Abstract
The Wheat@URGI portal has been developed to provide the international community of researchers and breeders with access to the bread wheat reference genome sequence produced by the International Wheat Genome Sequencing Consortium. Genome browsers, BLAST, and InterMine tools have been established for in-depth exploration of the genome sequence together with additional linked datasets including physical maps, sequence variations, gene expression, and genetic and phenomic data from other international collaborative projects already stored in the GnpIS information system. The portal provides enhanced search and browser features that will facilitate the deployment of the latest genomics resources in wheat improvement.
Similar content being viewed by others
Background
The International Wheat Genome Sequencing Consortium (IWGSC) [1] is an international collaborative group of growers, academic scientists, and public and private breeders that was established to generate a high-quality reference genome sequence of the hexaploid bread wheat, and to provide breeders with state-of-the-art tools for wheat improvement. The vision of the consortium is that the high-quality, annotated ordered genome sequence integrated with physical maps will serve as a foundation for the accelerated development of improved varieties and will empower all aspects of basic and applied wheat science to address the important challenge of food security. A first analysis of the reference sequence produced by the consortium (IWGSC RefSeq v1.0) was recently published [2].
To ensure that wheat breeding and research programs can make the most of this extensive genomic resource, the IWGSC endorsed the establishment of a data repository at URGI (Unité de Recherche Génomique Info/research unit in genomics and bioinformatics) from INRA (Institut National de la Recherche Agronomique/French national institute for agricultural research) to develop databases and browsers with relevant links to public data available worldwide. The IWGSC data repository is thus hosted by URGI to support public and private parties in data management as well as analysis and usage of the sequence data. Wheat functional genomics (expression, methylation, etc.), genetic, and phenomic data have increased concurrently, requiring the development of additional tools and resources to integrate different data for biologists and breeders. To manage this escalation of data, URGI has built this data repository for the wheat community with the following specific aims: (1) to store resources for which no public archive exists (e.g. physical maps, phenotype information); (2) to enable pre-publication access to specific datasets (e.g. sequence assemblies and annotations, physical maps, markers); and (3) to enable rapid release of integrated resources upon publication. The repository has been designed in accordance with the “FAIR” principles [3] to ensure that the data are Findable, Accessible, Interoperable, and Reusable. To address the challenge of integrating diverse data types from multiple sources, URGI employs solutions that provide enhanced features for data exploration, mining, and visualisation using the GnpIS information system [4] combined with a high level of data interoperability.
Here we describe the data and tools currently available through the Wheat@URGI portal [5], the primary resource for the reference sequence of the bread wheat genome (IWGSC RefSeq v1.0) and other IWGSC wheat genomic data. The links to functional genomics, genetic, and phenomic data from many other large wheat projects are also described.
A wealth of data is available throughout the Wheat@URGI portal
The data hosted by the Wheat@URGI portal are available through flat files stored in the IWGSC data repository and through the GnpIS information system [4]. GnpIS encompasses a set of integrated databases to manage genomic data using well-known tools such as the Basic Local Alignment Search Tool (BLAST), JBrowse, GBrowse, and InterMine, and an in-house database called GnpIS-coreDB developed by URGI to manage genetic and phenomic data.
IWGSC data
Through its concerted efforts to achieve a high-quality, functionally annotated reference wheat genome sequence, the IWGSC has developed a variety of resources for the bread wheat (Triticum aestivum L.) accession Chinese Spring. The IWGSC data hosted in the Wheat@URGI portal within the IWGSC data repository are shown in Table 1. They fall into four broad categories: (1) physical maps, (2) sequence assemblies and annotations, (3) gene expression data, and (4) variation data.
Physical maps
Physical maps assembled by IWGSC scientists for the 21 bread wheat chromosomes, based on high-information-content (fluorescence) fingerprinting (HICF) [6] or Whole Genome Profiling (WGP™) [7] of flow-sorted chromosome or chromosome-arm specific bacterial artificial chromosome (BAC) libraries, are stored and displayed. The positions of individual BAC clones, markers, and deletion bins are mapped onto physical contigs. The database maintains all released versions of each physical map with the software used to produce the BAC clone assemblies (FingerPrinted Contig (FPC) [8] or Linear Topological Contig (LTC) [9]), information from the group that produced the map, and a link to order the BAC clones from the French plant genomic resource centre [10].
Sequence assemblies and annotations
The IWGSC wheat genome sequence assemblies available for download, BLAST [11], and display in genome browsers include the draft survey sequence assemblies released in 2014 (IWGSC Chromosome Survey Sequencing (CSS) v1) and two improved versions (CSS v2 and v3) [12] and the chromosome 3B reference sequence (the first reference-quality chromosome sequence obtained by the consortium) [13]. Associated with these assemblies are the virtual gene order map generated for the CSS (Genome Zipper), the population sequencing (POPSEQ) data used to order sequence contigs on chromosomes [14], and mapped marker sets. The reference sequence of the bread wheat genome (IWGSC RefSeq v1.0, 14.5 Gb assembly with super scaffold N50 of 22.8 Mb) was obtained by integrating whole genome shotgun Illumina short reads assembled with NRGene’s DeNovoMAGIC™ software with the wealth of IWGSC map and sequence resources [2]. IWGSC RefSeq v1.0 is available for download, BLAST, and browser display. Users can access the whole genome, pseudomolecules of individual chromosomes or chromosome arms, and scaffolds with the structural and functional annotation of genes, transposable elements, and non-coding RNAs generated by the IWGSC. In addition, mapped markers as well as alignments of nucleic acid and protein evidence supporting the annotation are available. Updated versions of the annotation for genes belonging to specific gene families or regions of specific chromosomes that have been manually annotated (ca. 3685 genes) can be found in the IWGSC RefSeq v1.1 annotation.
In addition to the bread wheat sequence, the IWGSC also assembled seven diploid and tetraploid wheat-related species: Triticum durum cv. Cappelli, Triticum durum cv. Strongfield, Triticum durum cv. Svevo, Triticum monococcum, Triticum urartu, Aegilops speltoides, Aegilops sharonensis [12]. Download and BLAST are available for these data.
Expression data
RNA-Seq expression data are available as read counts and transcripts per kilobase million (TPM) for the IWGSC RefSeq v1.1 annotation. It is a transcriptome atlas developed from 850 RNA-Seq datasets representing a diverse range of tissues, development stages, and environmental conditions [15].
Variation data
These data consist of downloadable variant call format (VCF) files from genotyping by sequencing (GBS) and whole exome capture experiments of 62 diverse wheat lines [16] and of the IWGSC 3,289,847 single nucleotide polymorphisms (SNPs) [17]. Moreover, varietal SNPs aligned on IWGSC RefSeq v1.0 can be displayed in the browser and downloaded.
Wheat gene pool
As well as IWGSC resources, URGI also hosts other open access wheat sequence data to facilitate research into the wheat gene pool. Sequence assemblies available for download and BLAST include the bread wheat whole genome sequence assembly Triticum aestivum TGACv1 [18] and the diploid progenitor of Aegilops tauschii [19].
Genetic and phenomic data
In addition to sequence data, the Wheat@URGI portal hosts, within GnpIS-coreDB, several sets of genetic and phenomic wheat data [20] that have been produced from French, European, and international projects since 2000 [21]. A significant amount of these data is available without restriction. However, access to restricted data can be obtained through a material transfer or intellectual property agreement. Table 2 presents the types and number of genetic and phenomic data hosted in the GnpIS-coreDB database.
Genetic information corresponds to genetically mapped markers, quantitative trait loci (QTLs), genetic resources (germplasms), and genetic studies (genome-wide association studies (GWASs)). Genomic information consists of variations from SNP discovery experiments, genotyping, comparative genomics (synteny), and expression data (microarray, RNA-Seq). Phenomic data are available as whole trials including phenotypic and environmental observations recorded using variables from ontologies with Minimum Information About a Plant Phenotyping Experiment (MIAPPE) [22] compliant metadata.
Germplasm data were mainly provided by the French small grain cereals genebank maintained by INRA at Clermont-Ferrand [23] but also by partners of several European Union (EU) projects. They were linked together with related genotyping or phenotyping characterisation data. Generally, genetic and phenomic data have been produced by INRA and its partners in large collaborative projects.
Browsing and searching a large variety of integrated data
Data can be easily accessed through the Wheat@URGI portal [5] using (1) tabs at the top of the webpages allowing access in one click to the data, tools, and projects descriptions as well as the IWGSC data repository, (2) direct links from the home page to the different data types (e.g. clicking on “Physical maps” opens the physical maps browser), and (3) data discovery and InterMine [24] tools on the home page.
The IWGSC data repository [25] allows one to access consortium data by (1) clicking on a chromosome to open a pop-up menu with all related data (e.g. 3A, 3B, etc.), or (2) using the tabs on the left to access the data by type (e.g. Assemblies, Annotations, etc.) or useful links to the news, the BLAST tool, frequently asked questions, the access status of the data (e.g. open access), etc.
Physical maps browser
GBrowse [26] displays the physical maps generated by the IWGSC members [27]. A clickable image on the top of the browser gives access to all versions of the physical map for each chromosome. The browser displays physical contigs, BACs, deletion bins, and markers. From the BACs track, it is possible to order BAC clones directly at the INRA French plant genomic resource centre [10]. From the BACs and markers tracks, one can go directly to the corresponding region in the IWGSC RefSeq v1.0 browser.
Genome browser and BLAST
IWGSC RefSeq v1.0 is displayed in a dedicated JBrowse [28, 29]. The “markers track” provides links to additional genetic information stored in GnpIS-coreDB which includes access to the position of the marker in centimorgans (cM) on genetic maps and to the overlapping QTLs. The most popular tool of the IWGSC data repository is the BLAST search tool (476,000 BLAST searches launched in 2017, Additional file 1: Table S1, Additional file 1: Figure S1). All of the wheat sequences available on the Wheat@URGI portal are indexed for BLAST search (see [30] for the complete list). A set of databanks can be selected, e.g. IWGSC RefSeq v1.0 and IWGSC CSS v3 for a given chromosome. The result is presented in a classical tabular format with (1) links to download the data (matching contigs and high scoring pairs (HSP)), (2) links on the genome browsers directly zooming in on the matching region, and (3) external links to Ensembl Plants [31].
Genetic and phenomic data in GnpIS-coreDB
The IWGSC sequence data are linked to genetic and phenomic data within the GnpIS information system [4]. This integration is organised around key data, also called “pivot data” as they are pivotal objects which allow integration between data types. The key objects used to link genomic resources to genetic data are markers and QTLs. Markers are mapped on the genome sequences and provide information on neighbour genes and their function. They also have links to GnpIS-coreDB genetic maps, QTLs, genotyping, and GWAS data. Additional information on the marker itself can be found regarding the marker type (e.g. simple sequence repeats (SSRs), Diversity Arrays Technology (DArT)), the primer sequences for PCR amplification, and SNP details (including the flanking sequences) when relevant. QTLs link the genetic data to the phenomic data in GnpIS-coreDB and to synteny data displayed by the PlantSyntenyViewer tool [32, 33].
The accessions (i.e. germplasm) and the variables (i.e. observed traits), described with dedicated ontologies, are other important key data for genetic studies as they allow linking phenotype data to genetic associations and QTLs through traits. The genetic resources stored in GnpIS-coreDB displays the unambiguous identification of the accession used (with Digital Object Identifier (DOI)) and a rich set of associated data following the Multi-Crop Passport Descriptors (MCPD, [34]) standard: picture, synonyms, descriptors, geolocation of the sites (origin, collecting, and evaluation), the collections or panels it belongs to, and the holding stock centre with a link to order the accession when possible. The phenotype data include traceability on trials with timing (e.g. year, temporal series), location, and environment including soil and cultural practices. The phenotype and environment variables follow the Crop Ontology format [35], which includes unique identifiers for each variable, composed of a trait (e.g. grain yield, plant height, spike per plant, etc.), a method (e.g. measurement, computational), and a scale (e.g. International System of Units, notation scale). All these data are displayed in the GnpIS-coreDB web interface and can be downloaded in different file formats, all compliant with the MIAPPE standard [22].
Mining and data discovery tools
To complete these already rich integrated datasets, a gene-centric data warehouse, the WheatMine, has been set up using the well-established InterMine tool [24]. The gene card displays gene function, gene ontology terms, and overlapping genomic features. WheatMine [36] provides access to the IWGSC RefSeq v1.0 and v1.1 annotation data (genes, messenger RNA, polypeptides, transposable elements), markers and, through pivotal objects, to genetic data (QTL, metaQTL). It is also possible to navigate from a gene card to its position on the wheat genome browser or to relevant marker details in GnpIS-coreDB.
Figure 1 summarises the concept and the tools to navigate through the key data in GnpIS.
Finally, to facilitate data search and access to this wealth of data, we developed a data discovery tool, which, similar to a google search, allows the user to enter keywords or terms to find all the matching information in the various data warehouses. The results are presented in a table with details on the matches (database source, type, species, description) and a direct link to the feature (e.g. a gene in a browser, a marker page in GnpIS-coreDB, etc.).
Figure 2 highlights a practical use case describing how to use the Wheat@URGI portal to go from a gene sequence to find the related genetic studies.
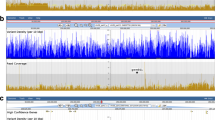
Screenshots of the web interfaces for a practical use case to explore all the genomic data in the vicinity of a dedicated gene and find out if there are genetic studies pointing to this genomic region. Search the gene name (e.g. TraesCS5A01G033100) in the data discovery tool (1A, [5]) or BLAST the sequence of the gene against IWGSC RefSeq v1.0 (1B, [30]). The results are displayed in Tables (2A, 2B) with links to JBrowse directly zooming in on the gene [48]. Explore the region around the gene to find a marker (3, e.g. wPt-4131_PURr). By clicking on the marker, display additional information stored in GnpIS-coreDB (4, [49]) showing that the marker is used in GWAS experiments (5A, [50]) and is linked to a QTL (5B, e.g. GY_ml.8.Np_5A, [51]). From the trait description of this QTL (6, [52]), display all the phenotyping experiments performed on this trait (7, e.g. grain yield, [53])
Conclusion and future directions
The Wheat@URGI portal hosts and gives access to essential, high-quality wheat data from the IWGSC, European, and international projects. Furthermore, its added value is that it integrates different data types altogether (genomics, genetics, and phenomics) and provides dedicated tools to explore them.
As new wheat resources such as GWASs, genomic selection, and pan-genome data are generated in the frame of ongoing projects, GnpIS will allow their management and integration with other data already available in the information system, linking new upcoming data to this central IWGSC genomic resource.
At a wider scale, an expert working group (EWG) of the international Wheat Initiative has built an international wheat information system, called WheatIS, with the aim of providing a single-access web-based system to all available wheat data resources and bioinformatics tools [37]. The Wheat@URGI portal is a major node of the WheatIS federation that exposes genomic, genetic, and phenomic integrated data to the community. The WheatIS data discovery tool allows a one-stop search in GnpIS [4] (including IWGSC browsers, InterMine and GnpIS-coreDB), from URGI; Ensembl Plants, from the European Bioinformatics institute (EMBL-EBI) [31]; CrowsNest [38], at the Plant Genome and Systems Biology (PGSB) group; CR-EST [39], GBIS [40] and MetaCrop [41], from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK); The Triticeae Toolbox (Triticeae Coordinated Agricultural Product); CIMMYT DSpace and Dataverse (International Maize and Wheat Improvement Center (CIMMYT)); Gramene [42], from Cold Spring Harbor Laboratory (CSH), Ohio State University (OSU), and EMBL-EBI; Cropnet, from the Institute of Plant Genetics of the Polish Academy of Sciences (IPGPAS); WheatPan [43], from the University of Western Australia (UWA); and GrainGenes [44], US Department of Agriculture (USDA).
Figure 3 presents the WheatIS ecosystem.
Data integration is fundamental for researchers and breeders who want to use genomic information to improve wheat varieties. However, the diversity of data types and the concomitant lack of data harmonisation and standards hamper cross-referencing and meta-analysis. A joint action between the WheatIS EWG and a group of linked data scientists created the Wheat Data Interoperability Working Group under the Research Data Alliance (RDA) umbrella [45] to help tackle this difficult issue [46]. The Wheat@URGI portal continuously evolves its repository to follow the standard recommendations [47].
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- BLAST:
-
Basic Local Alignment Search Tool
- CIMMYT:
-
International Maize and Wheat Improvement Center
- cM:
-
Centimorgan
- CSH:
-
Cold Spring Harbor Laboratory
- CSS:
-
Chromosome Survey Sequencing
- DArT:
-
Diversity Arrays Technology
- DOI:
-
Digital object identifier
- EMBL-EBI:
-
European Bioinformatics Institute
- EWG:
-
Expert working group
- FAIR:
-
Findable, accessible, interoperable, reusable
- FPC:
-
FingerPrinted Contig
- GBS:
-
Genotyping by sequencing
- GWAS:
-
Genome-wide association study
- HICF:
-
High-information-content fingerprinting
- HSP:
-
High scoring pairs
- INRA:
-
Institut National de la Recherche Agronomique/French national institute for agricultural research
- IPGPAS:
-
Institute of Plant Genetics of the Polish Academy of Sciences
- IPK:
-
Leibniz Institute of Plant Genetics and Crop Plant Research
- IWGSC:
-
International Wheat Genome Sequencing Consortium
- LTC:
-
Linear topological contig
- MCPD:
-
Multi-Crop Passport Descriptor
- MIAPPE:
-
Minimum Information About a Plant Phenotyping Experiment
- OSU:
-
Ohio State University
- PCR:
-
Polymerase chain reaction
- PGSB:
-
Plant Genome and Systems Biology (group)
- POPSEQ:
-
Population sequencing
- QTL:
-
Quantitative trait locus
- RDA:
-
Research Data Alliance
- RNA:
-
Ribonucleic acid
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
- TPM:
-
Transcripts per kilobase million
- URGI:
-
Unité de Recherche Génomique Info/research unit in genomics and bioinformatics
- USDA:
-
US Department of Agriculture
- UWA:
-
University of Western Australia
- VCF:
-
Variant call format
- WGP™:
-
Whole genome profiling
References
IWGSC website. http://www.wheatgenome.org/. Accessed 10 April 2018.
International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science, in press. DOI: https://doi.org/10.1126/science.aar7191.
Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3:160018.
Steinbach D, Alaux M, Amselem J, Choisne N, Durand S, Flores R, et al. GnpIS: an information system to integrate genetic and genomic data from plants and fungi. Database J Biol Databases Curation. 2013;2013:bat058.
Wheat@URGI portal. https://wheat-urgi.versailles.inra.fr. Accessed 10 April 2018.
Nelson WM, Bharti AK, Butler E, Wei F, Fuks G, Kim H, et al. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 2005;139:27–38.
Philippe R, Choulet F, Paux E, van Oeveren J, Tang J, Wittenberg AH, et al. Whole Genome Profiling provides a robust framework for physical mapping and sequencing in the highly complex and repetitive wheat genome. BMC Genomics. 2012;13:47.
Soderlund C, Humphray S, Dunham A, French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000;10:1772–87.
Frenkel Z, Paux E, Mester D, Feuillet C, Korol A. LTC: a novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes. BMC Bioinformatics. 2010;11:584.
French plant genomic resource centre. https://cnrgv.toulouse.inra.fr/en. Accessed 10 April 2018.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788.
Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345:1249721.
Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, et al. Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J Cell Mol Biol. 2013;76:718–27.
Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, Van Ex F, Pasha A, Khedikar Y, Robinson SJ, Cory AT, Florio T, Concia L, Juery C, Schoonbeek H, Steuernagel B, Xiang D, Ridout CJ, Chalhoub B, Mayer KFX, Benhamed M, Latrasse D, Bendahmane A, International Wheat Genome Sequencing Consortium, Wulff BBH, Appels R, Tiwari V, Datla R, Choulet F, Pozniak CJ, Provart NJ, Sharpe AG, Paux E, Spannagl M, Bräutigam A, Uauy C. 2018. The transcriptional landscape of polyploid wheat. Science, in press doi: https://doi.org/10.1126/science.aar6089.
Jordan KW, Wang S, Lun Y, Gardiner L-J, MacLachlan R, Hucl P, et al. A haplotype map of allohexaploid wheat reveals distinct patterns of selection on homoeologous genomes. Genome Biol. 2015;16:48.
Rimbert H, Darrier B, Navarro J, Kitt J, Choulet F, Leveugle M, et al. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS One. 2018;13:e0186329.
Clavijo BJ, Venturini L, Schudoma C, Accinelli GG, Kaithakottil G, Wright J, et al. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017;27:885–96.
Luo M-C, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, et al. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci U S A. 2013;110:7940–5.
GnpIS wheat data. https://wheat-urgi.versailles.inra.fr/Data. Accessed 10 Ap 2018.
Samson D, Legeai F, Karsenty E, Reboux S, Veyrieras J-B, Just J, et al. GénoPlante-Info (GPI): a collection of databases and bioinformatics resources for plant genomics. Nucleic Acids Res. 2003;31:179–82.
Ćwiek-Kupczyńska H, Altmann T, Arend D, Arnaud E, Chen D, Cornut G, et al. Measures for interoperability of phenotypic data: minimum information requirements and formatting. Plant Methods. 2016;12:44.
French small grain cereals genebank. https://www6.clermont.inra.fr/umr1095_eng/Teams/Research/Biological-Resources-Centre. Accessed 10 Ap 2018.
Kalderimis A, Lyne R, Butano D, Contrino S, Lyne M, Heimbach J, et al. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014;42:W468–72.
IWGSC data repository. https://wheat-urgi.versailles.inra.fr/Seq-Repository. Accessed 10 Apr 2018.
Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–610.
GnpIS: Physical map browser. https://urgi.versailles.inra.fr/gb2/gbrowse/wheat_phys_pub. Accessed 10 Apr 2018.
Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–8.
GnpIS: IWGSC RefSeq v1.0 browser. https://urgi.versailles.inra.fr/jbrowseiwgsc/gmod_jbrowse/?data=myData%2FIWGSC_RefSeq_v1.0. Accessed 10 Apr 2018.
GnpIS: IWGSC BLAST tool. https://urgi.versailles.inra.fr/blast_iwgsc/blast.php. Accessed 10 Apr 2018.
Bolser DM, Staines DM, Perry E, Kersey PJ. Ensembl Plants: integrating tools for visualizing, mining, and analyzing plant genomic data. Methods Mol Biol Clifton NJ. 2017;1533:1–31.
GnpIS PlantSyntenyViewer. https://urgi.versailles.inra.fr/synteny/synteny/viewer.do#form/datasetId=6. Accessed 10 Apr 2018.
Pont C, Murat F, Guizard S, Flores R, Foucrier S, Bidet Y, et al. Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J Cell Mol Biol. 2013;76:1030–44.
Multi-Crop Passport Descriptors V.2.1. https://www.bioversityinternational.org/e-library/publications/detail/faobioversity-multi-crop-passport-descriptors-v21-mcpd-v21/. Accessed 10 Apr 2018.
Shrestha R, Matteis L, Skofic M, Portugal A, McLaren G, Hyman G, et al. Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the crop ontology developed by the crop communities of practice. Front Physiol. 2012;3:326.
GnpIS: WheatMine tool. https://urgi.versailles.inra.fr/WheatMine. Accessed 10 Apr 2018.
WheaIS. http://www.wheatis.org/. Accessed 10 Apr 2018.
Spannagl M, Nussbaumer T, Bader KC, Martis MM, Seidel M, Kugler KG, et al. PGSB PlantsDB: updates to the database framework for comparative plant genome research. Nucleic Acids Res. 2016;44:D1141–7.
Künne C, Lange M, Funke T, Miehe H, Thiel T, Grosse I, et al. CR-EST: a resource for crop ESTs. Nucleic Acids Res. 2005;33:D619–21.
Oppermann M, Weise S, Dittmann C, Knüpffer H. GBIS: the information system of the German Genebank. Database J Biol Databases Curation [Internet]. 2015 [cited 2017 Sep 18];2015. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4423411/
Schreiber F, Colmsee C, Czauderna T, Grafahrend-Belau E, Hartmann A, Junker A, et al. MetaCrop 2.0: managing and exploring information about crop plant metabolism. Nucleic Acids Res. 2012;40:D1173–7.
Tello-Ruiz MK, Stein J, Wei S, Preece J, Olson A, Naithani S, et al. Gramene 2016: comparative plant genomics and pathway resources. Nucleic Acids Res. 2016;44:D1133–40.
Montenegro JD, Golicz AA, Bayer PE, Hurgobin B, Lee H, Chan C-KK, et al. The pangenome of hexaploid bread wheat. Plant J Cell Mol Biol. 2017;90:1007–13.
Carollo V, Matthews DE, Lazo GR, Blake TK, Hummel DD, Lui N, et al. GrainGenes 2.0. An improved resource for the small-grains community. Plant Physiol. 2005;139:643–51.
Wheat Data Interoperability Working Group of the Research Data Alliance. https://rd-alliance.org/groups/wheat-data-interoperability-wg.html . Accessed 10 Apr 2018.
Dzale Yeumo E, Alaux M, Arnaud E, Aubin S, Baumann U, Buche P, et al. Developing data interoperability using standards: a wheat community use case. F1000Research. 2017;6:1843.
Wheat Data Interoperability Working Group guidelines. https://ist.blogs.inra.fr/wdi/. Accessed 10 Apr 2018.
Use case: JBrowse link. https://urgi.versailles.inra.fr/jbrowseiwgsc/gmod_jbrowse/?data=myData%2FIWGSC_RefSeq_v1.0&loc=chr5A%3A30211546..30218715&tracks=DNA%2CHighConfidenceGenesv1.0%2CLowConfidenceGenesv1.0%2CrepeatRegion%2CrepeatMasker%2CDART_PUBLIC_SUMMARY&highlight=chr5A%3A30214481..30215784%20(−%20strand)%20(TraesCS5A01G033100). Accessed 11 July 2018.
Use case: marker link. https://urgi.versailles.inra.fr/GnpMap/mapping/id.do?action=MARKER&id=40393. Acccessed 11 July 2018.
Use case: GWAS link. https://urgi.versailles.inra.fr/association/association/viewer.do?#results/markerIds=40393. Accessed 11 July 2018.
Use case: QTL link. https://urgi.versailles.inra.fr/GnpMap/mapping/id.do?action=QTL&id=59588. Accessed 11 July 2018.
Use case: trait link. https://urgi.versailles.inra.fr/GnpMap/mapping/id.do?action=TRAIT&id=255. Accessed 11 July 2018.
Use case: phenotyping link. https://urgi.versailles.inra.fr/ephesis/ephesis/viewer.do#dataResults/traitCode=WIPO:0000217. Accessed 11 July 2018.
Acknowledgements
The authors would like to thank for their help or advice at various stages of the project, the following people from INRA-URGI: Véronique Jamilloux, Joëlle Amselem, Dorothée Charruaud, Guillaume Cornut, Laura Burlot, Florian Philippe, Nicolas Francillonne, Loïc Couderc, Daphné Verdelet, Baptiste Brault, Kirsley Chennen; from INRA-GDEC: Jacques Le Gouis, Gilles Charmet, François Balfourier, Pierre Sourdille, Catherine Ravel, François-Xavier Oury, Audrey Didier; from INRA-DIST: Esther Dzale, Sophie Aubin, Odile Hologne; and from INRA-Agronomie: Arnaud Gauffreteau.
Thanks to Isabelle Caugant (IWGSC), Hélène Lucas (Wheat Initiative), the International Wheat Genome Sequencing Consortium and its sponsors, the WheatIS expert working group, the URGI platform, and all the data submitters.
Funding
The development of the information system and the integration of wheat data were supported by INRA and several projects: BreedWheat (ANR-10-BTBR-03, France Agrimer, FSOV), Whealbi (EU FP7–613556), TriticeaeGenome (EU FP7-KBBE-212019), 3BSEQ (ANR-09-GENM-025, FranceAgrimer), and TransPLANT (EU FP7–283496).
Availability of data and materials
The open access data (including all the IWGSC data) are available through the Wheat@URGI portal [5].
Author information
Authors and Affiliations
Consortia
Contributions
MA, JR, TL, FA, and KE designed, developed, and filled the IWGSC data repository. MA, TL, RF, FA, CP, NM, SD, EK, CM, CG, MLo, MLa, DS, AFAB, and HQ designed, developed, and filled the GnpIS information system. FC, HR, PL, NG, JS, CF, IWGSC, and EP generated and submitted the data and gave feedback on the tools. MA, JR, EP, KE, AFAB, and HQ drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Supplementary data on software technologies and usage metrics. (PDF 541 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Alaux, M., Rogers, J., Letellier, T. et al. Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol 19, 111 (2018). https://doi.org/10.1186/s13059-018-1491-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13059-018-1491-4