan synthesis [35]. The amino acid sequences of FKS1 and FKS2 from P. pastoris X33 showed a sequence identity of 74.95% and 54.99% respectively, when compared to FKS1 from S. cerevisiae (Fig. 6c). A high-resolution cryo-electron microscopy structure of S. cerevisiae FKS1 was recently reported, and it was confirmed to participate in β-1,3-glucan synthesis as well as affect the lipid composition, resulting in drug-resistance of fungi [36]. Here, we also obtained similar results, whereby the upregulation of FKS in the ΔPAS_0305 strain elicited cell wall remodeling by increasing the β-1,3-glucan and lipid content, thus improving yeast robustness. Notably, stress resistance may be the result of fungal responses to external signals, transduced either through the CWI pathway or the pbs2-Mkk1 mediated HOG pathway. In a previous study, activation of the CWI pathway by different transcriptional factors was confirmed to improve the robustness of S. cerevisiae [36]. This mechanism could also explain why the ΔPAS_0305 strain exhibited a remodeled cell wall composition and improved robustness after activating cell wall sensors.
Discussion
Microorganism-derived SCPs can be produced in abundance and contain essential amino acids, making them a promising alternative protein source that does not affect the supply of food sources such as soy, meat, milk, and fish [37]. SCP produced from methanol using P. pastoris offers an attractive alternative to animal-derived protein due to the rapid production rate, lower space requirements, independence of climate or seasons and more sustainable production process [37]. Moreover, methanol can be industrially produced from CO2 by photocatalytic or electrochemical reduction, which may contribute to SCP overproduction with a (nearly) zero CO2 footprint. Stoichiometric models of P. pastoris showed that 18% of the used methanol was dissimilated by yeast cells [38], while the value could reach 70–80% in fed-batch fermentations [6]. However, excessive dissimilation of methanol greatly increases carbon losses. The complexity of the metabolic network of P. pastoris made it difficult to transform a single metabolic pathway without affecting another. It has been reported that simply knocking out the dissimilation pathway may not be the best solution, because this would lead to a significant loss of cell robustness [39]. Therefore, the construction of SCP overproduction strains based on P. pastoris is time-consuming and laborious since it requires multiple rounds of genetic manipulation, which face the challenge of balancing methanol metabolism and biomass biosynthesis.
In this study, cell wall sensor activation was used as an effective breeding strategy for improving the methylotrophic performance of P. pastoris. The global metabolism of P. pastoris was successfully reprogrammed for SCP overproduction, which provided a new theoretical basis for understanding the mechanisms of methanol utilization and carbon loss in P. pastoris via unlocking cell wall sensors. This research provides new insights into cellular mechanisms underlying the response to environmental stress, which resulted in a superior methylotrophic fermentation performance of P. pastoris. Thus, the novel strategy can provide a new perspective for the construction of versatile P. pastoris cell factories to produce a variety of chemicals based on the flux balance response to cell wall stress.
Currently, the production of well-balanced animal feed heavily relies on corn kernel and soybean proteins, with almost all imported soybean in China being used as a protein source for animal feed. However, this reliance on plant-based proteins poses challenges due to the low energy efficiency of plant photosynthesis, as well as significant freshwater consumption and increasing land use. To address these challenges, the internal metabolism of P. pastoris has been thoroughly reprogrammed to maximize the flux towards biomass accumulation. This optimization resulted in impressive crude protein and cell dry weight yields of 67.21% and 121.2 g/L, respectively. The single-cell protein (SCP) produced by the ΔPAS_0305 strain is competitive with soy (38.6%), fish (17.8%), meat (21.2%), and whole milk (3.28%) [40]. Notably, the methanol conversion rate of the engineered P. pastoris reached the state-of-the-art level of 0.46 g/g, amounting to 92% of the maximum theoretical yield. This biotechnological strategy resulted in a significant advancement in the directed conversion of methanol into biomass. The impressive advancements in the space–time productivity of SCP in P. pastoris have positioned it as a cost-effective option for the industrial production of protein.
Further analysis of the strain has shed new light on the mechanisms involved in unlocking cell wall sensors to maximize the flux from methanol towards biomass accumulation. This novel strategy has revealed the potential of manipulating cellular signaling pathways to optimize metabolic networks and achieve exceptional phenotypic characteristics, thereby providing new strategies for constructing versatile cell factories in P. pastoris. By addressing the issues of methanol fermentation efficiency and carbon loss in the directed conversion of methanol into SCP, this approach represents a great advance in cost-competitive industrial bio-manufacturing of microbial protein, thereby contributing to global food security.
Materials and methods
Strains and culture conditions
All strains and plasmids used in this study are listed in Additional file 2: Extended Data Table S2. Unless otherwise specified, P. pastoris strains were cultivated in YPD medium, which was supplemented with 100 mg/L zeocin to screen transformants. Delft basic salt medium containing 7.5 g/L (NH4)2SO4, 14.4 g/L KH2PO4, 0.5 g/L MgSO4•7H2O, 1 mL/L vitamin solution, and 2 mL/L PTM1 trace salts solution was used for cell cultivation with methanol as sole carbon sources [41]. E. coli DH5α used as the host for plasmid construction and was cultured at 37 °C in LB medium.
Transmission electron microscopy
To assess the thickness of the cell wall by transmission electron microscopy, P. pastoris cells were grown at 30 °C and 220 rpm to the exponential phase in Delft basic salt medium containing 0.5% or 3% methanol. The cells were harvested by centrifugation at 5000×g for 10 min, washed three times, and resuspended in phosphate buffer (pH = 7.4, 50 mM). The suspension was premixed with an equal volume of 2.5% glutaraldehyde for 12 h at 4 °C and subsequently dehydrated with a graded series of 25, 50, 70, 80, 95, and 100% ethanol solution. Pure ethanol was then changed to propylene oxide and specimens were gradually infiltrated with increasing concentrations (30%, 50%, 70% and 100%) of Agar 100 epoxy resin mixed with propylene oxide for a minimum of 3 h per step. Samples were embedded in pure, fresh Agar 100 epoxy resin and polymerized at 60 °C for 48 h. Ultrathin sections were stained for 3 min with lead citrate and viewed with an JEM-1230 electron microscope.
Illumina sequencing and analysis of significantly differentially expressed genes
The total RNA was extracted from the cells grown to the exponential phase under the two indicated culture conditions using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), after which the two sequencing libraries were constructed using the TruSeq™ Stranded Total RNA Library Prep Kit for Illumina (Illumina, San Diego, CA, USA) following the manufacturer’s protocol. After cluster generation, the library preparations were sequenced using the Illumina HiSeq 2000 platform, which generated 150 bp paired-end reads. The significantly differentially expressed genes were identified using the inclusion criteria |log2FC| ≥ 1 and P < 0.05.
Construction of strains and plasmids
The method used for chromosomal gene deletion in P. pastoris was based on the CRISPR–Cas9 meditated genome editing system as described previously [11]. The plasmid pPICZ-Cas9-gGUT1, which expresses Cas9 and GUT1-gRNA under the control of the bidirectional promoter PHTX1 [11], was used as template to construct the genome editing vector pPICZCas9-g (target genes) by Gibson assembly. A PAS_0305-complementation strain, ΔPAS_0305-cp, was generated by reintegrating the knocked-out ΔPAS_0305 gene into its original position in the genome of strains ΔPAS_0305. The mutants were screened by colony PCR and further confirmed by gene sequencing. All primers used for strain and plasmid construction are listed in Additional file 3: Extended Data Table S3.
To detect the secretion of a heterologously expressed protein for verifying cell wall permeability, the gene encoding endoglucanase HgCel5A from Humicola grisea (Accession number: KX096883.1) was synthesized by Genewiz Biotech Co., Ltd. (Suzhou, China), and cloned into the expression vector with PAOX promoter and TGAP terminator.
Transformation and selection of P. pastoris clones
The fragment was linearized by AvrII restriction digestion and then electroporated into P. pastoris X33 using a MicroPulser electroporator (BIO-RAD). Positive clones were selected on YPD plates with 100 mg/L zeocin for 48–96 h at 30 °C. The recombinant clones were screened by colony PCR and confirmed by DNA sequencing, followed by sub-culture on YPD plates with increasing zeocin concentrations (100, 200, 300, 400, and 500 mg/L) for 48 h at 30 °C each. The colonies that grew well under high selection pressure were selected for shake-flask fermentation.
Fed-batch fermentation
After overnight cultivation in YPD medium at 30 °C, the seed cultures were inoculated (8%, v/v) into a 5-L bioreactor containing 3 L of basal salt medium (BSM) supplemented with 4.35 mL/L of PTM1 trace salts. The fermentation process consisted of three stages. In Stage I, the focus was on cell growth, and the dissolved oxygen (DO) level was maintained above 30% of air saturation by adjusting the agitation rate (200–800 rpm). In Stage II, a 50% (w/w) glycerol solution (containing 12 mL/L of PTM1 solution) was continuously fed to the bioreactor to maintain the dissolved oxygen level above 30%. The temperature was controlled at 30 °C using recirculating water, and the pH was maintained at 5.5 by adding 25% ammonium hydroxide during stages I and II.
In Stage III, protein production was induced by adding methanol (containing 12 mL/L of PTM1 solution). The culture temperature was lowered to 28 °C, the pH was adjusted to 6.0, and the DO level was maintained above 20% of air saturation with an agitation rate of 800 rpm.
Biomass and crude protein analysis
The biomass was characterized by measuring the dry cell weight (DCW, gL−1) and absorbance at 600 nm [42]. Cell pellets from 3 mL culture were washed with sterile water three times and resuspended in 1 mL of sterile water. The tubes containing centrifuged cells were dried at 105 °C until a constant weight was maintained. The nitrogen content of the homogenized residues was analyzed using the Kjeldahl method [43]. A conversion factor of 6.25 was used to calculate the theoretical protein content from the nitrogen content.
Enzyme activity assays
CMCase activity was measured colorimetrically, using a 3,5-dinitrosalicylic acid (DNS) assay with carboxymethyl cellulose (CMC) as the substrate [44].
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
The extracellular proteins were analyzed using SDS-PAGE. The purified proteins were separated using 12% separating gel with 0.1% SDS and 5% stacking gel. The protein bands were stained with Coomassie brilliant blue R-250.
Analysis of cell wall components in recombinant P. pastoris
The total cellular β-1,3 glucan and chitin contents of P. pastoris cells were quantitatively analyzed according to the NREL Laboratory Analytical Procedures for biomass using a two-step acid method. Lipids were extracted and quantified by (UPLC)-Q Exactive HF MS (Thermo Fisher Scientific).
Disaccharide identification and trehalose measurement
The contents of osmolytes was determined using high-performance liquid chromatography (Shimadzu, Kyoto, Japan) with a refractive index detector (Shimadzu) on an Aminex HPX-42A column (Bio-Rad, Hercules, CA, USA) as the mobile phase at a flow rate of 0.5 mL/min at 35 °C. The disaccharides were analyzed using an ultra-performance liquid chromatography (UPLC) system (Nexera 30A, Shimadzu, Kyoto, Japan) coupled with a mass spectrometer (TripleTOF™ 5600, Applied Biosystem Sciex, United States) in negative electrospray ionization (ESI) mode. The disaccharide was identified using an LC equipped with a SeQuant ZIC-HILIC column (100 × 2.1 mm, 3.5 μm, Merck, Germany) [45].
The intracellular trehalose content was measured using the modified anthrone method as described previously [46]. Aliquots of 200 µL were mixed with 1 mL of cold anthrone reagent and incubated at 100 °C for 10 min prior to the measurement of A620.
Propidium iodide and Nile red staining
To verify cell wall permeability, P. pastoris cells were grown in YPD to an OD600 of approximately 1.0, washed twice with phosphate-buffered saline (PBS, pH 7.4) and resuspended in the same buffer [47]. After 5 min at room temperature, 200 μL of the cell suspension were added in triplicate to a 96-well fluorescence plate, followed by the addition of propidium iodide or Nile red stock solution (0.5 mg/mL) prepared in advance according to the calculated volume (5 μL). Then, the initial fluorescence was measured and the black fluorescence plate placed on a shaker at 150 rpm and 30 °C. Propidium iodide fluorescence was measured at an excitation wavelength of 535 nm and an emission wavelength of 615 nm, while Nile red was measured at an excitation wavelength of 540 nm and an emission wavelength of 630 nm.
Methanol tolerance assay
Pichia pastoris cells were grown in liquid YPD to an OD600 of approximately 0.4, after which equal volumes of the culture (10 μL) were transferred to a 96-well fluorescence plate containing YPD medium with 1.5%, 2%, 2.5%, and 3.5% methanol, respectively. The fluorescence plate was then incubated in a 30 °C shaker at 150 rpm. Starting at 24 h, the OD600 was measured every 1 h to plot growth curves [48].
SDS and CR sensitivity assay
The strains were grown in liquid YPD at 30 °C overnight. Then, 6.1 × 107 cells from each strain were subjected to tenfold serial dilution in sterile water. Aliquots (1 μL) from each dilution were then spotted onto YPD agar plates containing 50 μg/mL Congo red (CR) or 50 μg/mL sodium dodecyl sulfate (SDS) and cultured at 30 °C for 48 h [49].
Numerical analysis of P. pastoris growth
The optimal growth rate and flux distribution were calculated based on the biomass objective function using the genome-scale metabolic model iMT1026 v3.0 of P. pastoris by conducting flux balance analysis (FBA). The COBRApy toolbox was used to perform FBA in Python [50]. CPLEX (IBM, Armonk, NY, USA) was used as the linear programming solver. For the simulation, glycerol was used as supplementary carbon source, and the uptake rate was set at 0.502 mmol gDCW−1 h−1. The sources of nitrogen, oxygen, phosphorus and sulfur were not constrained.
Protein docking
The homology modeling of Mkk1 and Pbs2 was performed using SWISS-MODEL (https://swissmodel.expasy.org/). A rigid docking method was used to determine the spatial position and pose of the protein molecules. Both PDB structures for the proteins were submitted to the ClusPro online server (https://cluspro.bu.edu/), and the highest-scoring initial docking result was chosen [51]. Fine docking was performed using RosettaDock for molecular contact localization.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
References
Ritala A, Häkkinen ST, Toivari M, Wiebe MG. Single cell protein-state-of-the-art, industrial landscape and patents 2001–2016. Front Microbiol. 2017; 8:2009.
Ghasemi Y, Rasoul-Amini S, Morowvat MH. Algae for the production of SCP. Bioprocess sciences and technology. Hauppauge: Nova Science Publishers, Inc.; 2011. p. 163–84.
Zhu T, Zhao T, Bankefa OE, Li Y. Engineering unnatural methylotrophic cell factories for methanol-based biomanufacturing: challenges and opportunities. Biotechnol Adv. 2020;39:107467.
Klankermayer J, Wesselbaum S, Beydoun K, Leitner W. Selective catalytic synthesis using the combination of carbon dioxide and hydrogen: catalytic chess at the interface of energy and chemistry. Angew Chem Int Ed. 2016;55:7267–7267.
Schrader J, Schilling M, Holtmann D, Sell D, Fiho MV, Marx A, Vorholt JA. Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 2009;27:107–15.
Vanz AL, Lünsdorf H, Adnan A, Nimtz M, Gurramkonda C, Khanna N, Rinas U. Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes. Microb Cell Fact. 2012;11:103.
Zhan C, Li X, Baidoo EEK, Yang Y, Sun Y, Wang S, Wang Y, Wang G, Nielsen J, Keasling J D, Chen Y, Bai Z. The glyoxylate-serine pathway enables conversion of Saccharomyces cerevisiae to a synthetic methylotroph. SSRN J. 2021.
Zepeda AB, Pessoa A Jr, Farías JG. Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Braz J Microbiol. 2018;49:119–27.
Way JC, Collins JJ, Keasling JD, Silver PA. Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell. 2014;157:151–61.
Yadav VG, De Mey M, Lim CG, Ajikumar PK, Stephanopoulos G. The future of metabolic engineering and synthetic biology: towards a systematic practice. Metab Eng. 2012;14:233–41.
Cai P, Duan XP, Wu XY, Gao LH, Ye M, Zhou YJ. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021;49:7791–805.
Cai P, Liu YX, Zhai XX, Yao L, Ma XJ, Jia LY, Zhou YJ. Microbial synthesis of long-chain α-alkenes from methanol by engineering Pichia pastoris. Bioresour Bioprocess. 2022;9:58.
Moser JW, Prielhofer R, Gerner SM, Graf AB, Wilson I, Mattanovich D, Dragosits M. Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris. Microb Cell Fact. 2017;16:49.
Gao J, Li Y, Yu W, Zhou YJ. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nat Metab. 2022;4:932–43.
Guo YG, Liao Y, Wang J, Ma C, Qin JL, Feng J, Li Y, Wang X, Chen KQ. Methylotrophy of Pichia pastoris: current advances, applications, and future perspectives for methanol-based biomanufacturing. ACS Sustain Chem Eng. 2022;10:1741–52.
Klis FM, Boorsma A, De Groot PWJ. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202.
Yu J, Tran V, Li M, Huang X, Niu C, Wang D, Zhu J, Wang J, Gao Q, Liu J. Both phthiocerol dimycocerosates and phenolic glycolipids are required for virulence of Mycobacterium marinum. Infect Immun. 2021;80:1381–9.
Lee SY. Metabolic engineering and synthetic biology in strain development. ACS Synth Biol. 2012;1:491–2.
Benaroudj N, Lee DH, Goldberg AL. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J Biol Chem. 2001;276:24261–7.
Wang P, Zhang L, Fisher R, Chen M, Liang S, Han S, Zheng S, Sui H, Lin Y. Accurate analysis of fusion expression of Pichia pastoris glycosylphosphatidylinositol-modified cell wall proteins. J Ind Microbiol Biotechnol. 2017;44:1355–65.
Bulawa CE. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol Cell Biol. 1992; 12.
Zou CJ, Wang P, Liang SL, Han SY, Zheng SP, Lin Y. Deletion of the GCW13 gene derepresses Gap1-dependent uptake of amino acids in Pichia pastoris grown on methanol as the sole carbon source. Biochem Biophys Res Commun. 2018;501:226–31.
Gao Y, Li D, Liu Y. Production of single cell protein from soy molasses using Candida tropicalis. Ann Microbiol. 2012;62:1165–72.
Pires JF, Ferreira GMR, Reis KC, Schwan RF, Silva CF. Mixed yeasts inocula for simultaneous production of SCP and treatment of vinasse to reduce soil and fresh water pollution. J Environ Manag. 2016;182:455–63.
Yap SM, Lan JC, Kee PE, Ng HS, Yim HS. Enhancement of protein production using synthetic brewery wastewater by Haematococcus pluvialis. J Biotechnol. 2022;350:1–10.
Tomàs-Gamisans M, Ferrer P, Albiol J. Fine-tuning the P. pastoris MT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carb on sources. Microb Biotechnol. 2018;11:224–37.
Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–8.
Dikicioglu D, Kirdar B, Oliver SG. Biomass composition: the “elephant in the room” of metabolic modelling. Metabolomics. 2015;11:1690–701.
Mensonides FIC, Brul S, Klis FM, Hellingwerf KJ, Mattos MJTD. Activation of the protein kinase C1 pathway upon continuous heat stress in Saccharomyces cerevisiae is triggered by an intracellular increase in osmolarity due to trehalose accumulation. Appl Environ Microbiol. 2005;71:4531–8.
Rodríguez-Peña JM, García R, Nombela C, Arroyo J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast. 2010;27:495–502.
Jimenez-Gutierrez E, Alegrıa-Carrasco E, Alonso-Rodrıguez E, Fernandez-Acero T, Molina M, Martın H. Rewiring the yeast cell wall integrity (CWI) pathway through a synthetic positive feedback circuit unveils a novel role for the MAPKKK Ssk2 in CWI pathway activation. FEBS J. 2020;287:4881–901.
Pierce B, Weng Z. A combination of rescoring and refinement significantly improves protein docking performance. Proteins. 2008;72:270–9.
Mattison CP, Spencer SS, Kresge KA, Lee J, Ota IM. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol Cell Biol. 1999;19:7651–60.
Sanz AB, García R, Rodríguez-Peña JM, Arroyo J. The CWI pathway: regulation of the transcriptional adaptive response to cell wall stress in yeast. J Fungi. 2018;4:1.
Hu XL, Yang P, Chai CD, Liu J, Sun HH, Wu YN, Zhang MJ, Zhang M, Liu XT, Yu HJ. Structural and mechanistic insights into fungal β-1, 3-glucan synthase FKS1. Nature. 2023;616:190–8.
Son SH, Kim JE, Oh SS, Lee JY. Engineering cell wall integrity enables enhanced squalene production in yeast. J Agric Food Chem. 2020;68:4922–9.
Zhang L, Hu YY, Badar IH, Xia XF, Kong BH, Chen Q. Prospects of artificial meat: opportunities and challenges around consumer acceptance. Trends Food Sci Technol. 2021;116:434–44.
Tortajada M, Llaneras F, Picó J. Validation of a constraint-based model of Pichia pastoris metabolism under data scarcity. BMC Syst Biol. 2010;4:115.
Rohlhill J, Sandoval NR, Papoutsakis ET. Sort-Seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol. ACS Synth Biol. 2017;6:1584–95.
Salazar-López NJ, Barco-Mendoza GA, Zuñiga-Martínez BS, Domínguez-Avila JA, Robles-Sánchez RM, Villegas Ochoa MA, González-Aguilar GA. Single-cell protein production as a strategy to reincorporate food waste and agro by-products back into the processing chain. Bioengineering. 2022;9:623.
Zhou YJ, Buijs NA, Zhu Z, Qin J, Siewers V, Nielsen J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709.
Wang D, Chen M, Zeng X, Li W, Liang S, Lin Y. Improving the catalytic performance of Pichia pastoris whole-cell biocatalysts by fermentation process. RSC Adv. 2021;11:36329–39.
Marco A, Rubio R, Compano R, Casals I. Comparison of the Kjeldahl method and a combustion method for total nitrogen determination in animal feed. Talanta. 2002;57:1019–26.
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–8.
Chang ZS, Dai W, Mao YF, Cui ZZ, Zhang ZD, Wang ZW, Ma HW, Chen T. Enhanced 3-hydroxypropionic acid production from acetate via the malonyl-CoA pathway in Corynebacterium glutamicum. Front Bioeng Biotechnol. 2021;9:808258.
Benjaphokee S, Hasegawa D, Yokota D, Asvarak T, Auesukaree C, Minetaka S, Kaneko Y, Boonchird C, Harashima S. Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. New Biotechnol. 2012;29:379–86.
Li Q, Zhou M, Fan X, Yan J, Li W, Xie J. Mycobacteriophage SWU1 gp39 can potentiate multiple antibiotics against Mycobacterium via altering the cell wall permeability. Sci Rep. 2016;6:28701.
Meng J, Liu SF, Gao L, Hong K, Liu SG, Wu X. Economical production of Pichia pastoris single cell protein from methanol at industrial pilot scale. Microbial Cell Factories. 2023;22:198.
Inokuma K, Kitada Y, Bamba T, Kobayashi Y, Yukawa T, den Haan R, van Zyl WH, Kondo A, Hasunuma T. Improving the functionality of surface-engineered yeast cells by altering the cell wall morphology of the host strain. Appl. Microbiol. Biotechnol. 2021;105:5895–904.
Heirendt L, Arreckx S, Pfau T, Mendoza SN, Richelle A, Heinkenb A, Haraldsdóttir HS, Wachowiak J, Keating SM, Vlasov V. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat Protoc. 2019;14:639–702.
Desta IT, Porter KA, Xia B, Kozakov D, Vajda S. Performance and Its limits in rigid body protein-protein docking. Structure. 2020; 28:1071–81.
Acknowledgements
Not applicable.
Funding
This work was supported by the National key R&D program of China (2021YFD1301002), Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA28030301).
Author information
Authors and Affiliations
Contributions
LG and XW conceived the study. LG and JM wrote the manuscript. LG and WLD performed most of the experiments. ZKZ and SGL performed fed-batch fermentation experiments. QQY performed model calculations. HFD performed protein docking experiments. ZWY and XW reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Identification of significantly differentially expressed genes by RNA-seq in response to changes of methanol stress.
Additional file 2
. Strains and plasmids used in this study.
Additional file 3
. Primers used for strain and plasmid construction in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gao, L., Meng, J., Dai, W. et al. Deciphering cell wall sensors enabling the construction of robust P. pastoris for single-cell protein production. Biotechnol Biofuels 16, 178 (2023). https://doi.org/10.1186/s13068-023-02428-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-023-02428-7