Abstract
Background
Catalytic RNAs, or ribozymes, are regarded as fossils of a prebiotic RNA world that have remained in the genomes of modern organisms. The simplest ribozymes are the small self-cleaving RNAs, like the hammerhead ribozyme, which have been historically considered biological oddities restricted to some RNA pathogens. Recent data, however, indicate that small self-cleaving ribozymes are widespread in genomes, although their functions are still unknown.
Results
We reveal that hammerhead ribozyme sequences in plant genomes form part of a new family of small non-autonomous retrotransposons with hammerhead ribozymes, referred to as retrozymes. These elements contain two long terminal repeats of approximately 350 bp, each harbouring a hammerhead ribozyme that delimitates a variable region of 600–1000 bp with no coding capacity. Retrozymes are actively transcribed, which gives rise to heterogeneous linear and circular RNAs that accumulate differentially depending on the tissue or developmental stage of the plant. Genomic and transcriptomic retrozyme sequences are highly heterogeneous and share almost no sequence homology among species except the hammerhead ribozyme motif and two small conserved domains typical of Ty3-gypsy long terminal repeat retrotransposons. Moreover, we detected the presence of RNAs of both retrozyme polarities, which suggests events of independent RNA-RNA rolling-circle replication and evolution, similarly to that of infectious circular RNAs like viroids and viral satellite RNAs.
Conclusions
Our work reveals that circular RNAs with hammerhead ribozymes are frequently occurring molecules in plant and, most likely, metazoan transcriptomes, which explains the ubiquity of these genomic ribozymes and suggests a feasible source for the emergence of circular RNA plant pathogens.
Similar content being viewed by others
Background
The hypothesis of a prebiotic RNA world, where the first living organisms were based on RNA as both the genetic material and as catalyst [1–3], was strongly supported by the landmark discovery of ribozymes [4, 5]. It is thought that a few of those ancient ribozymes have remained in contemporary organisms performing key biological functions like the peptide bond formation by the ribosome [6], tRNA maturation by RNAse P [5] or mRNA splicing by the spliceosome [7], among others. In addition, there is an enigmatic group of small self-cleaving ribozymes that have been historically regarded as molecular oddities of some infectious RNA genomes, but recently have been found widespread in DNA genomes from all life kingdoms (for reviews see [8–10]). Among the few naturally occurring self-cleaving RNAs discovered, the hammerhead ribozyme (HHR) was the first and one of the best known members of the family. It is composed of a catalytic core of 15 conserved nucleotides surrounded by three double helixes (I to III), which adopt a γ-shaped fold where helix I interacts with helix II through tertiary interactions required for efficient in vivo activity [11–13]. Depending on the open-ended helix that connects the HHR motif to the flanking sequences, there are three possible circularly permuted forms named type I, II or III (Fig. 1). Originally described in small circRNA plant pathogens, like viral satellite RNAs [14] and viroids [15], the HHR catalyzes a self-cleavage transesterification reaction required during rolling-circle replication. A few HHRs were also described in DNA genomes of some unrelated eukaryotes like plants [16, 17], invertebrates [18, 19] and even vertebrates [20, 21], and were found mostly associated to repetitive DNAs. More recently, the widespread presence of HHRs in genomes from bacteria to eukaryotes has been reported [22–25], including human genomes [26], unveiling the HHR as an ubiquitous catalytic RNA motif [9, 10]. Similar results have been reported for other small small self-cleaving RNAs like the human hepatitis-δ [27] or twister ribozymes [28], which indicates that small ribozymes are frequent motifs encoded by DNA genomes. Although the precise biological functions of these genomic self-cleaving RNAs are yet unknown, their involvement in DNA retrotransposition would seem to be a frequent trend in eukaryotes [29, 30]. Retrotransposons are the major components of most eukaryotic genomes, especially in the plant kingdom, where long terminal repeat (LTR)retrotransposons can make up more than 70 % of the genome [31]. These retroelements encode the protein factors required for their own mobilization. Plant genomes, however, often contain many small non-autonomous LTR-retrotransposons, like the terminal-repeat retrotransposons in miniature (TRIMs) [32] or the small LTR-retrotransposons (SMARTs) [33], which do not encode any protein and whose mobility depends on autonomous LTR-retrotransposons. In general, all these retrotransposons and other mobile elements remain heavily silenced at the transcriptional level, and they are only expressed under certain conditions. In this work, following in silico, in vitro and in vivo approaches, we reveal that genomic HHRs in plants are part of an atypical family of non-autonomous LTR-retrotransposons that accumulate in the cell transcriptomes as abundant RNA circles.
Representation of the three possible hammerhead ribozyme (HHR) topologies. The conserved nucleotides involved in the catalytic center are boxed. Conserved loop-loop interactions are also indicated. Dotted and continuous lines refer to non-canonical and Watson-Crick base pairs, respectively. The three HHR types have been reported in prokaryotic/phage genomes, whereas only types I and III have been described in plants. Metazoan genomes mostly show type I HHRs
Results
Genomic HHRs in plants are embedded in the LTRs of a new form of non-autonomous retroelement: retrozymes
The occurrence of HHRs has been previously reported in some plant genomes [9, 16, 17, 22]. In this work, we performed extensive bioinformatic searches for HHR motifs in plant genomes (see Methods section), which were mostly found in eudicots (42 species), notably among rosids, together with isolated examples in monocots, ferns and algae (Additional file 1). Type III HHRs were the most frequent motifs found, whereas only a few examples corresponded to type I HHRs. The number of ribozyme motifs detected per genome varied from the absence of any recognizable HHR in many species to more than 100 bona fide ribozymes in some others (Additional file 1).
As previously noticed [22], we confirmed that plant HHRs most frequently occur as isolated motifs, but also as close tandem repeats of two, three or, rarely, even four HHRs. Sequence repeats between HHR motifs were sized from 400 to 1000 bp, lacked any detectable protein-coding capacity and did not show clear sequence identity among different plant species. For each plant genome, isolated HHR motifs were usually found embedded within sequences of about 300–400 bp.
Analysis of the elements containing tandem HHR copies showed that ribozymes were embedded in direct repeats of about 300–400 bp, which delimited a central region of about 300–600 bp without any coding potential (see Fig. 2a) [32]. These elements were flanked by target site duplications (TSDs) of 4 base pairs characteristic of LTR-retrotransposons. Most of these features are in common with those of TRIM and SMART retrotransposons (Additional file 2). However, given the peculiarities of these new retroelements, like the presence of catalytic RNA motifs in their LTRs, their slightly larger sizes and high sequence heterogeneity, as well as their singular transcriptional activity (see below), we named them retrozymes (after retroelements with hammerhead ribozymes).
Sequence features of genomic retrozymes and Ty3-gypsy retrotransposons. a Schematic representation of a full genomic retrozyme element. Target site duplications (TSDs) delimiting the retrozyme are shown in grey. The 5′ and 3′ ends of the long terminal repeats (LTRs) are shown in blue and red, respectively, with the small conserved boxes highlighted in darker colors. The positions of the conserved primer binding site (PBS), the polypurine tract (PPT) and the hammerhead ribozymes (HHR) are indicated. The self-cleavage (SC) sites delimiting the retrozyme RNA are indicated with arrows. b Schematic representation of the transcribed and self-cleaved retrozyme RNA. c Alignment of the conserved 3′ and 5′ boxes (PBSs, PPTs and LTR ends) of representative retrozymes (top) and the equivalent regions of Ty3-gypsy retrotransposons (bottom). Consensus sequence of the motif is shown in the middle: totally conserved positions are shown as uppercase letters (either A, C, G, U, Y or R), whereas positions conserved in between 70–100 % of the sequences are shown as lowercase letters, and positions below 70 % in conservation are shown as dots. d Schematic representation of a genomic Ty3-gypsy retrotransposon (top) and its transcribed RNA (bottom). LTRs are shown in light grey. Small conserved 5′ and 3′ boxes are shown in blue and red respectively. Characteristic unique (U5, U3) and repeated (R) domains of the LTRs are indicated. The typical open reading frames of Ty3-gypsy retrotransposons are indicated as gag and pol. e Sequence alignment of selected HHRs from 25 plant species. The helixes and loops of the HHR are depicted at the bottom. Self-cleavage site is indicated with an arrow. Consensus sequences shown at the bottom were obtained as in panel c
We deduced that transcription of genomic retrozymes followed by self-processing through HHR motifs would result in RNA transposition intermediates (hereafter, retrozyme RNAs) of about 600–1000 nt, depending on the plant species (Fig. 2b). These retrozyme RNAs lack the characteristic repeated regions (R, Fig. 2b and d) of the transposition intermediates of LTR-retrotransposons required for retrotranscription of the full retroelement [34–37].
Homology of retrozyme sequences was evident between plants within the same genus, despite some clear heterogeneity. However, sequence identity was almost absent between retrozymes of less related plant species, with the exception of two small boxes of about 25 nt, referred to as 3′ and 5′ boxes (Fig. 2c), and the HHR motifs (Fig. 2e). To identify potential autonomous LTR-retrotransposons responsible for retrozyme mobilization, these small conserved boxes were used as queries to search against autonomous retroelements. We found that the conserved 3′ box in retrozymes is almost identical to the LTR 3′ end and the primer binding site (PBS, tRNAMet) of the Ty3-gypsy retrotransposons (Fig. 2c). The retrozyme 5′ box, in turn, is also very similar to the polypurine tract (PPT) and the LTR 5′ end of the same family of retrotransposons [38] (Fig. 2c). No other sequence similarities were detected between retrozymes and Ty3-gypsy or any other family of retrotransposons.
Overall, these data indicate that retrozymes constitute a new group of non-autonomous LTR retroelements that may use the machinery of plant Ty3-gypsy retrotransposons for their genomic mobilization in the same way as other non-autonomous retrotransposons do.
Genomic retrozymes in the physic nut Jatropha curcas
Jatropha curcas or physic nut plant has a genome of about 410 Mb that has been recently sequenced [39–41]. In silico analysis of the available J. curcas sequences (70 % of the total genome) with RNAMotif revealed up to 30 bona fide type III HHRs, which showed some sequence heterogeneity for the same secondary structure (Additional file 3). Blast homology searches resulted in more than 70 HHR-like sequences (about 90 % identity for 90 nt), with 48 of these motifs occurring as tandem dimeric copies. These dimeric arrangements corresponded to 24 different retrozymes like the ones described above, whereas the rest of the HHRs motifs mostly corresponded to LTR sequences with no adjacent internal sequence (solo LTRs).
J. curcas retrozymes were flanked by TSDs of 4 bp with the consensus sequence WWRR (where W stands for A or T and R for a purine). LTRs were about 330 bp long, and the two HHR self-cleavage sites encompassed a non-coding region of 697–776 bp. At least four genomic retrozyme sequences were detected embedded within 18S rRNA gene sequences, whereas four others were found close (less than 2 kb) to LTR-retrotransposon sequences and the rest were detected within intergenic regions, in a similar way as described for other non-autonomous retroelements like TRIMs and SMARTs [32, 33].
To ascertain the activity of the HHRs contained in these retrozymes, in vitro transcription of a cloned J. curcas retrozyme fragment covering the 5′ LTR and the internal region was carried out. RNA self-cleaving activity was observed in the polarity containing the ribozyme motif (hereafter, the plus polarity of the retrozyme RNA) but not in the complementary (hereafter, the minus polarity) (Additional file 4). The amount of transcript processed by the HHR during transcription was 60 %, an efficiency similar to that reported for viroid and satellite HHRs [42].
Different genomic retrozymes are transcribed, processed and accumulated as circRNAs in J. curcas tissues
Transcription is the first step in the replication cycle of retrotransposons and, under natural conditions, it is tightly repressed to avoid the mutational effects of transposon insertions in the host genome. To explore the transcriptional activity of retrozymes, RNA extracts from J. curcas leaves were initially analysed by native PAGE and Northern blot hybridization with probes complementary to the RNA intermediate of both polarities. A hybridization signal corresponding to the positive polarity of the retrozyme RNA (i.e. harbouring the HHR) was observed as a substantial band or bands around the 700 nt region (Fig. 3a). No band was detected for the negative polarity of the retrozyme (data not shown). These results indicate that at least some of the genomic retrozymes were transcribed and, likely, self-processed by the HHRs in leaves of J. curcas.
Retrozymes in the genome of the physic nut (Jatropha curcas). a Ethidium bromide staining of an RNA extract (~20 μg) from J. curcas leaves and a 100–1000 bp DNA ladder run in a 5 % native polyacrylamide gel (left) and its corresponding Northern blot hybridization (right) using as a probe a digoxigenin-labelled J. curcas retrozyme fragment of negative polarity. Approximate positions of the bands corresponding to the DNA marker are indicated on the Northern blot. b Northern blot analysis of an RNA extract (~30 μg) from J. curcas leaves carried out as in panel a. Doublet bands can be seen for linear as well as circular RNA molecules. 0.1 ng of a (+)RNA transcript of J. curcas retrozyme was included as a marker. Approximate positions of the bands corresponding to the RNA marker are indicated for clarity. c Northern blot analysis of RNA extracts (~30 μg each) from J. curcas leaves, flowers, seedlings and seeds. Samples were run in a 5 % denaturing polyacrylamide gel and detected as in panel a. The positions of the linear and circular RNAs are indicated. Ethidium bromide staining of the 5S rRNA band is shown at the bottom of panels b and c as a loading control. d Minimum free energy secondary structure prediction for a retrozyme RNA from J. curcas leaves (clone Jc_066, 753 nt). Numbering starts at the self-cleavage site of the HHR. The HHR motif is highlighted with purple letters, whereas the conserved 5′ and 3′ ends of the LTRs are shown in blue and red, respectively, and PPT and PBS regions in cyan and orange, respectively. A region of 45 nt usually absent in smaller variants is shown in green letters. Positions showing sequence heterogeneity among cloned variants from different plant tissues are indicated within circles (triangle corresponds to deleted nucleotides, whereas arrows correspond to inserted nucleotides). e Schematic representation of the circular retrozyme RNA and the position of the oligos used for RT and PCR experiments
When the same RNA extracts from J. curcas leaves were analysed by denaturing PAGE followed by Northern blot hybridization, the bands in the 700 nt region appeared as a clear doublet (Fig. 3b), possibly corresponding to transcribed and HHR-processed RNAs from genomic retrozymes of different size. Interestingly, an additional doublet that did not appear in native gels was observed in denaturing gels with an apparent higher molecular weight (>3 kb) (Fig. 3b). This behaviour suggested the existence of a mixture of circular and linear RNAs that co-migrate in native gels, as has been observed during plant infection by pathogenic circRNAs such as viroids and virus satellites [43]. In order to confirm this finding, RNA samples were analysed by double PAGE: RNA extracts were first run in a native gel, and then the region of 600–800 nt was cut out and placed on a denaturing gel followed by Northern blot hybridization (Additional file 5A). This experiment confirmed that circular and linear RNAs co-migrated in the native gel and only became separated under denaturing conditions.
The transcriptional activity of genomic retrozymes in different tissues and developmental stages of J. curcas was analysed by Northern blot (Fig. 3c and Additional file 5B). RNA extracts from young seedlings and leaves showed the presence of two circular and their corresponding two linear bands. In flowers and seeds extracts, however, only the faster migrating circular and linear RNAs were observed. This result indicates that retrozymes are differentially transcribed in different tissues of J. curcas.
Purified circRNAs from J. curcas seeds, young seedlings and leaves were retrotranscribed, cloned and sequenced (Additional file 6). Two types of variants were detected in leaves, one of 753 nt approximately and a second one of about 708 nt, which are in agreement with the size estimated for the two linear bands detected by denaturing Northern blots. Prediction of minimum free energy secondary structures for the cloned retrozyme RNAs revealed a highly structured architecture with an elevated degree of self-complementarity (about 70 % of nucleotides are paired), similar to that reported for circRNA pathogens with HHRs [43] (Fig. 3d and Additional file 7). In the predicted structures, the LTR region adopts a long and stable hairpin structure with most of the HHR motif paired with a highly complementary sequence that prevents the hammerhead fold from forming and, consequently, its self-cleavage.
The obtained cDNA clones showed sequence variability between them (Fig. 3d), but also with respect to any of the genomic copies detected in the databases (Additional file 6). Such a high sequence heterogeneity together with the similarities between retrozyme and plant pathogenic RNAs (circular and highly structured RNA molecules, small size and presence of HHRs) suggested the possibility that retrozyme circRNAs may follow RNA-RNA replication through a rolling-circle mechanism similar to that described for viroids and viral satellite RNAs [44]. If that were the case, there would exist replication intermediates of negative polarity, either as circular or multimeric linear RNAs. To explore this possibility, we carried out RT-PCR experiments with adjacent primers outside of the LTRs to avoid the amplification of negative polarity RNAs resulting from transcription of genomic retrozyme copies (see Methods). Positive results were obtained with RNA extracts from J. curcas seeds (Additional file 8A). Northern blot analysis of RNA-enriched extracts from J. curcas tissues, however, did not reveal the presence of negative polarity RNAs of the retrozyme.
Retrozyme-derived circRNAs accumulate to high levels in strawberry
HHR motifs have been previously reported in diverse genomic sequences of strawberry (Fragaria x ananassa) [22]. Thanks to the recently published genome of F. ananassa [45], our bioinformatic searches for HHRs revealed the presence of about 90 bona fide ribozyme motifs (Additional file 1) and up to 6 potential retrozyme RNAs of sizes ranging from 673 to 701 nt.
In vitro transcription of a cloned genomic retrozyme fragment from F. ananassa carrying a single HHR motif showed a clear self-cleaving activity (48 % processed transcript) (Fig. 4a). Northern blot hybridization of RNA extracts revealed that retrozyme RNAs of positive polarity accumulated at high levels, with up to 0.1 ng of retrozyme RNAs per μg of total RNA from leaves (about 0.1 %), and slightly lower amounts from flowers (Fig. 4a). Northern blot analysis of the negative polarity did not reveal any clear band, but just a weak smear (Fig. 4b). However, in a similar way as found for J. curcas seeds, RT-PCR experiments with RNA extracts from F. ananassa leaves revealed the presence of retrozyme RNAs of negative polarity (Additional file 8B). Cloning and sequencing of the cDNAs from both positive and negative RNA polarities showed again a heterogeneous population of highly structured retrozyme RNAs (Fig. 4c).
Retrozymes in the genome of the strawberry (Fragaria ananassa). a Northern blot analysis of RNA extracts (~20 μg) of F. ananassa flowers and leaves carried out as described in Fig. 3. One ng of a (+)RNA transcript of F. ananassa was included as a marker. The positions of the linear and circular RNAs are indicated. Approximate positions of the bands corresponding to the RNA marker are indicated for clarity. b Northern blot analysis of RNA extracts (~20 μg) of F. ananassa flowers and leaves run in a 5 % denaturing polyacrylamide gel and detected using a F. ananassa retrozyme fragment of the positive polarity as a probe. 0.1 ng of a (–)RNA transcript of F. ananassa was included. Ethidium bromide staining of the 5S rRNA band is shown at the bottom of panels a and b as a loading control. c Minimum free energy secondary structure prediction for a retrozyme RNA from F. ananassa leaves (clone Fa_049, 674 nt). Numbering, colors and natural sequence heterogeneity are shown as in Fig. 3. d Schematic representation of the circular retrozyme RNA and the position of the oligos used for RT and PCR experiments
To ascertain the presence of a mixture of circular and linear retrozyme RNAs in the plant, we carefully checked the migration properties of strawberry RNAs under native and denaturing conditions in the presence of appropriate markers obtained by in vitro transcription of a full genomic retrozyme of F. ananassa (Additional file 5C). The retrozyme RNA resulting from double self-cleavage (679 nt) was purified and circularized in vitro using a Solanum melongena tRNA ligase as previously described [46] (Additional file 5D). Purified linear and circularized retrozyme RNAs were run in a native PAGE together with an RNA extract of F. ananassa. Northern blot hybridization revealed a single band of about 700 nt in the three cases (Additional file 5E). When these three samples were run in a denaturing PAGE, the RNA extract of F. ananassa showed the typical duplet of bands, whereas the linear retrozyme RNA migrated as a 679-nt band and the circular RNA run with an apparent size of 3 kb. Circularized and linear RNA markers perfectly matched the two bands detected in the RNA extract, which confirms the presence of a mixed population of circular and linear retrozyme RNAs in the plant.
Retrozymes in eucalyptus and citrus trees
In order to generalize the data obtained for the physic nut and strawberry, retrozymes of several woody plants were also investigated. Our bioinformatic analysis of the genomes of Eucalyptus camaldulensis [47] and E. grandis [48] detected more than 100 copies of bona fide HHRs in each of these genomes (Additional file 1). Dozens of ribozymes occurred in tandem copies of two, three and even four HHRs (Additional file 2D), suggesting large retrozyme RNAs of about 900–1050 bp.
In vitro transcription of a cloned retrozyme fragment from E. camaldulensis showed clear levels (54 % processed transcript) of HHR self-cleavage (Fig. 5a). Northern blot analysis of RNA extracts from different E. camaldulensis tissues confirmed the presence of a retrozyme RNA of about 1 kb, although the band detected in flowers was of slightly lower molecular weight than the one found in leaves and young sprouts (Fig. 5a). The minimum free energy secondary structure of a genomic retrozyme RNA displayed a similar architecture to J. curcas and strawberry retrozymes: a long stable arm composed of the LTR sequence, and a ramified region of hairpins corresponding to the rest of the element (Fig. 5b).
Retrozymes in the genome of the eucalyptus (Eucalyptus camaldulensis). a Northern blot analysis of RNAs (~20 μg) obtained from E. camaldulensis leaves, sprouts and flowers carried out as described in Fig. 3. 0.1 ng of a (+)RNA transcript of a genomic retrozyme from E. camaldulensis was included. The positions of the linear and circular RNAs are indicated. Ethidium bromide staining of the 7.5S rRNA band is shown at the bottom as a loading control. b Minimum free energy secondary structure prediction for a retrozyme RNA of 979 nt derived from a genomic retrozyme of E. camaldulensis (entry BADO01091291.1). Numbering and colors are shown as in Fig. 3
We also analysed the genomic HHRs present in a number of citrus species. The genome of the sweet orange (Citrus sinensis, cv. Valencia) [49] only showed 10 bona fide HHRs, but up to 11 retrozyme-like elements able to encode putative retrozyme RNAs of 665–694 nt (Additional file 1). Many of the HHRs within putative retrozymes, however, showed punctual mutations that are expected to deeply affect their self-cleaving activity.
In vitro transcription of a retrozyme fragment carrying a bona fide HHR showed an intense RNA self-cleavage (about 75 % processed transcript) (Fig. 6a and b). However, RNA extracts from different C. sinensis tissues did not show the presence of circular or linear retrozyme RNAs, neither by Northern blot (detection limit of 0.1 pg) nor by RT-PCR analysis (data not shown). The genome of the related species Citrus x clementina revealed similar HHRs (up to 19 bona fide ribozymes) and 13 genomic retrozyme copies (Additional file 1). When RNA extracts of C. clementina were analysed by Northern blot, circular and linear RNAs were detected in ovaries and whole flowers, but not in leaves (Fig. 6a). In the case of lemon tissues (Citrus x limon), Northern blot analysis revealed the presence of circular and linear retrozyme RNAs in leaves, flowers and seeds (Fig. 6b). The minimum free energy secondary structure prediction for a genomic retrozyme RNA from C. clementina resulted again in a highly structured RNA (Fig. 6c).
Retrozymes in the genome of different citrus trees. a Northern blot analysis of RNA extracts of C. clementina ovaries, flowers and leaves carried out as described in Fig. 3. 1 pg of a (+)RNA transcript of C. sinensis was included. b Northern blot analysis of RNA extracts of C. limon leaves, flowers and seeds carried out as described in Fig. 3. 0.1 ng of a (+)RNA transcript of C. sinensis was included. The positions of the linear and circular RNAs are indicated. Ethidium bromide staining of the 5S rRNA band is shown at the bottom of panels a and b as a loading control. c Minimum free energy secondary structure prediction for a retrozyme RNA of 689 nt derived from a genomic retrozyme of Citrus clementina (entry NW_006262201). Numbering and colors are shown as in Fig. 3
Finally, comparative analysis of C. clementina (13 retrozymes) and C. sinensis (11 retrozymes) genomes gave us another indication of the mobile nature of retrozymes. Alignment of two orthologous genomic regions of around 5 kb (Additional file 9) showed that while C. clementina contains a typical retrozyme element flanked by a duplicated CTAT sequence (TSDs), the equivalent region in C. sinensis genome did not show any retrozyme sequence, but showed a single CTAT sequence at this particular position.
Other putative retrozyme elements in plant and metazoan genomes
Our bioinformatic analyses revealed the presence of putative retrozymes in the genomes of more than 40 plant species (Additional file 1). Again, the retrozyme sequences in each plant genome showed a noticeable variability. Moreover, sequence identity of retrozymes from evolutionarily distant plant species was nearly absent, with the exception of the small conserved 5′ and 3′ boxes and the HHR motif (Fig. 2c and e). However, secondary structure prediction of minimal free energy for different plant retrozymes revealed a similar architecture, with a long arm corresponding to the LTR region harbouring the HHR in a blocked conformation, and a stable but more ramified structure corresponding to the rest of the RNA (Figs. 3d, 4c, 5b, 6c and Additional file 10).
Genomic HHRs have also been reported previously in metazoans like newts [20], schistosomes [18] and cave crickets [19] and are widespread in a large set of animal genomes [9, 22, 24, 26]. Many of these type I HHR motifs occur as multimeric tandem repeats separated by a few hundred base pairs (about 150–400 bp) similar to those described above for plant retrozymes. However, our sequence analysis of metazoan repeats with HHRs did not detect any PBS or PPT motif, with the exception of a full tRNAGln in the case of Schistosome repeats [50]. Moreover, we found that Nematostella vectensis repeats were flanked by large TSDs (15 bp) similar to those described in the mobilization of long interspersed element (LINE) retrotransposons [51]. We also noticed that the monomeric RNA intermediates resulting from transcription and HHR processing of metazoan repeats show highly stable secondary structures (Fig. 7 and Additional file 11) similar to those found for plant retrozymes. We are aware that in silico RNA secondary structure prediction, especially for large RNA molecules, is not an accurate determination of the in vivo structure of the molecule. However, the stability and degree of self-complementarity deduced for these RNAs (above 70 % of the nucleotides are double-stranded) is much higher than the one observed for an RNA without any selection pressure on its secondary structure.
Minimum free energy secondary structure prediction for putative retrozyme RNAs deduced from different metazoan genomes: a the anemone Nematostella vectensis, b the trematode Schistosoma mansoni (tRNAGlu sequence is shown in blue), c the tick Argas monolakensis and d the newt Cynops pyrrhogaster. Insets in each panel show the structure of the type I HHR. Similar representative examples of structures of metazoan retrozyme RNAs are shown in Additional file 11. HHR sequences are shown in purple, and the self-cleavage site is indicated with an arrow. Numbering starts at the self-cleavage site of the HHR
Discussion
In this work, we have described the retrozymes, a new and atypical group of non-autonomous retroelements with self-cleaving ribozymes. At the genomic level, retrozymes highly resemble other small non-autonomous LTR-retrotransposons of plants like TRIMs [32] and SMARTs [33] (Fig. 2 and Additional file 2), but differ in some peculiarities that make them a unique class of retroelements. As non-autonomous retrotransposons, retrozymes do not show protein-coding regions but, in contrast, do encode active self-cleaving HHR motifs in their LTRs. These ribozymes catalyze the self-processing of the retrotransposon RNA intermediate, which accumulates in vivo as circular and linear non-coding RNAs of the precise size encompassed by the HHRs.
Genomic retrozymes show a patchy distribution among plants, occurring numerously in different species, but being absent in some others. For example, the eggplant (Solanum melongena) contains more than 150 HHRs and 18 different retrozymes, whereas the genomes of related Solanum species, like tomato or potato, do not show a single example. An illuminating case is found in the cassava genomes [52]. There are 34 full retrozymes in the wild variety (Manihot esculenta ssp. flabellifolia) but only 9 retrozyme copies in the genome of the domesticated one (M. esculenta Crantz), which suggests a negative selection pressure over these retroelements during plant domestication.
Another prominent feature of retrozymes is the high accumulation levels of heterogeneous circular and linear RNA intermediates in most of the plant tissues analysed. Retrotransposons are mostly quiescent in somatic cells, but activate under different stress conditions [38, 53, 54]. Our results indicate that, under natural conditions, some of the genomic retrozymes are either actively transcribed or weakly transcribed into highly stable covalently closed RNA circles. There is even the intriguing possibility that these circRNAs may undergo autonomous replication by plant polymerases as suggested by (1) the evident similarity of retrozymes with small circRNA pathogens with ribozymes (Additional file 6) [43], (2) the presence of multimeric retrozyme RNAs of the opposite polarity (Additional file 8) and (3) the observed sequence heterogeneity at RNA level (Additional file 7) indicative of replication events by error-prone RNA polymerases [44]. However, other explanations different from RNA replication are also possible, like a genomic origin of the minus RNAs, or sequence heterogeneity due to RNA hyperediting like that observed for some intronic circRNAs in animals [55]. Future research will be required to clarify these observations.
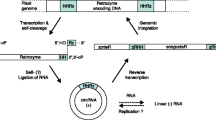
Regarding the retrotransposition mechanism of retrozymes, the most plausible model would involve the circRNAs as the final template for retrotranscription, whereas linear retrozyme RNAs would just be intermediaries and/or by-products of the circRNAs. This assumption is based on what we know about the retrotranscription of LTR retroelements, where the RNA template is a linear RNA carrying a characteristic repeated domain (R) at both ends that is necessary for 5′ to 3′ strand transfer and retrotranscription completion (Fig. 2d) [34, 37]. In retrozyme RNAs, processing by the self-cleaving ribozymes in the two LTRs produces a linear RNA with no repeated R domain (Fig. 2b). However, and as summarized in Fig. 8, covalent circularization of the self-cleaved RNAs through either the HHR itself or a host RNA ligase factor [43] would result in stable circRNAs able to bind a tRNA through their PBS. Retrotransposon-encoded retrotranscriptases could then be able to produce cDNAs of different lengths thanks to the circular nature of the RNA template (either solo LTR, full or multimeric retrozymes) (see Additional file 2). Finally, the resulting cDNAs would be integrated in new genomic locations through the machinery of the autonomous LTR-retrotransposons (Fig. 8).
Model of the life cycle of retrozymes. A full genomic retrozyme containing at least two HHRs (top) is transcribed. The resulting RNA would self-process through the HHRs to give a linear RNA with 5′-OH and 2′-3′-cyclic phosphate ends, which would be circularized. The resulting circRNA(+) would be template for either endogenous RNA polymerases (replication cycles) and/or retrotranscriptases encoded by LTR-retrotransposons. In the latter case, the cDNAs resulting from retrotranscription of a circular RNA template could have different lengths (solo LTR, monomeric or multimeric) depending on the processivity of the retrotranscriptase. Finally, the machinery of the LTR-retrotransposon would integrate the retrozyme DNAs at a new genomic location
Conclusion
In summary, our work reveals that genome-encoded circRNAs carrying a self-cleaving ribozyme like the HHR are frequent molecules in plant transcriptomes, and constitute a feasible source for the origin of some virus satellites and viroids. In this regard, host RNAs derived from Ty3-gypsy retroelements are known to be efficiently encapsidated by the coat protein of a plant virus [56], and also Ty1-copia retrotransposons have already been proposed as the origin of non-HHR viroids of the family Pospiviroidae [57]. Finally, a plethora of splicing-derived circRNAs with diverse biological functions have been recently reported in eukaryotes [58–64] and, consequently, future research will be focused on deciphering the possible roles and biotechnological applications of genome-encoded circRNAs with HHRs.
Methods
Bioinformatics
RNAMotif [65] was used for the detection of canonical type I and type III HHR motifs in DNA sequences and whole genomes previously downloaded from public repositories (phytozome.jgi.doe.gov, ftp.ncbi.nlm.nih.gov). The hits obtained were inspected for the presence of tertiary interactions between helixes I and II to ensure they were bona fide HHRs. Sequence homology searches through BLAST, BLASTX [66] and BLAT [67] tools were carried out against sequences of the GenBank and Whole Genome Shotgun (WGS) sequence databases. Sequence alignments were performed with ClustalX and Jalview software [68]. Secondary RNA structures of minimum free energy were calculated with the RNAfold program from the ViennaRNA Package [69] and depicted with RnaViz [70].
DNA and RNA extraction
DNA from leaves of the different plants analysed was extracted following the CTAB-chloroform protocol [71] with some modifications. Briefly, the leaves were homogenized in CTAB extraction buffer with a Polytron (Kinematica) homogenator and incubated at 60 °C for 60 min. The homogenate was mixed with an equal volume of chloroform and isoamyl alcohol (24:1 v/v). DNA in the aqueous phase was precipitated with 2.5 volumes of 100 % ethanol and 0.1 volume of 3 M sodium acetate, dissolved in MilliQ water and quantified in a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific).
For RNA extractions, the CTAB-chloroform method [72] was used with some modifications, followed by purification with silica [73]. Briefly, the frozen material (seeds, seedlings, flowers, sprouts or leaves) was homogenized in CTAB extraction buffer with a Polytron homogenator (Kinematica) and incubated at 65 °C for 30 min. The homogenate was extracted twice with an equal volume of chloroform and isoamyl alcohol (24:1 v/v). RNA in the aqueous phase was purified by adding 0.5 volume of 100 % ethanol, one volume of 6 M NaI and 0.175 volume of 100 % SiO2 (pH 2). The slurry was incubated for 30 min at room temperature and then washed four times with a buffer containing 10 mM Tris-HCl, 0.5 mM EDTA, 50 mM NaCl and 50 % ethanol. The RNA was eluted in MilliQ water by incubating 4 min at 70 °C, and finally it was concentrated by precipitation with ethanol and quantified as described above.
J. curcas seeds of two different origins (called Jc India and Jc Malaysia) were provided by SLF seeds (Dehradun, UL 248002 India). Molecular analyses were performed with material obtained from Jc India seeds, with the exception of those analyses shown for Jc Malaysia in Additional files 5B and 6.
PCR, RT-PCR and molecular cloning of retrozyme fragments
Genomic retrozyme fragments containing one of the HHRs and one LTR sequence plus the central variable region were amplified by PCR. The proofreading enzyme PrimeSTAR HS DNA Polymerase (Takara) was used following the manufacturer’s instructions, together with adjacent degenerate primers designed to target conserved retrozyme regions (Additional file 12). Amplification products of the adequate size were extracted from native 5 % PAGE gel slices with phenol:chloroform:isoamyl alcohol (25:24:1) and concentrated by ethanol precipitation as described above. The purified amplicons were inserted between the XbaI and BamHI restriction sites of the plasmid pBlueScript KS+, and were sequenced automatically with an ABI Prism DNA sequencer (Perkin-Elmer). The resulting plasmids were used for analysis of ribozyme self-cleavage and probe synthesis for Northern blot.
Retrozyme RNAs, of both positive and negative polarity, were reverse-transcribed and PCR-amplified with divergent (adjacent and facing away from each other) primers (Additional file 12). RNA extracts were run in native 5 % polyacrylamide gels with 1× TAE, and a gel section of the appropriate retrozyme size was excised. RNA was purified from gel slices by phenol extraction and ethanol precipitation, and was digested with DNaseI (Roche Diagnostics GmbH). The enriched RNA extracts were used for reverse transcription (typically 100 ng RNA in a 20 μl reaction with SuperScript II, Invitrogen) and PCR (5 μl of retrotranscription products in a 50 μl reaction with PrimeSTAR HS DNA Polymerase), both performed following the instructions of the manufacturers. Amplicons of the adequate size were purified, cloned and sequenced as described above.
Analysis of ribozyme self-cleavage and riboprobe synthesis
Retrozyme RNAfragments harbouring one HHR motif were synthesized by in vitro run-off transcription of pBlueScript KS+ plasmids containing the corresponding retrozyme insert previously linearized with EcoRI (for T7 RNA polymerase) or XbaI (for T3 RNA polymerase). For ribozyme self-cleavage analysis, transcription reactions contained: 40 mM Tris-HCl (pH 8), 6 mM MgCl2, 2 mM spermidine, 0.5 mg/ml RNase-free bovine serum albumin, 0.1 % Triton X-100, 10 mM dithiothreitol, 1 mM each of ATP, CTP and GTP, 0.1 mM UTP plus 0.5 μCi/μl [α-32P]UTP, 0.4 U/μl of porcine liver ribonuclease inhibitor (Takara), 20 ng/μl of plasmid DNA and 4 U/μl of T7 (Takara) or T3 (Roche Diagnostics GmbH) RNA polymerases. After incubation at 37 °C for 1–2 h, products were fractionated by polyacrylamide gel electrophoresis (PAGE) in 5 % gels with 8 M urea, and detected by phosphorimaging (FLA-5100 phosphorimager with BAS-MP 2040S imaging plates, Fujifilm). For the synthesis of DIG-labelled riboprobes of positive and negative polarity, transcription reactions were carried out in the same conditions as described above, except that radiolabelled UTP was replaced by 0.5 mM digoxigenin-11-UTP (Roche Diagnostics GmbH) and reactions were incubated at 37 °C for 4 h.
Northern blot hybridization
For Northern blot analysis, from 5 up to 100 μg of purified RNA from different plant tissues were examined in 5 % polyacrylamide gels containing 8 M urea and 1× TBE (89 mM Tris/89 mM boric acid/2.5 mM EDTA, pH 8.3). For double PAGEs, nucleic acids enriched in RNAs of the appropriate retrozyme size were obtained by cutting a section from nondenaturing 5 % polyacrylamide gels. These RNAs were examined in denaturing 5 % polyacrylamide gels containing 8 M urea and 0.25× TBE (22.5 mM Tris/22.5 mM boric acid/2.5 mM EDTA, pH 8.3). After ethidium bromide staining, RNAs were electroblotted to nylon membranes (Amersham Hybond-N, GE Healthcare) and UV-fixed with a crosslinker (UVC 500, Hoefer). Prehybridization, hybridization (at 68 °C in 50 % formamide for 16 h) and washing (twice with 0.1× SSC at 68 °C for 15 min) was done following the instructions of the manufacturer (GE Healthcare). The DIG-labelled probes were detected with an anti-digoxigenin antibody conjugated with alkaline phosphatase (anti-digoxigenin-AP Fab fragments, 1:104 dilution in blocking solution; Roche Diagnostics GmbH). The chemiluminiscence produced in the presence of the substrate CDP-Star (1:200 dilution in 0.1 M Tris-Cl, 0.1 M NaCl, pH 9.5; Roche Diagnostics GmbH) was finally visualized in a LAS-3000 Imaging System (Fujifilm).
Cloning, transcription and circularization of a full genomic retrozyme from F. ananassa
The genomic retrozyme with the highest sequence homology to most of the cloned F. ananassa retrozyme RNAs [GenBank:BATT01039028.1:c14263-13183] was amplified by PCR using primers designed to bind outside the sequence of the retrozyme (Fa92D and Fa92R, Additional file 12). The PCR product was cloned in the XbaI and BamHI sites of pBlueScript KS+, and the resulting plasmid was XbaI-linearized for run-off transcription with T3 RNA polymerase. The full uncleaved transcript and the retrozyme RNA resulting from self-cleavage at both HHR motifs were fractioned in 5 % polyacrylamide gels containing 8 M urea and 1× TBE, and extracted from gel slices as described above.
The retrozyme RNA, with 5′-hydroxyl and 2,3′-phosphodiester termini, was circularized using the chloroplastic isoform of a tRNA ligase from Solanum melongena [46], kindly provided by Drs. J. A. Daròs and R. Flores. The circularization reactions contained 1 μg of retrozyme RNA, about 1 μg of purified tRNA ligase, 50 mM Tris-HCl (pH 8), 50 mM KCl, 4 mM MgCl2, 5 mM DTT and 1 mM ATP, in a final volume of 50 μl. Reactions were incubated for 2 h at 30 °C and stopped by phenol extraction followed by ethanol precipitation. The circularized RNA was then separated in 5 % polyacrylamide gels containing 8 M urea and 1× TBE, and phenol-extracted from gel slices.
Abbreviations
circRNA, circular RNA; HHR, hammerhead ribozyme; LINE, long interspersed element; LTR, long terminal repeat; MITE, miniature inverted-repeat transposable element; PBS, primer binding site; PPT, polypurine tract; RT, retrotranscriptase; SINE, small interspersed nucleotide element; SMART, small LTR-retrotransposon; TRIM, terminal-repeat retrotransposon in miniature; TSD, target site duplication
References
Crick FH. The origin of the genetic code. J Mol Biol. 1968;38:367–79.
Orgel LE. Evolution of the genetic apparatus. J Mol Biol. 1968;38:381–93.
Woese CR. The fundamental nature of the genetic code: prebiotic interactions between polynucleotides and polyamino acids or their derivatives. Proc Natl Acad Sci U S A. 1968;59:110–7.
Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–57.
Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–57.
Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–30.
Valadkhan S, Manley JL. Splicing-related catalysis by protein-free snRNAs. Nature. 2001;413:701–7.
Webb CH, Luptak A. HDV-like self-cleaving ribozymes. RNA Biol. 2011;8:719–27.
Hammann C, Luptak A, Perreault J, De la Peña M. The ubiquitous hammerhead ribozyme. RNA. 2012;18:871–85.
Garcia-Robles I, Sanchez-Navarro J, De la Peña M. Intronic hammerhead ribozymes in mRNA biogenesis. Biol Chem. 2012;393:1317–26.
De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–70.
Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–12.
Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–20.
Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–80.
Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–40.
Daros JA, Flores R. Identification of a retroviroid-like element from plants. Proc Natl Acad Sci U S A. 1995;92:6856–60.
Przybilski R, Graf S, Lescoute A, Nellen W, Westhof E, Steger G, et al. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005;17:1877–85.
Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18:3880–8.
Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, et al. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res. 2000;28:4037–43.
Epstein LM, Gall JG. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987;48:535–43.
Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902.
De la Peña M, Garcia-Robles I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA. 2010;16:1943–50.
Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C. From alpaca to zebrafish: hammerhead ribozymes wherever you look. RNA. 2011;17:21–6.
Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, et al. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol. 2011;7:e1002031.
Jimenez RM, Delwart E, Luptak A. Structure-based search reveals hammerhead ribozymes in the human microbiome. J Biol Chem. 2011;286:7737–43.
De la Peña M, Garcia-Robles I. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep. 2010;11:711–6.
Webb CH, Riccitelli NJ, Ruminski DJ, Luptak A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953.
Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol. 2014;10:56–60.
Cervera A, De la Peña M. Eukaryotic Penelope-like retroelements encode hammerhead ribozyme motifs. Mol Biol Evol. 2014;31:2941–7.
Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol. 2010;30:3142–50.
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–5.
Witte CP, Le QH, Bureau T, Kumar A. Terminal-repeat retrotransposons in miniature (TRIM) are involved in restructuring plant genomes. Proc Natl Acad Sci U S A. 2001;98:13778–83.
Gao D, Chen J, Chen M, Meyers BC, Jackson S. A highly conserved, small LTR retrotransposon that preferentially targets genes in grass genomes. PLoS One. 2012;7:e32010.
Sandmeyer S, Patterson K, Bilanchone V. Ty3, a position-specific retrotransposon in budding yeast. Microbiol Spectr. 2015;3:MDNA3-0057-2014.
Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532.
Sabot F, Schulman AH. Parasitism and the retrotransposon life cycle in plants: a hitchhiker’s guide to the genome. Heredity (Edinb). 2006;97:381–8.
Finnegan DJ. Retrotransposons. Curr Biol. 2012;22:R432–437.
Gorinsek B, Gubensek F, Kordis D. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol. 2004;21:781–98.
Sato S, Hirakawa H, Isobe S, Fukai E, Watanabe A, Kato M, et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18:65–76.
Wu P, Zhou C, Cheng S, Wu Z, Lu W, Han J, et al. Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant J. 2015;81:810–21.
Zhang L, Zhang C, Wu P, Chen Y, Li M, Jiang H, et al. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to salt stress. PLoS One. 2014;9:e97878.
De la Peña M, Flores R. An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: implications for the design of more efficient ribozymes. J Biol Chem. 2001;276:34586–93.
Flores R, Grubb D, Elleuch A, Nohales MA, Delgado S, Gago S. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme. RNA Biol. 2011;8:200–6.
Gago S, Elena SF, Flores R, Sanjuan R. Extremely high mutation rate of a hammerhead viroid. Science. 2009;323:1308.
Hirakawa H, Shirasawa K, Kosugi S, Tashiro K, Nakayama S, Yamada M, et al. Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res. 2014;21:169–81.
Nohales MA, Molina-Serrano D, Flores R, Daros JA. Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae. J Virol. 2012;86:8269–76.
Hirakawa H, Nakamura Y, Kaneko T, Isobe S, Sakai H, Kato M, et al. Survey of the genetic information carried in the genome of Eucalyptus camaldulensis. Plant Biotech. 2011;28:471–80.
Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, et al. The genome of Eucalyptus grandis. Nature. 2014;510:356–62.
Xu Q, Chen LL, Ruan X, Chen D, Zhu A, Chen C, et al. The draft genome of sweet orange (Citrus sinensis). Nat Genet. 2013;45:59–66.
Copeland CS, Marz M, Rose D, Hertel J, Brindley PJ, Santana CB, et al. Homology-based annotation of non-coding RNAs in the genomes of Schistosoma mansoni and Schistosoma japonicum. BMC Genomics. 2009;10:464.
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–82.
Wang W, Feng B, Xiao J, Xia Z, Zhou X, Li P, et al. Cassava genome from a wild ancestor to cultivated varieties. Nat Commun. 2014;5:5110.
Okamoto H, Hirochika H. Silencing of transposable elements in plants. Trends Plant Sci. 2001;6:527–34.
Feschotte C, Jiang N, Wessler SR. Plant transposable elements: where genetics meets genomics. Nat Rev Genet. 2002;3:329–41.
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–7.
Ghoshal K, Theilmann J, Reade R, Maghodia A, Rochon D. Encapsidation of host RNAs by cucumber necrosis virus coat protein during both agroinfiltration and infection. J Virol. 2015;89:10748–61.
Kiefer MC, Owens RA, Diener TO. Structural similarities between viroids and transposable genetic elements. Proc Natl Acad Sci U S A. 1983;80:6234–8.
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733.
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57.
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8.
Talhouarne GJ, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA. 2014;20:1476–87.
Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–35.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–64.
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91.
Lorenz R, Bernhart SH, Honer zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26.
De Rijk P, Wuyts J, De Wachter R. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300.
Dhakshanamoorthy D, Selvaraj R. Extraction of genomic DNA from Jatropha sp. using modified CTAB method. Rom J Biol Plant Biol. 2009;54:117–25.
Sangha JS, Gu K, Kaur J, Yin Z. An improved method for RNA isolation and cDNA library construction from immature seeds of Jatropha curcas L. BMC Res Notes. 2010;3:126.
Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503.
Acknowledgments
We would like to thank A. Ahuir for her excellent technical assistance, the Instituto Valenciano de Investigaciones Agrarias (IVIA) for kindly providing us with Citrus plant materials and the Molina family and ’El campito’, who kindly grew and provided us with J. curcas and strawberry plant materials.
Funding
Funding for this work was provided by the Ministerio de Economía y Competitividad of Spain (grants BFU2011-23398 and BFU2014-56094-P). Support of the publication fee was provided by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Availability of data and materials
Sequences corresponding to cloned retrozyme circRNAs have been deposited in the GenBank database (accession numbers KX273065-KX273079 for J. curcas retrozymes and KX281154-KX281154 for F. ananassa retrozymes, respectively).
Authors’ contributions
AC and MdlP designed the experiments. AC, DU and MdlP performed the experiments. AC and MdlP wrote and edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Ethics approval was not needed for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at http://dx.doi.org/10.1186/s13059-017-1175-5.
Additional files
Additional file 1: Table S1.
Bioinformatic compilation of putative HHRs and retrozymes found in Phytozome v10.0 and GenBank (15 Aug 2014) sequence databases of plants (see Methods section for more information). (PDF 76 kb)
Additional file 2:
Schematic representation of small plant non-autonomous LTR-retrotransposons. A: TRIM (top) and SMART (bottom) retroelements. B: Truncated solo-LTR. C: Full-copy retrozyme. D: Multimeric retrozyme. LTRs are shown in blue and the approximated sizes of the different elements and regions are indicated. (PDF 86 kb)
Additional file 3:
Hammerhead ribozymes in the genome of Jatropha curcas. A: Sequence alignment of the 30 bona fide HHRs detected in the genome of J. curcas (Bioprojects: PRJNA63485 and PRJNA279873). Only 5 out of the 30 HHRs show identical sequences, whereas the other 25 motifs showed one or more changes. B: Secondary structure of the J. curcas HHRs. Variable positions are indicated within circles (single changes) or boxes (covariations). Conserved tertiary interactions between loops 1 and 2 are shown with dotted lines. (PDF 128 kb)
Additional file 4:
Catalytic functionality of J. curcas hammerhead ribozymes. Schematic representation (top) and autoradiography (bottom) of run-off transcriptions in the presence of [α-32P]-UTP of the positive and negative polarities of a genomic retrozyme fragment from Jatropha curcas. The transcribed RNAs covered approximately from position 574 to position 573 of the circular RNA depicted in Fig. 3a, and were separated by denaturing 5 % PAGE. Quantification of the bands indicated that 60 % of the transcript was processed by the ribozyme. The position of T7 and T3 RNA promoters, primer binding site (PBS), polypurine tract (PPT) and HHR self-cleavage site (arrowhead) are indicated. (PDF 108 kb)
Additional file 5:
Circular and linear retrozyme RNAs in J. curcas and F. ananassa. A: Double PAGE analysis of an RNA extract (~20 μg) from J. curcas leaves. A gel stripe from a 5 % native polyacrylamide gel containing RNAs of 600–900 nt (left) was cut out and run on top of a second 5 % denaturing polyacrylamide gel (center). The corresponding Northern blot (right), using a digoxigenin-labelled J. curcas retrozyme fragment as a probe, revealed the presence of both circular and linear RNA forms. B: Northern blot analysis of RNA extracts (~30 μg each) from J. curcas leaves, young seedlings and seeds. Samples were run on a 5 % denaturing PAGE and were detected using the same probe as in panel A. C: An in vitro transcription of a previously cloned full genomic retrozyme of F. ananassa was run on a 5 % denaturing PAGE. The RNAs corresponding to the uncleaved full RNA (1134 nt) and double self-cleaved retrozyme RNA (679 nt) were cut out and purified (marked in red). D: Purified retrozyme RNA (679 nt) was circularized with a tRNA ligase for 2 h and run on a 5 % denaturing PAGE. The circularized RNA (upper band) was cut out and purified. E: Northern blot hybridization of a F. ananassa RNA extract (~30 μg) and previously purified RNA markers (linear, circular and uncleaved full retrozyme RNAs, ~1 ng each) run on a 5 % native PAGE. F: Northern blot hybridization of the same RNA samples as in panel E run on a 5 % denaturing PAGE. Ethidium bromide staining of the 5S rRNA is shown at the bottom as a loading control. (PDF 728 kb)
Additional file 6:
Sequence variability of genomic retrozymes and retrozyme RNAs. Sequence alignment of the 24 different genomic retrozymes found in J. curcas (names in black) against 16 selected clones of circRNAs from J. curcas tissues (names in red; 8 clones for each of the two major variants detected). Only the nucleotides encompassed by the HHR self-cleavage sites (equivalent to an HHR-processed retrozyme RNA) were used in the alignment. SM and SI correspond to circRNAs from J. curcas seeds, whereas HM and HI were obtained from leaves, and PM and PI from young seedlings. Based on their highest degree of similarity with the cloned RNAs, two genomic retrozyme sequences (at the top and at the 9th position of the alignment) were chosen as the tentative parental elements. For clarity, the full sequence of the two tentative parental retrozymes is shown, whereas for all the other sequences, only the non-conserved positions are depicted. (PDF 110 kb)
Additional file 7:
Minimum free energy secondary structure predictions for the viral satellite RNA sTRSV (top, entry M14879.1) and the viroids ELVd (middle, entry AJ536612.1) and CChMVd (bottom, entry AJ878085.1). HHR sequences are shown in purple (positive polarity) and grey (negative polarity). Self-cleavage sites are indicated with arrows. Numbering starts at the self-cleavage site of the positive polarity HHR. (PDF 86 kb)
Additional file 8:
RT-PCR amplification of negative polarity retrozyme RNAs from J. curcas and F. ananassa. A: RT-PCR experiment carried out with RNA extracts from J. curcas seeds using the direct primer Jc60D for RT to make a cDNA of the minus polarity, and the adjacent primers Jc60D and Jc60fullR (left) or Jc77D and Jc77R (right) for the PCRs (see Additional file 12). B: RT-PCR experiments performed with two different RNA extracts from F. ananassa leaves using the direct primer Fa87D for RT to make a cDNA of the minus polarity, and the adjacent primers Fa87D and Fa85R for PCR (see Additional file 12). PCR products and 100 bp DNA marker were separated by native 5 % PAGEs and stained with ethidium bromide. Controls (RT-) were the same RT-PCR experiments done without adding retrotranscriptase, whereas Controls (PCR) were PCRs done without any template. Schematic representations of a full genomic retrozyme and the positions of the oligos used for RT and PCR experiments are shown below each gel picture. (PDF 649 kb)
Additional file 9:
A retrozyme insertion in the genome of C. clementina is absent from the orthologous region in the genome of C. sinensis. A: Comparative analysis of two orthologous genomic regions of around 5 kb from C. clementina and C. sinensis revealed the presence in the former but not in the latter genome of a typical retrozyme element (not to scale) flanked by the target site duplications (TSDs, in grey letters). The position of hammerhead ribozymes, the primer binding site and the polypurine tract are indicated. B: Sequence alignment of the two orthologous genomic regions of C. clementina and C. sinensis. Differences between both sequences are highlighted in red. The sequence corresponding to the genomic retrozyme is underlined. (PDF 118 kb)
Additional file 10:
Minimum free energy secondary structure prediction of five plant circRNA retrozymes deduced from the corresponding genomic retrozyme sequences (GenBank entries are indicated below the species names). HHR sequences are shown in purple letters and the self-cleavage site is indicated with an arrow. Numbering starts at the self-cleavage site of the HHR. (PDF 100 kb)
Additional file 11:
Minimum free energy secondary structure prediction of putative retrozyme RNAs of metazoan genomes from different phyla, like A: rotifers, B: corals, C: arthropods, D: mollusks and E: vertebrates, deduced from the corresponding sequence repeats with HHRs. HHR sequences are shown in purple letters and the self-cleavage sites are indicated with an arrow. Numbering starts after the HHR self-cleavage site. (PDF 104 kb)
Additional file 12: Table S2.
Oligonucleotide compilation. (PDF 58 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Cervera, A., Urbina, D. & de la Peña, M. Retrozymes are a unique family of non-autonomous retrotransposons with hammerhead ribozymes that propagate in plants through circular RNAs. Genome Biol 17, 135 (2016). https://doi.org/10.1186/s13059-016-1002-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13059-016-1002-4