Abstract
Background
Duckweed is considered a promising feedstock for bioethanol production due to its high biomass and starch production. The starch content can be promoted by plant growth regulators after the vegetative reproduction being inhibited. Maleic hydrazide (MH) has been reported to inhibit plant growth, meantime to increase biomass and starch content in some plants. However, the molecular explanation on the mechanism of MH action is still unclear.
Results
To know the effect and action mode of MH on the growth and starch accumulation in Spirodela polyrrhiza 7498, the plants were treated with different concentrations of MH. Our results showed a substantial inhibition of the growth in both fronds and roots, and increase in starch contents of plants after MH treatment. And with 75 µg/mL MH treatment and on the 8th day of the experiment, starch content was the highest, about 40 mg/g fresh weight, which is about 20-fold higher than the control. The I2-KI staining and TEM results confirmed that 75 µg/mL MH-treated fronds possessed more starch and big starch granules than that of the control. No significant difference for both in the photosynthetic pigment content and the chlorophyll fluorescence parameters of PII was found. Differentially expressed transcripts were analyzed in S. polyrrhiza 7498 after 75 µg/mL MH treatment. The results showed that the expression of some genes related to auxin response reaction was down-regulated; while, expression of some genes involved in carbon fixation, C4 pathway of photosynthesis, starch biosynthesis and ABA signal transduction pathway was up-regulated.
Conclusion
The results provide novel insights into the underlying mechanisms of growth inhibition and starch accumulation by MH treatment, and provide a selective way for the improvement of starch production in duckweed.
Similar content being viewed by others
Background
Duckweeds (Lemnaceae) are among the known smallest flowering plants, which are aquatic, propagate mainly via asexual reproduction, and most importantly grow more rapidly than other higher plants [1,2,3]. These tiny plants usually have high starch content and, therefore, have been developing into very promising bioenergy resources [3, 4]. It has been reported that the starch content of duckweed can be elevated by subjecting the plants to nutrient starvation or treating them with plant growth regulators, which results in inhibition or delay of asexual propagation [5,6,7,8,9,10].
Maleic hydrazide (MH) is one of the plant growth regulators and has been shown to inhibit plant growth without causing obvious morphological abnormalities [11, 12]. Therefore, it has been widely used as a temporary growth inhibitor in many plants. For example, it has been reported to delay the development of buds, so that vegetables can last longer in storage and arrive fresher to the consumer [13, 14] and was also used for control of axillary growth (suckers) in tobacco production, to alter cured-leaf quality, and to increase yield [15].
Despite many studies of MH application, the mode of its action is less investigated. In this paper, we explored the effect and mechanism of MH on inhibiting growth in plants of Spirodela polyrrhiza 7498, the first duckweed species used for genome sequencing, as an appropriate concentration can promote starch content, which is very feasible in the application of bioenergy production of duckweed plants [3, 16, 17, 30].
Results
Effect of MH on vegetative growth
Plants were inoculated into flasks with fresh medium containing different concentration of MH, respectively, and cultivated under conditions described in the “Material and methods” part. On the 8th day of experiment, the effect of MH on plant growth was observed. While the control plants, cultivated on the medium without MH, grew very quickly, the growth of plants on medium containing MH was inhibited in a concentration-dependent manner. As shown in Fig. 1, both total frond number and fresh weight were decreased by MH treatment in the concentration range from 25 to 200 µg/mL. MH-treated plants also had fewer and shorter roots (rhizoid) than the controlled plants (Additional file 1: Figure S1). Vegetative reproduction was almost completely inhibited at 75 µg/mL MH and higher concentrations. However, dry weight was not significantly affected even by 100 µg/mL MH (Fig. 1d).
Effect of MH on the growth of S. polyrrhiza 7498. Six groups of duckweed plants (total 24 fronds) were inoculated into the flasks with different medium, respectively. On the 8th day of experiment, photos (a) were taken, and frond numbers (b) were accounted. Fresh and dry weight (c, d) were measured every 2 days. In a a to f show plants incubated in 0 µg/mL, 25 µg/mL, 50 µg/mL, 75 µg/mL, 100 µg/mL and 200 µg/mL MH, respectively. * indicates statistically significance on the level of P < 0.05; and ** indicates statistically significance on the level of P < 0.01, in comparison with the control (0 µg/mL). Each point in b–d represents mean ± SE of 3 biological replicates
Effect of MH on starch accumulation
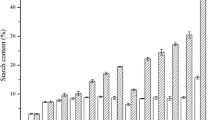
We measured the starch content of S. polyrrhiza 7498 during a cultivation period of 12 days. While the starch content in the plants grown on medium without MH was maintained in a similarly low level, dramatical increases of starch content were found in plants grown on medium with all concentrations of MH (Fig. 2a, b). The highest starch content was demonstrated in plants grown on medium with 75 µg/mL MH at the 8th day of the experiment (Fig. 2a, b). In these plants, the starch content was about 60 mg per gram fresh weight, which was about 20-fold higher than that of the control, and the total starch yield was about sevenfold higher than that of the control. This dramatic increase in starch content was further confirmed by staining the fronds with I2-KI (Fig. 2c, d). Then, Transmission Electron Microscope (TEM) was used to investigate the starch granules in mesophyll cells. It was found that there were more big starch granules in the chloroplasts of MH-treated fronds than that of the control (Fig. 2e, f).
Starch accumulation in S. polyrrhiza 7498 plants after MH treatment. Starch content was calculated based on fresh weight (a); or each flask (b). Plants cultivated on the medium without (c and e) or with 75 µg/mL MH (d and f) were collected on the 8th day of the experiment and stained with I2-KI (c and d) or subjected to starch granule observation (e and f) with TEM. C, V and S represents chloroplast, vacuole and starch granule, respectively. * indicates statistically significance on the level of P < 0.05; and ** indicates statistically significance on the level of P < 0.01. Each point in A and B represents mean ± SE of 3 biological replicates
Effect of MH on parameters related to light reactions
To figure out the reasons for starch accumulation after MH treatment, we investigated parameters related to light reactions of photosynthesis. The results showed that there was no significant difference in photosynthetic pigment contents, but the chlorophyll fluorescence parameters PSII and Fv/Fm were increased in fronds treated by 75 µg/mL MH (Fig. 3a–e). Additionally, the rate of photosynthetic oxygen evolution from the MH-treatment fronds was also significantly higher than that of the control (Fig. 3f).
Comparison of photosynthetic parameters in S. polyrrhiza 7498 plants. Photosynthetic parameters were determined on the 8th day of experiment with 75 µg/mL MH. These include pigment content (a–c), fluorescence parameters of PSII (d and e) and rate of photosynthetic oxygen evolution (f). ** in f indicates statistically significance (P < 0.01). Each point in the figure represents mean ± SE of 3 biological replicates
Effect of MH on morphology, structure and stomatal opening of fronds
To the naked eye, the MH-treated fronds looked thicker than that of the controls; therefore, fronds were subjected to dissecting microscope observation and the results confirmed this phenotype change (Fig. 4a, b). Then, paraffin sections (transverse sections) of fronds were prepared and observed with the light microscope. As shown in Fig. 4e, f, the size of the mesophyll cells and vascular bundle sheath cells was larger in the MH-treated fronds than that in the controls. Bigger chloroplasts in the mesophyll cells of MH-treated fronds were also observed in transmission electron microscope (TEM) samples (Fig. 4c, d). The upper surface of the fronds was also observed by scanning electronic microscope (SEM), and the results showed that the opening of stomata in the MH-treated fronds was larger than that of the control (Fig. 4g–j).
Phenotypic and microscopic observation of S. polyrrhiza 7498 plants. Phenotypic and microscopic observation were conducted on the 8th day of the experiments. Plants cultivated on medium without (a, c, e, g and h) or with 75 µg/ml MH (b, d, f, i and j) were collected at about 4 pm in the day time for observation with a dissecting microscope (a and b), for frond mesophyll cell observation with TEM (c and d) and frond stomatal observation with SEM (g to j). Paraffin slices of fronds were stained by Safranin and Fast Green and observed with light microscope (e and f). MC indicates mesophyll cell and BSC indicates bundle sheath cell
Transcriptomics response to MH treatment
To investigate the transcriptional response of S. polyrrhiza 7498 plants to MH treatment, RNA-Seq analysis was conducted using RNA samples isolated from fronds treated with 75 µg/mL MH for 8 days, by which fronds grown on medium without supplementation of MH were used as the control.
The KEGG Pathway functional enrichment of differentially expressed genes (DEGs) indicates that MAPK pathway, endocytosis and pathways related to carbon metabolism are mostly affected by MH treatment (Additional file 2: Figure S2). Expression analysis of genes involved in developmental processes by GO classification shows that 39 genes involved in the developmental progress (including many transcriptional factors, some of them being the growth-regulating factors, such as Spo003981, Spo012722 and Spo013751) are down-regulated (Additional file 3: Table S1).
As MH treatment inhibited growth and simultaneously promoted starch accumulation (Figs. 1a, b, 2a, b, d, f), we then mainly focused on photosynthesis, starch biosynthesis and processes related to phytohormone IAA and ABA.
Photosynthesis is divided into two phases: the light-dependent reactions and the carbon fixation. By light-dependent reactions, ATP and NADPH are generated, which are then used for carbon fixation. As shown in Additional file 4: Table S2, the expression of genes related to light reactions was not significantly affected by MH treatment, except the gene encoding thioredoxin reductase. However, carbon fixation pathways are significantly affected by 75 µg/mL MH treatment. While the expression of some genes involved in Calvin cycle was increased, some other genes involved in the cycle decreased their expression after MH treatment (Table 1). The MH up-regulated genes include those encoding phosphoglycerate kinase [EC:2.7.2.3], glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12], triosephosphate isomerase (TIM) [EC:5.3.1.1], ribose 5-phosphate isomerase A [EC:5.3.1.6] and one of the two ribulose-phosphate 3-epimerase [EC:5.1.3.1]. The MH down-regulated genes include those encoding ribulose-bisphosphate carboxylase small chain [EC:4.1.1.39], glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) [EC:1.2.1.13], fructose-1,6-bisphosphatase I [EC:3.1.3.11], transketolase [EC:2.2.1.1], the other ribulose-phosphate 3-epimerase [EC:5.1.3.1] and phosphoribulokinase [EC:2.7.1.19]. Besides, it was found, surprisingly, that the expression of some marker genes of C4 pathway was also significantly up-regulated by MH treatment (Table 1). Duckweed plants are classified as C3 plants [18]. There is no report demonstrating that duckweed has C4 pathway. To confirm our finding, we performed qRT-PCR. The results showed that the expression of the two marker genes encoding PEPC (phosphoenolpyruvate carboxylase) and MDH (NADP-malate dehydrogenase), respectively, were all up-regulated significantly by MH treatment (Fig. 5a, b). We also analyzed the enzyme activity of PEPC and MDH in the plant tissues of S. polyrrhiza 7498, which showed also a significant increase after MH treatment (Fig. 5c, d).
Our RNA-seq data showed that the expression of some transcripts encoding key enzymes involved in the starch biosynthesis pathway could be affected by MH treatment (Fig. 6a). Therefore, we further detected the expression of these genes by qRT-PCR, the results of which confirmed the tendency of the expression of these genes. In detail, MH treatment up-regulated the expression of APL3 encoding ADP-Glc pyrophosphorylase, GBSS encoding granule bound starch synthase, SSS encoding soluble starch synthase and DBE1 encoding starch debranching enzyme, but down-regulated the expression of APL1, APL2 and SBE1 encoding starch branching enzymes (Fig. 6b–h).
Expression of genes in starch biosynthetic pathway in S. polyrrhiza 7498. a Expression heatmap, b–h. qRT-PCR results. **indicates statistically significance (P < 0.01). Each point in b–h indicates mean ± SE of 3 biological replicates. Plants were cultivated in the medium supplemented with or without 75 µg/mL MH and sampled on the 8th day of cultivation
Besides, as shown in gene expression heatmaps of KEGG Pathway functional enrichment (Additional file 5: Figure S3, the genes information in the heatmap A, B, C, D and E was listed in the Additional files 10–14, respectively), MH treatment up-regulated the expression of many genes involved in carbon metabolism and lipid degradation.
Auxin is known to regulate many aspects of plant growth and development, and MH is a type of auxin inhibitor. The RNA-Seq results show that the expression of 11 Auxin-related genes was down-regulated and that of 4 genes was up-regulated (Table 2). Analysis of KEGG Orthology shows that these down-regulated DEGs include those encoding the auxin response protein or auxin-induced protein. The phytohormone ABA is well known to be involved in many aspects of plant growth and development, and it has been reported to participate in the regulation of starch biosynthesis [19]. Analysis of DEGs shows that the expression of 9 genes responsible for ABA transport and ABA signaling was up-regulated, and that the expression of 2 and 3 genes involved in ABA regulation was down- or up-regulated, respectively (Table 3).
Discussion
Mechanisms underlying growth inhibition by MH
It has been reported that MH, as an inhibitor of auxin, inhibited plant vegetative growth [20,21,22], which is confirmed in our present research with S. polyrrhiza (Fig. 1). The inhibition effect of MH on vegetative growth is very similar to the reported delay of sprouting in onions [21] and yam tubers [20, 22].
Our RNA-Seq analysis for the auxin signal transduction showed that 11 genes expression of auxin response protein or auxin-induced protein was down-regulated (Table 2), which all play a pivotal role in auxin signaling. Among these, the decrease in the expression of genes of Aux/IAA family that encode short-lived nuclear proteins could be related to growth inhibition by MH treatment. MH is an inhibitor of auxin. It has been reported that the expression of genes of the Aux/IAA family are mostly rapidly induced by auxin [23], and mediate the responses of auxin-regulated gene expression [24]. It has also been demonstrated that auxin-mediated transcriptional regulation is exclusively dependent on the functions of Aux/IAA [25]. We also found that IAA22-like, IAA27 and IAA30 (IAA30-like) were all down-regulated in S. polyrrhiza 7498 after MH treatment (Table 2). Decreasing expression level of these IAA genes may be resulted in the growth inhibition in S. polyrrhiza 7498. Additionally, four genes encoding proteins belonging to the SAURs family were detected to be MH-down-regulated in S. polyrrhiza 7498. SAURs are primary auxin response genes involved in the auxin signaling pathway. Their expression could be induced within 2–5 min by active auxin. The regulation of SAURs may occur at the transcriptional, post-transcriptional and protein levels [26, 27]. Inhibition of growth by MH resulted in the decrease in frond propagation (Fig. 1a, b), the production of daughter fronds. It has been reported that the high content of IAA in buds during active growth stage might take the responsibility to the up-regulation of most CsSAURs in Camellia sinensis [28]. Accordingly, MH inhibited the growth and propagation of daughter fronds at least partly by down-regulating the expression of SAUR in S. polyrrhiza 7498. The expression of one GH3 (Gretchen Hagen 3) gene was also down-regulated by MH treatment. The GH3 gene family contains important auxin response genes that help maintaining hormonal homeostasis by conjugating excess indole-3-acetic acid (IAA), salicylic acid (SA), and jasmonic acid (JA) to amino acids via hormone- and stress-related signaling pathways [29].
In addition, the expression of some key genes of ABA signaling pathway and ABA response was up-regulated after MH treatment in S. polyrrhiza (Table 3). ABA is well known for its inhibition of the plant growth. When ABA treatment was used to induce turion formation (inhibition of growth) in S. polyrrhiza, the expression of many genes regulated by ABA, such as large subunit ribosomal protein L6, centromeric-specific histone H3, late embryogenesis abundant protein LEA were affected [16, 30]. The changes of the expression of these genes were similar in fronds after MH treatment (Table 3), suggesting that ABA signaling also plays a role in MH inhibition of vegetative growth.
MH treatment could also affect the expression of genes related to other phytohormone (Additional file 6: Figure S4), and therefore, the involvement of the signaling of those phytohormone should be considered and investigated in the future.
Mechanisms underlying the starch accumulation after treatment of MH
Higher starch content in tissues of plants, such as cotton, tobacco and Abelmoschus esculentus were found after MH treatment [15, 31, 32], which is in accordance with our results (Fig. 2).
The synthesis of starch is inseparable from photosynthesis. The comparison of chlorophyll content and PSII fluorescence parameters suggests that MH does not affect the chlorophyll synthesis and PSII activity in S. polyrrhiza, which is in agreement with the results of the report from Koske and Svec [33].
After MH treatment, the fronds of S. polyrrhiza looked much thicker than the control, which is also similar to the reports from leaves of bean, sunflower, cotton and Croft Easter Lilies following MH application [34, 35]. Besides, we observed larger opening of the stomata in the MH-treated fronds, indicating that the MH-treated fronds have favorable structural features for higher photosynthetic capability. Intriguingly, larger vacuole was observed in S. polyrrhiza fronds, being similarly to the characteristics found in Bienertia cycloptera, a kind of single-celled C4 species [36, 37]. The large vacuole could separate the inner and outer cytoplasm, and is thought to be the diffusive barrier that slows CO2 leakage out of the core cytoplasm in single-celled C4 plant [37]. In addition, increased vein density was reported as a factor for efficient photosynthesis [38]. And the veins were reported colorless in C3 and dark green in C4 [39, 40]. After MH treatment, the veins of the frond also turned a little bit darker, and the expression of genes for C4 pathway increased significantly. Therefore, functioning of C4 pathway in MH-treated fronds could be possible. This is not surprising, as some studies have revealed that C3 plant could exhibit C4 pathway in different organ or different stage of development or under stress [41], and the polyphyletic evolution of the C4 pathway suggested that the transition from C3 to C4 was relatively simple [42]. Increase or activation of C4 pathway could contribute to an increase in carbon fixation. ABA signaling may be involved in the induction of the C4 pathway. In this concern, large quantities of accumulation of key enzymes of the C4 pathway were observed at the appropriate sites of the cells in the C3 plant Eleocharis vivipara after 5 µM ABA treatment [43]. However, the Kranz-leaf anatomy was not found in the MH-treated fronds of S. polyrrhiza 7498.
Calvin cycle is responsible for the biosynthesis of sugars from photosynthetic carbon, and therefore, increased expression of the Calvin cycle genes (Table 1) after MH treatment is very understandable. Based on our finding, we propose that MH treatment can not only promote Calvin cycle, but also induce and/or strengthen the operating of the C4 photosynthetic pathway, probably by modifying the ABA signaling in S. polyrrhiza 7498, as shown in Fig. 7.
Calvin (left) and C4-Dicarboxylic acid cycle (right) in S. polyrrhiza 7498 treated by MH for 8 day. Expression variations of some related transcripts are displayed in carbon fixation pathway. Red boxes indicate the up-regulated enzymes in response to MH, green for down-regulated, orange means the enzyme has more transcripts, one or more up-regulated or down-regulated. And white means this enzyme was not found in this study. TKT: transketolase [EC:2.2.1.1], SEBP: sedoheptulose-bisphosphatase [EC:3.1.3.37], PGK: phosphoglycerate kinase [EC:2.7.2.3], GAPDH: glyceraldehyde 3-phosphate dehydrogenase [EC:1.2.1.12], TPI(TIM): triosephosphate isomerase [EC:5.3.1.1], ALDO: fructose-bisphosphate aldolase, class I [EC:4.1.2.13], FBP: fructose-1,6-bisphosphatase I [EC:3.1.3.11], RPI: ribose 5-phosphate isomerase A [EC:5.3.1.6], RPE: ribulose-phosphate 3-epimerase [EC:5.1.3.1]. RBCS: ribulose-bisphosphatecarboxylase small chain [EC:4.1.1.39], GAPA: glyceraldehyde-3-phosphate dehydrogenase (NADP +) (phosphorylating) [EC:1.2.1.13], RPK: phosphoribulokinase [EC:2.7.1.19]. xfp(xpk): xylulose-5-phosphate/fructose-6-phosphate phosphoketolase [EC:4.1.2.9], [4.1.2.22], SHPK: sedoheptulokinase [EC:2.7.1.14], PEPC: phosphoenolpyruvate carboxylase [EC:4.1.1.31], MDH: malate dehydrogenase [EC:1.1.1.37], PPDK: pyruvate, orthophosphate dikinase [EC:2.7.9.1], GOT1: aspartate aminotransferase, cytoplasmic [EC:2.6.1.1], ALT: alanine transaminase [EC:2.6.1.2], phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49], ME(maeB): malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+) [EC:1.1.1.40], [EC:1.1.1.39]: malate dehydrogenase (decarboxylating), [EC:1.1.1.82]: malate dehydrogenase (NADP+)
As energy substances, sugars can be used to support plant growth, or converted to starch for storage when growth is inhibited, especially when the photosynthesis is in full function (Figs. 1, 2 and 3; Table 1). Therefore, large amount of starch accumulated in plant tissues after MH treatment required the increase in the activity of many enzymes of starch biosynthesis, which might trigger the elevation of the expression of their encoding genes, such as APL3, SSS and DBE (Fig. 6).
ABA has also been reported to be involved in regulating starch biosynthesis in duckweeds [44], and the starch of fronds was highly accumulated in S. polyrrhiza, after ABA treatment for 8 days [6]. The expression of starch biosynthetic gene was found to be closely related with ABA signaling transduction [45, 46]. The expression pattern of APS1, APL1 and APL3 in the MH-treated fronds was found to be similar with the results of 10 µM ABA-induced starch accumulation in turions of S. polyrrhiza 7498 [30, 46]. Therefore, ABA signaling could play an important role during the induction of starch accumulation of fronds after MH treatment. In support of this view, we found that the expression of many key genes of ABA signaling, including several protein phosphatase 2C, protein ABSCISIC ACID-INSENSITIVE 5 and ABA-responsive element binding factor, were up-regulated by MH in S. polyrrhiza 7498 (Table 3). Besides, MH and ABA modified similarly the expression of genes related to carbon, glycolysis gluconeogenesis and glycerolipid metabolism (Additional file 5: Figure S3 A–C), as well as the alpha-dioxygenase (Table 3) [30]. Alpha-dioxygenase is a key enzyme responsible for the degradation of lipids, which probably allocate carbon to starch rather than fatty acids.
Previous studies have suggested that there are several kinds of transcription factors that are involved in regulating starch biosynthesis and accumulation, including NAC, WRKY, bZIP, et al. [47,48,49,50]. We also found that many genes encoding NAC domain-containing protein and WRKY were up-regulated by MH treatment in S. polyrrhiza 7498 (Tables 2 and 3), suggesting that they probably play an important regulating role in the starch synthetic pathway.
Conclusion
75 µg/mL MH can effectively prevent vegetative growth, and at the same time induce starch accumulation in S. polyrrhiza 7498. MH could inhibit the frond multiplication in S. polyrrhiza by down-regulating the expression of key genes involved in auxin signal transduction and up-regulating the expression of key genes involved in ABA signal transduction. MH could promote the expressions of some genes participating in carbon fixation and starch accumulation, probably through the modification of ABA signaling and regulation of some transcription factors.
This study sheds light on the mechanism(s) of MH on inhibiting vegetative reproduction and inducing starch accumulation in S. polyrrhiza 7498, which is closely related with ABA signaling. And the research provides a selective way for increasing duckweed starch accumulation as biofuel source in bioenergy production. The regulated genes by MH treatment would provide good candidates for improving the starch content by genetic engineering in duckweed.
Materials and methods
Duckweed strain and treatment
S. polyrrhiza (L.) Schleid. 7498 were grown aseptically on Datko medium as described by Wang and Kandeler [51] under long-day conditions (16-h light and 8-h dark) with a light intensity of ∼ 45 µmol m−2 s−1 at the plant level. The temperature of the culture room was kept at 22 ± 2 °C. S. polyrrhiza plants were transferred to fresh medium every 15 days, to minimize the effect of nutrient shortage. For experiments on the effect of MH, six whole S. polyrrhiza plants were transferred to medium with different concentration of MH. The plants were cultivated in the same long-day light period condition and temperature, with a light intensity of 100 µmol·m−2 s−1 at the plant level. MH was from Cool Seoul Bio, Anhui, China.
Analysis of fronds numbers and weight
For the counting of fronds, fronds were counted with or without MH treatment. Both bigger and smaller fronds were counted and analyzed. For the measurements of fresh weight and dry weight, duckweed plants treated with different concentrations of MH were harvested at different time points. Plants were collected by filtering to separate them from liquid medium, and free water was removed with paper towels. The fresh weight (FW) was measured immediately after which the plants were dried at 85 °C in aluminum foil and weighed every 30 min until the values remained stable, which was recorded as the dry weight (DW).
Measurements of photosynthetic parameters
Chlorophyll was extracted with 95% ethanol. After centrifugation, the supernatant was collected and used to measure chlorophyll content according to the method of Wintermans and De Mots [52]. Chlorophyll fluorescence parameters were measured with Mini-PAM-II (WALZ, Germany). The value Fv/Fm and Y(II) of photosystem II was measured according to WALZ protocol. Rate of photosynthetic oxygen evolution was measured with Chlorolab 2 (Hansatech, England), according to Hansatech manual.
Measurements of starch content and enzyme activity
Duckweed plants were sampled after designed time at about 4:00 p.m. in the day time, about 0.1 g fresh weight samples were ground in liquid nitrogen with a mortar and pestle. The starch was then extracted and measured using the starch extraction and measurement kit according to the manufacturer’s protocol (BC0700, Solarbio Biological &Technology Co. Ltd., Beijing, China). For the analysis of PEPC and MDH enzyme activity, we carried out according to the procotol BC2190 and BC1040, respectively (Solarbio Biological &Technology Co. Ltd., Beijing, China). The determination of the starch content and enzyme activity was done in Thermo Multiskan GO (Thermo, USA).
Observation of fronds morphology and ultrastructure
For observation of the morphology and ultrastructure of frond, the duckweeds were sampled at about 4:00 p.m. in the day time, similar with the starch extracting time after 8 days of cultivation with or without the presence of MH. For the I2-KI staining, the duckweed was completely decolorized with 95% ethanol, then stained with I2-KI solution for 20 min. Frond morphological and I2-KI staining results were observed with dissecting microscope with Leica M165 FC (Germany). Fronds were also prepared in paraffin slices to observe and compare the tissue structure by optical microscopy after staining with Safranin and Fast Green according to the method described by Li [53]. SEM and TEM were prepared and observed by Hangzhou Yanqu Information & Technology Co. Ltd.
For SEM samples, plant tissues were collected and fixed overnight with 2.5% glutaraldehyde solution at 4 °C. The fixed solution was poured out, rinsing the sample with 0.1 M, pH7.0 phosphoric acid buffer for three times, each time for 15 min. The samples were fixed with 1% osmium solution for 1–2 h. Osmium acid waste liquid was carefully taken out, rinsing the sample with 0.1 M, pH7.0 phosphoric acid buffer for three times, each time for 15 min. The samples were dehydrated with ethanol solution of gradient concentration (including 30%, 50%, 70%, 80%, 90% and 95%). Each concentration was treated for 15 min, and then treated with 100% ethanol twice for 20 min. The mixture of ethanol and isoamyl acetate (v/v = 1/1) was used to treat the sample for 30 min; then isoamyl acetate was used to treat the sample for 1 h or overnight. The treated samples were observed with Hitachi SU8010 (Japan) after critical point drying and coating.
For the preparation of TEM samples. The former several steps were same with the SEM samples preparation until treating with 100% ethanol twice for 20 min and finally treated with pure acetone for 20 min. The mixture of embedment and acetone (v/ v = 1/1) was used to treat the sample for 1 h, then the mixture of embedment and acetone (v/v = 3/1) was used to treat the sample for 3 h. The sample treated with pure embedding agent was buried overnight; the embedded sample was embedded and heated at 70 °C overnight, then sliced in Leica EM uc7 ultra-thin microtome, and 70–90 nm sections were obtained. The sections were stained with lead citrate solution and 50% alcohol saturated solution of uranyl acetate for 5–10 min respectively, and then observed with HITACHI H-7650 (Japan).
RNA-Seq methods and sequence analysis
As described in method, duckweed strain and treatment, there are 3 flasks repeat cultures for control and MH treatment samples prepared for the RNA-Seq, respectively. Whole plants were ground in liquid nitrogen after 8 days culturing, and total RNA was extracted using RNeasy® Plant Mini Kit (Qiagen) according to the manufacturer’s protocol. Integrity of the RNA was confirmed by gel electrophoresis and total RNA was then submitted to Beijing Genomics Institute (BGI-Shenzhen), Shenzhen, China (http://www.genomics.cn) for RNA-Seq, library construction and sequencing. The sample RNA was sequenced with the DNBSEQ platform, the number of detected genes and average sample-to-genome alignment rate was shown in Additional file 7: Table S3. Reference Genome Alignment Analysis software was HISAT (Hierarchical Indexing for spiced Alignment of Transcripts (version: V2.1.0). Clean reads were compared to reference gene sequences using BOWTIE2 (version: V2.2.5), and then RSEM was used to calculate gene and transcript expression levels. The analysis of the filtered Reads quality shown in Additional file 8: Table S4. Differential expressed genes between samples were identified, and clustering analysis and functional annotations were done according to the annotation information from S. polyrrhiza kindly shared by Professor Wenqing Wang [54]. The significance of the differential expression of genes was defined by the bioinformatics service of BGI according to the combination of the absolute value of log2-Ratio ≥ 1 and false discovery rate (FDR) ≤ 0.001. KOG functional classification, Gene Ontology (GO) and pathway annotation and enrichment analyses were based on the Gene Ontology Database (http://www.geneontology.org/) and KEGG pathway database (http://www.genome.jp/kegg/), respectively. The software Cluster and Java Treeview were used for hierarchical cluster analysis of gene expression patterns [55, 56].
RNA extraction and qRT-PCR
RNA was isolated from Spirodela polyrrhiza 7498 which were treated with 75 μmol/L MH for different days, using an Eastep Super Total RNA Extraction Kit (Promega, shanghai, China). After reverse transcription was performed, the cDNAs for INO1 (Inositol-3-phosphate synthase, used as a control), APL1, APL2, APL3, GBSS, SSS, SBE, DBE, PEPC and MDH were amplified using qRT-PCR with specific primers (in Additional file 9: Table S5). qRT-PCR was performed on an iCycler Thermal Cycler (Bio-Rad iQ5, Hercules, CA, USA) using TB Green Premix Ex TaqII (Code No. RR420A:TB Green® Premix Ex Taq™, Dalian Takara, Dalian, China) according to the manufacturer’s protocol. The reaction mixture was heated to 95 °C for 30 s followed by 40 PCR cycles at 95 °C for 5 s, 58 °C for 30 s and 72 °C for 30 s. All primer pair efficiencies were between 95 and 105%, and the individual efficiency values were considered in the calculation of normalized relative expression. The difference in the relative expression levels of APL1, APL2, APL3, GBSS, SSS, SBE, DBE, PEPC and MDH was calculated using the 2−ΔΔCT method after the data were normalized to INO1. All values are shown as the mean ± standard error of the mean using at least three biological replicates.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- MH:
-
Maleic hydrazide
- ABA:
-
Abscisic acid
- 6-BA:
-
6-Benzylamino-purine
- AIP:
-
2-Aminoindan-2-phosphonic acid
- DEGs:
-
Differentially expressed genes
- AUX1/LAX:
-
Auxin influx carrier
- Aux/IAA:
-
Auxin/indole-3-acetic acid
- SAUR:
-
Small auxin up-regulated RNA
- ATP:
-
Adenosine triphosphate
- NADPH:
-
Reduced form of nicotinamide adenine dinucleotide phosphate
- TIM:
-
Triosephosphate isomerase
- PEPC:
-
Phosphoenolpyruvate carboxylase
- MDH:
-
NADP-malate dehydrogenase
- AGPase:
-
ADP-Glc pyrophosphorylase
- GH3:
-
Gretchen Hagen 3
- FW:
-
Fresh weight
- DW:
-
Dry weight
- FDR:
-
False discovery rate
- GO:
-
Gene Ontology
- INO1 :
-
Inositol-3-phosphate synthase
- TEM:
-
Transmission electron microscope
- SEM:
-
Scanning electronic microscope
References
Hillman WS, Culley DD Jr. The Uses of Duckweed: The rapid growth, nutritional value, and high biomass productivity of these floating plants suggest their use in water treatment, as feed crops, and in energy efficient farming. Am Sci. 1978;66:442–51.
Lemon GD, Posluszny U, Husband BC. Potential and realized rates of vegetative reproduction in Spirodela polyrhiza, Lemna minor, and Wolffia borealis. Aquat Bot. 2001;70(1):79–87. https://doi.org/10.1016/S0304-3770(00)00131-5.
Cheng JJ, Stomp AM. Growing duckweed to recover nutrients from wastewaters and for production of fuel ethanol and animal feed. Clean: Soil, Air, Water. 2009;37(1):17–26.
Land R, Fao WDD, Sess AGL. Food and Agriculture Organization of the United Nations, FAO, Statistics Division. http://faostat3.fao.org/. Accessed 10 Jan 2015.
Tao X, Fang Y, Xiao Y, Jin Y, Ma X, Zhao Y, He K, Zhao H, Wang H. Comparative transcriptome analysis to investigate the high starch accumulation of duckweed (Landoltia punctata) under nutrient starvation. Biotechnol Biofuels. 2013;6:72–87.
Wang X, Cui W, Hu W, Feng C. Abscisic acid-induced starch accumulation in bioenergy crop duckweed Spirodela polyrrhiza. Bioenerg Res. 2017;10:417–26.
McCombs P, Ralph R. Protein, nucleic acid and starch metabolism in the duckweed Spirodela oligorrhiza treated with cytokinins. Biochem J. 1972;129:403–17.
Janas KM, Osiecka R, Zoń J. Growth-retarding effect of 2-aminoindan-2-phosphonic acid on Spirodela punctate. J Plant Growth Regul. 1998;17:169–72.
Liu Y, Fang Y, Huang M, Jin Y, Sun J, Tao X, Zhang G, He K, Zhao Y, Zhao H. Uniconazole-induced starch accumulation in the bioenergy crop duckweed (Landoltia punctata) II: transcriptome alterations of pathways involved in carbohydrate metabolism and endogenous hormone crosstalk. Biotechnol Biofuels. 2015;8(64):1–12.
Liu Y, Chen X, Wang X, Fang Y, Zhang Y, Huang M, Zhao H. The influence of different plant hormones on biomass and starch accumulation of duckweed: a renewable feedstock for bioethanol production. Renew Energy. 2019;138:659–65.
Schoene DL, Hoffmann OL. Maleic hydrazide, a unique growth regulant. Science. 1949;109:588–90.
Naylor AW, Davis EA. Maleic hydrazide as a plant growth inhibitor. Bot Gaz. 1950;112:112–26.
Lee W, Tsung-Lin L, Pi-Chiou C, Shin-Shou C. High performance liquid chromatographic determination of maleic hydrazide residue in potatoes. J Food Drug Anal. 2001;9:167–72.
Renaud J, Keller Y, Vuillaume G. Determination of maleic hydrazide residues in cured tobacco by gas chromatography. J Chromatogr. 1992;604:243–6.
Crafts-Brandner SJ, Sutton TG. Effect of maleic hydrazide1 on photosynthesis, carbohydrate metabolism, and senescence of burley tobacco (Nicotiana tabacum L.). Field Crop Res. 1994;37(2):129–35.
Wang W, Haberer G, Gundlach H, Gläßer C, Nussbaumer T, Luo MC, Lomsadze A, Borodovsky M, Kerstetter RA, Shanklin J, Byrant D, Mockler T, Appenroth KJ, Grimwood J, Jenkins J, Chow J, Choi C, Adam C, Cao XH, Fuchs J, Schubert I, Rokhsar D, Schmutz J, Michael TP, Mayer KFX, Messing J. The Spirodela polyrhiza genome reveals insights into its neotenous reduction, fast growth and aquatic lifestyle. Nat Comm. 2014a;5:3311–34.
Djandja OS, Yin L, Wang Z, Guo Y, Zhang X, Duan P. Progress in thermochemical conversion of duckweed and upgrading of the bio-oil: A critical review. Sci Total Environ. 2021;769:144660.
Xu H, Yu Q, Shi Y, Hua X, Tang H, Yang L, Ming R, Zhang J. PGD: pineapple genomics database. Horticult Res. 2018;66(5):1–9.
Chen J, Huang B, Li Y, Du H, Gu Y, Liu H, Zhang J, Huang Y. Synergistic influence of sucrose and abscisic acid on the genes involved in starch synthesis in maize endosperm. Carbohydr Res. 2011;346(13):1684–91.
Doroy FS, Acedo AL Jr. Effects of preharvest spray with maleic hydrazide, ethephon, and paclobutrazol on yam tuber dormancy ishs. Acta Horticult. 2013. https://doi.org/10.17660/ActaHortic.2013.989.46.
Ilić ZS. Maleic hydrazide residues in the onion bulbs induce dormancy and hamper sprouting for long periods. J Food Agric Environ. 2011;9(1):113–8.
Roberts BR, Domir SC. The influence of daminozide and maleic hydrazide on growth and net photosynthesis of silver maple and American sycamore seedlings. Sci Hortic. 1983;19(3–4):367–72.
Yamamoto M, Yamamoto KT. Differential effects of 1-naphthaleneacetic acid, indole-3-acetic acid and 2,4-dichlorophenoxyacetic acid on the gravitropic response of roots in an auxin-resistant mutant of arabidopsis, aux1. Plant Cell Physiol. 1998;39(6):660–4.
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13(12):2809–22.
Lavy M, Estelle M. Mechanisms of auxin signaling. Development. 2016;143:3226–9.
Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA. 2006;103:236–41.
Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM. The SAUR19subfamily of small auxin-up RNA genes promote cell expansion. Plant J. 2012;70:978–90.
Hao X, Tang H, Wang B, Wang L, Cao H, Wang Y, Zeng J, Fang Sh, Chu J, Yang Y, Wang X. Gene characterization and expression analysis reveal the importance of auxin signaling in bud dormancy regulation in tea plant. J Plant Growth Regul. 2019;38:225–40.
Yuan H, Zhao K, Lei H, Shen X, Liu Y, Liao X, Li T. Genome wide analysis of the GH3 family in apple (Malus x domestica). BMC Genomics. 2013;14:297.
Wang W, Wu Y, Messing J. RNA-Seq transcriptome analysis of Spirodela dormancy without reproduction. BMC Genomics. 2014;15:60.
Mcllrath WJ. Response of the cotton plant to maleic hydrazide. Am J Bot. 1950;37:816–9.
Mondal S, Sukul Chunari S, Sukul NC. A new homeopathic potency promotes growth and yield of rice: a field trial. Int J High Dilution Res. 2013;12(44):115.
Koske TJ, Svec LV. Some effects of maleic hydrazide on light reactions of photosynthesis in isolated chloroplasts from Phaseolus vulgaris plants. Can J PlantSci. 1975;55:145–9.
Greulach VA. The effect of maleic hydrazide on some water relation of plants. J Elisha Mitchell Sci Soc. 1951;67:179–80.
Struckmeyer BE. The effect of maleic hydrazide on the anatomical structure of croft easter lilies. Am J Bot. 1953;40(1):25–9.
Voznesenskaya EV, Franceschi KO, Freitag H, Edwards GE. Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nature. 2001;414:543–6.
Voznesenskaya EV, Franceschi VR, Kiirats O, Artyusheva EG, Freitag H, Edwards GE. Evidence for C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae). Plant J. 2002;31:649–62.
McKown AD, Dengler NG. Shifts in leaf vein density through accelerated vein formation in C4 Flaveria (Asteraceae). Ann Bot. 2009;104:1085–98.
Dengler N, Nelson T. Leaf structure and development in C4 plants. In: Sage RF, Monson RK, editors. C4 plant biology. San Diego: Academic Press; 1999.
Maai E, Shimada S, Yamada M, Sugiyama T, Miyake H, Taniguchi M. The avoidance and aggregative movements of mesophyll chloroplasts in C4 monocots in response to blue light and abscisic acid. J Exp Bot. 2011;62:3213–21.
Reiskond JB. Evidence that inducible C4 type photosynthesis is a chloroplastic CO2-concentrating mechanism in Hydrilla, a submersed monocoat. Plant Environ. 1997;20:211–20.
Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of ricerationale and feasibility. Curr Opin Plant Biol. 2008;11:228–31.
Ueno O. Induction of Kranz anatomy and C4-like biochemical characteristics in a submerged amphibious plant by abscisic acid. Plant Cell. 1998;10:571–83.
Zeevaart JAD, Creelman RA. Metabolism and physiology of abscisic acid. Ann Rev Plant Phys. 1988;39:439–73. https://doi.org/10.1146/annurev.arplant.
Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26(4):421–33.
Wang W, Messing J. Analysis of ADP-glucose pyrophosphorylase expression during turion formation induced by abscisic acid in Spirodela polyrhiza (greater duckweed). BMC Plant Biol. 2012;12:5.
Zhang J, Chen J, Yi Q, Hu Y, Liu H, Liu Y, Huang Y. Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol Biol. 2014;84:359–69.
Huang H, Xie S, Xiao Q, Wei B, Zheng L, Wang Y, Cao Y, Zhang X, Long T, Li Y, Hu Y, Yu G, Liu H, Liu Y, Huang Z, Zhang J, Huang Y. Sucrose and ABA regulate starch biosynthesis in maize through a novel transcription factor, ZmEREB156. Sci Rep. 2016;6:27590.
Chen J, Yi Q, Cao Y, Wei B, Zheng L, Xiao Q, Xie Y, Gu Y, Li Y, Huang H, Wang Y, Hou X, Long T, Zhang J, Liu H, Liu Y, Yu G, Huang Y. ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J Exp Bot. 2016;67:1327–38.
Zhang Z, Dong J, Chen J, Wu Y, Messing J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. PNAS. 2019;116(23):11223–8.
Wang Y, Kandeler R. Promotion of flowering by a tumor promoter. J Plant Physiol. 1994;144:710–3.
Wintermans JFGM, De Mots A. Spectrophotometric characteristics of chlorophyll a and b and their pheophytins in ethanol. Biochimica Biophysica Acta. 1965;109:448–53.
Li Z. Plant Slicing technology. Beijing: Science Press; 1987.
Ana D, Zhou Y, Li C, Xiao Q, Wang T, Zhang Y, Wu Y, Li Y, Chao DY, Messing J, Wang W. Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. PNAS. 2019;116(38):18893–9.
de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. https://doi.org/10.1093/bioinformatics/bth078.
Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. https://doi.org/10.1093/bioinformatics/bth349.
Acknowledgements
We thank a lot for nice help of Professor Wenqing Wang. She kindly shared the information of the genome and genes annotation of S. polyrrhiza 7498 with us. We would also like to deeply thank Pro. Henrik V. Scheller for revising the manuscript.
Funding
The research was supported by the Projects of National Natural Science Foundation of China (NSFC, No. 31970365).
Author information
Authors and Affiliations
Contributions
YZ designed the whole study and edited the entire draft manuscript. XL, XG participated in all experiments, data collection and manuscript editing. JS, XJ, GF, GS, BX participated in some experiments or data analysis. YW participated in helpful discussion the manuscript and helped in its editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors agree to submission and publication of this manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Comparison of root length in S. polyrrhiza 7498.
Additional file 2: Figure S2.
Pathway functional enrichment of DEGs.
Additional file 3: Table S1.
Expression analysis of genes involved in developmental progress by GO classification.
Additional file 4: Table S2
. Expression analysis of genes up-regulated in light reactions.
Additional file 5: Figure S3.
Main pathway expression heatmap of DEGs classified by KEGG.
Additional file 6: Figure S4.
Plant hormone signal transduction induced by MH in S. polyrrhiza.
Additional file 7: Table S3.
Number of detected genes and alignment rate of RNA-Seq analysis.
Additional file 8: Table S4.
Summary of sequence read alignments to reference genome.
Additional file 9: Table S5.
Primers used in the qRT-PCR.
Additional file 10: Table S6.
Gene information of carbon metabolism pathway expression heatmap.
Additional file 11: Table S7.
Gene information of glycolysis gluconeogenesis.
Additional file 12: Table S8.
Gene information of glycerolipid metabolism pathway expression heatmap.
Additional file 13: Table S9.
Gene information of MAPK pathway expression.
Additional file 14: Table S10.
Gene information of carbon fixation pathway.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhu, Y., Li, X., Gao, X. et al. Molecular mechanism underlying the effect of maleic hydrazide treatment on starch accumulation in S. polyrrhiza 7498 fronds. Biotechnol Biofuels 14, 99 (2021). https://doi.org/10.1186/s13068-021-01932-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-021-01932-y