Abstract
Background
Peroxisome proliferator-activated receptor gamma (PPARγ) is a major regulator in sepsis. Our previous study identified the enhancer polymorphism rs10865710C/G to be associated with susceptibility to sepsis in trauma patients. We performed two-stage cohort studies integrating biological experiments of potential functional variants that modify susceptibility to traumatic sepsis.
Methods
Improved multiplex ligation detection reaction (iMLDR) was used to genotype rs10865710 in 797 Han Chinese trauma patients in Chongqing. Clinical relevance was validated in 334 patients in Guizhou. The potential function of rs10865710 in transcriptional regulation was explored through a dual luciferase reporter assay and electrophoretic mobility shift assay (EMSA). Expression of PPARγ was assessed by expression quantitative trait locus (e-QTL) and western blot analyses.
Results
The association results confirmed rs10865710 to be significantly strongly associated with sepsis risk in trauma patients of the Chongqing and Guizhou cohorts (OR = 1.41 (1.11–1.79), P = 0.004 and OR = 1.45 (1.01–2.09), P = 0.046, both for allele-dose effect, respectively). A meta-analysis of both cohorts and a previous study indicated strong evidence for this association (OR = 1.41 (1.17–1.71), P = 0.0004 for the dominant model, OR = 1.78 (1.34–2.36), P < 0.0001 for the recessive model and OR = 1.38 (1.20–1.58), P < 0.0001 for the allelic model). Functional experiments verified that rs10865710 was a causative variant influencing enhancer activity (G vs. C, 0.068 ± 0.004 vs. 0.096 ± 0.002, P = 0.0005) and CREB2 binding. Expression analysis also indicatevd rs10865710 genotypes to be associated with levels of PPARγ expression (P = 9.2 × 10−5 for dominant effect and P = 0.005 for recessive effect).
Conclusions
Our study provides evidence that the enhancer-region polymorphism rs10865710 might influence transcription factor binding and regulate PPARγ expression, thus conferring susceptibility to traumatic sepsis.
Trial registration
ClinicalTrials.gov, NCT01713205. Registered 18 October 2012, retrospectively registered.
Similar content being viewed by others
Introduction
Trauma is one of the leading causes of death worldwide. One of the most serious complications following major trauma is sepsis and sequential dysfunction of vital organs [1]. Individuals differ greatly in their response to infection, likely due to external (e.g., the site of infection and virulence and type of pathogenic bacteria) and host (e.g., sex, age, concomitant disease, and resistance) factors. Importantly, host genetic factors are also recognized as having a great impact on the occurrence and progression of sepsis [2]. In general, knowledge of genetic factors will allow for early identification of high-risk trauma patient populations with sepsis, facilitating the prompt initiation of anti-infection therapy and treatment to support organ function and thereby improving prognosis.
A tremendous amount of inflammatory mediator production is an important feature of traumatic sepsis [3]. Peroxisome proliferator-activated receptor gamma (PPARγ) is a ligand-dependent transcription factor of the nuclear receptor superfamily with several biological functions, including control of the inflammatory response [4]. PPARγ is widely expressed in various immune cells, such as neutrophils, lymphocytes, monocytes/macrophages, and dendritic cells [5]. Previous studies have indicated that PPARγ exerts potent anti-inflammatory effects by modulating neutrophil migration and activation; enhancing macrophage phagocytosis; regulating inflammatory mediator production such as tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), IL-6, IL-8, and IL-10; and activating oxygen/nitrogen species [6]. For example, the level of PPARγ expression was found to be reduced in freshly isolated mononuclear cells from sepsis patients [7] as well as in the lungs in sepsis murine models [8]. Numerous in vivo and in vitro studies have shown that administration of PPARγ agonists can suppress the inflammatory responses associated with sepsis [9], acute lung injury [10], and inflammation [11], increasing the host’s ability to kill and clear pathogenic bacteria and improving prognosis. Accordingly, PPARγ and its ligands have become novel therapeutic targets for the treatment of sepsis and other inflammatory diseases. Although PPARγ is consistently annotated as a potent anti-inflammatory molecule, the functional variants and the underlying mechanisms involved have not yet been identified.
Patients carrying a mutated PPARG gene might be susceptible to excessive inflammatory responses, sepsis, and/or poor prognosis. Our previous pilot study [12] revealed strong clinical relevance for rs10865710C/G, with a higher incidence of sepsis and multiple organ dysfunction (MOD) scores with the variant G allele in trauma patients. Further analysis indicated that rs10865710G was significantly associated with high levels of the pro-inflammatory cytokine TNF-α in the plasma of trauma patients in response to lipopolysaccharide (LPS) stimulation. Given the central role of PPARG in the pathogenesis of traumatic sepsis, we hypothesize that rs10865710 polymorphism may impact PPARG expression and the occurrence of sepsis in trauma patients. Thus, we evaluated two additional populations to further confirm the clinical association of rs10865710 with the risk of developing sepsis in trauma patients. Additionally, we explored whether there is a functional link between rs10865710 and PPARG transcription and protein expression. Such a mechanistic characterization of this sepsis-associated functional polymorphism might provide new opportunities for the development of targeted therapy for traumatic sepsis.
Methods
Study populations
Two unrelated cohorts of major trauma patients from Chongqing (internal test cohort) and Guizhou (external validation cohort) in China were assessed in this study. Major trauma patients were consecutively recruited if they met previously described criteria [13] from the department of trauma surgery or intensive care unit (ICU) at Daping Hospital and Chongqing Emergency Medical Center (Chongqing) from 2014 to 2016 and Department of Trauma and Emergency at the Affiliated Hospital of Guizhou Medical University (Guizhou) between 2010 and 2013. Briefly, the inclusion criteria were as follows: (1) the age of trauma patients is between 18 and 65 years, (2) patients with Injury Severity Score (ISS) more than 16, and (3) patients surviving more than 48 h after injury. Patients were not eligible if they had penetrating injuries, severe brain injury (Abbreviated Injury Scale of the head ≥ 3), preexisting organ dysfunction, or immune diseases. Whole blood specimens were collected within 2 h of admission to the hospital. Ethics approval for this study was obtained from the Ethical and Protocol Review Committees of Army Medical University, Chongqing Emergency Medical Center, and Guizhou Medical University. Informed written consent was obtained from the patients and their next of kin before enrollment, including explicit permission for DNA analysis and the collection of relevant clinical data. Patient confidentiality was preserved according to the guidelines of the Declaration of Helsinki.
Clinical evaluation
All patients requiring surgical intervention received standard surgical care and postoperative ICU treatment. The Injury Severity Score (ISS) was calculated according to the Abbreviated Injury Scale developed in 2005. Sepsis was diagnosed for trauma patients who had a rapid change of SOFA score ≥ 2 with confirmed or suspected infection during the hospital period according to “Sepsis-3.0” [14]. Infection was defined as a clinically obvious source or positive bacterial culture. Daily physiologic and laboratory data were obtained during the hospital stay, and clinical events were recorded thereafter until death or discharge from the hospital. MOD scores were calculated as the sum of the simultaneously obtained individual organ scores on each hospital day [15]. MOD scores and the presence of sepsis were determined by individuals who were unaware of the genotyping results.
Genotyping
Genomic DNA was extracted from whole blood using a Wizard genomic DNA purification kit (Promega, USA), and rs10865710 genotyping was performed using the improved multiplex ligation detection reaction (iMLDR) technique (Genesky Biotechnologies Inc., China), as described by our previous study [12]. Genotyping was performed in a blinded fashion without knowledge of the patients’ clinical data, and 10% of randomly selected samples were further confirmed by direct sequencing.
Bioinformatics analysis
As rs10865710 is located in the intron region of the PPARG gene and previous evidences [16] have suggested that intron might function as enhancer, we speculated that rs10865710 may affect enhancer activity and further influence expression of PPARG. We used the database Ensembl (http://aisa.ensembl.org/index.html) to search for any enhancer segment in the rs10865710 region and then used TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) to predict putative transcription factor binding sites and perform validation. Furthermore, we conducted expression quantitative trait locus (e-QTL) analysis using data from the NCBI PheGenI database (https://www.ncbi.nlm.nih.gov/gap/phegeni).
PPARɣ protein expression
The protein levels of PPARɣ from trauma patients were detected by western blotting according to a standard protocol. Briefly, white cells were lysed in RIPA buffer, and the protein concentration of the supernatant was quantified using BCA Protein Assay Kit (Beyotime, China). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose (NC) membranes (Millipore, USA) membrane. Following incubation for 1 h with 5% nonfat milk at room temperature, the membranes were incubated with a specific primary antibody against PPARɣ (Cell Signaling Technology, USA) overnight at 4 °C. The membranes were then incubated with the secondary antibody for 1 h at room temperature. Signals were detected by an Odyssey infrared scanner (LI-COR Biosciences, USA). The intensity of the bands was determined by densitography, and β-actin was used as a loading control.
Enhancer activity assays
The transcriptional regulation of rs10865710 in the enhancer region of the PPARG gene was revealed by a reporter gene assay system [17]. As previously reported [13], a 1000-bp regulatory region of the PPARG enhancer with the rs10865710 C or G allele was inserted into the pGL3-promotervector (Promega, USA) to construct two different plasmids. The reporter constructs were introduced into HEK293T cells using an NEPA21 electroporator at a 125-V poring pulse voltage and 5.0-ms poring time (Nepa Gene, Japan). A total of 5 × 105 cells were resuspended in 100 μl electroporation solution and mixed with 10 μg reporter plasmid DNA and 200 ng internal control plasmid (pRL-CMV). Transiently transfected cells were grown for 48 h before assaying with a Dual-Luciferase Reporter Assay System (Promega, USA) and Luminoskan Ascent luminometer (Thermo Labsystems, Finland). Relative luciferase activity was calculated as the ratio of luminescence for the experimental reporter to that of the internal control plasmid (pRL-CMV). Statistical significance was calculated using the two-tailed t test with three biological replicates.
Electrophoretic mobility shift and super-shift assays
Nucleoprotein was extracted from the mononuclear cell line THP-1 using the NE-PER nuclear and cytoplasmic extraction kit (ThermoFisher Scientific, USA) according to the manufacturer’s instructions. Infrared dye DY-682-labeled (Eurofins Genomics, Germany) and unlabeled complementary oligonucleotides flanking rs10865710 were annealed to generate a double-stranded EMSA probe (5′TTGGCATTAGATGCTGTTTTGTCTT[C/G]ATGGAAAATACAGCTATTCTAGGAT3′-biotin). Nuclear protein was incubated with the labeled probes, and DNA-protein complexes were resolved by electrophoresis on a 6% polyacrylamidegel (Life Technologies, USA). Gels were imaged using Odyssey Fc Infrared Imaging System (LI-COR Biosciences, USA). For competition experiments, 10, 200-fold excess unlabeled competitor probe was pre-incubated with the nuclear extract before biotin-labeled probes were added. For the super-shift assay, 2 μg or 4 μg of rabbit anti-human CREB2 monoclonal antibody (Abcam, UK) was added to the binding reaction. Biotin-labeled DNA-protein complexes were detected by chemiluminescence.
Statistical analysis
Continuous data are presented as the mean ± standard deviation (SD). Categorical data are presented as counts and percentages. Hardy-Weinberg equilibrium (HWE) was tested using χ2 analyses. Three different genetic models, including dominant, recessive, and allele-dose models, were applied to analyze the relevance of rs10865710C/G for clinical outcomes. Multivariable logistic regression models were used to calculate odds ratios (OR) with 95% confidence intervals (CIs) to assess sepsis risk, adjusting for age, sex, and injury severity for confounding effects. The association of the polymorphism with MOD scores was determined by linear regression analysis, adjusting by age, sex, and injury severity for confounding effects. Meta-analysis of the combined two validated cohorts and our previous study was performed using the Cochran-Mantel-Haenszel statistical test, and heterogeneity was assessed by calculating I2, as described by Higgins et al. [18]. For each comparison, the exact P values, considered significant at < 0.05, are reported. Data were analyzed using SPSS 11.5 statistical software (SPSS Inc., USA).
Results
Characteristics of the study population
Patients in the internal test (Chongqing, n = 797) and external validation (Guizhou, n = 334) cohorts were predominantly male and middle aged (Additional file 2: Table S1). The internal test and external cohorts were newly enrolled ethnic Han Chinese individuals, similar to our previous pilot study cohort [12]. The patients had sustained severe injuries (mean ISS 22.6 ± 7.9 and 23.4 ± 8.2). The median periods from injury to admission to hospital were 12 h (1–48 h) and 16 h (1–48 h) in the two cohorts, respectively. Sepsis morbidity rates were 34.9% and 37.1% in the two cohorts, respectively. Median time point for sepsis occurrence was 6 days (interquartile range 4.5–8.5 days) and 5 days (interquartile range 4.0–7.0 days). Organ dysfunction occurred in 34.4% and 40.4%, respectively, among whom 87 (31.8%) and 47 (34.8%) exhibited dysfunction in more than two organs. The median time point for MODS occurrence was found to be 7 days (interquartile range 5.5–10.0 days) and 6 days (interquartile range 4.5–8.5 days), respectively.
Associations of rs10865710 in the internal test trauma cohort
First, we selected successful genotyping 797 trauma patients from the Chongqing district to investigate associations of rs10865710 (Additional file 1: Fig. S1). The minor allele frequency (MAF) of rs10865710 was 33.9%, and the genotype distribution was consistent with HWE (P = 0.73, Additional file 3: Table S2). There were no significant differences in sex ratio, age, and ISS among the patients when stratified according to the different genotypes of rs10865710. As expected, trauma patients carrying the rs10865710G allele had a significantly increased risk of sepsis and higher MOD scores than those carrying the C allele (odds ratio (OR) = 1.44, 95% confidence interval (CI) = 1.07–1.94, P = 0.016 for sepsis risk and P = 0.005 for MOD scores in the case of a dominant effect). Data from multiple regression analyses further indicated that this association had a significant allele-dose effect on the sepsis morbidity rate (OR = 1.41, 95% CI = 1.11–1.79, P = 0.004) and MOD score (P = 0.002) (Table 1).
Validation of associations for rs10865710 in Guizhou trauma patients
We further confirmed the results in an additional trauma cohort (Guizhou). Due to the similar locations of the two cohorts (both in the southwestern region of China), clinical characteristics and genotype distribution were similar (Additional file 2: Table S1 and Additional file 3: Table S2). As shown in Table 1, we found similar associations with the risk of posttraumatic sepsis in the validation cohort. The sepsis morbidity rate was significantly higher in patients carrying the variant G allele than the wildtype C allele (OR = 1.65, 95% CI = 1.05–2.61, P = 0.03 for sepsis incidence and P = 0.009 for MOD score in the dominant model). Data from multiple regression analyses also showed that the association of this polymorphism with a higher risk of sepsis (OR = 1.45, 95% CI = 1.01–2.09, P = 0.046) and a higher MOD score (P = 0.002) in Guizhou trauma patients was significant.
Results of meta-analysis
Meta-analysis of the results from the Chongqing and Guizhou cohorts and our previous study was performed for rs10865710 in a total of 1865 trauma patients (Table 2). For traumatic sepsis, no obvious evidence of heterogeneity was observed using any of the three genetic models (I2 = 0). Thus, we selected a fixed-effects model to pool the combined OR. In the dominant model (GG vs. CG + CC), the overall pooled OR for the three studies combined was 1.41 (95% CI = 1.17–1.71) (P = 0.0004). Similarly, the recessive and allelic models demonstrated significant associations with sepsis risk (recessive model OR = 1.78, 95% CI = 1.34–2.36, P < 0.0001; allelic model OR = 1.38, 95% CI = 1.20–1.58, P < 0.0001). Regarding the MOD score, the meta-analysis also supported the association of rs10865710 in both dominant (MD = 0.70 score, 95% CI = 0.09–1.31, P = 0.02) and recessive (MD = 0.91 score, 95% CI = 0.56–1.27, P < 0.0001) models.
Results of bioinformatics analysis
We used the database Ensembl to search the fragment with potential enhancer function containing rs10865710, and the fragment chr3:12311574-12312573 showed enhancer activity. We analyzed expression data from the NCBI PheGenI database and found that rs10865710 genotype was significantly associated with PPARG mRNA expression (Pdominant = 1.2 × 10−3), with individuals carrying the CG (6.58 ± 0.15) or GG (6.20 ± 0.12) genotype having a lower level of PPARG in human peripheral leukocytes than individuals carrying the CC (6.82 ± 0.16) genotype.
Association of rs10865710 with PPARG expression
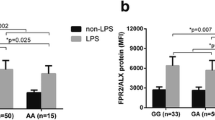
To explore the link between rs10865710 and PPARG expression, the protein level of PPARγ was detected in leukocytes exposed ex vivo to LPS stimulation, which were random selected from trauma patients with different genotype in Chongqing district, respectively (CC: n = 10, CG: n = 10, GG: n = 10). The results well associated rs10865710 of different genotypes, showing a significant difference in the case of dominant (P = 9.2 × 10−5) and recessive (P = 0.005) effects (genotype relative gray ratio: CC, 0.026 ± 0.013; CG, 0.018 ± 0.014; GG, 0.015 ± 0.006; Fig. 1).
Rs10865710 is associated with lower LPS-induced PPARG protein expression. Whole blood samples were collected from 30 trauma patients (CC: n = 10, CG: n = 10, GG: n = 10, respectively). PPARG expression by peripheral leukocytes was detected using western blotting and is presented as a gray value. *P = 9.2 × 10−5 for a dominant association (CG + GG vs. CC), #P = 0.005 for a recessive association (GG vs. CG + CC). Student’s t test was used to assess statistical significance
Influence of rs10865710 on transcriptional regulation of PPARG mRNA
We next conducted functional analysis of the rs10865710 polymorphism. Because rs10865710 is located in intron 1 of PPARG, we examined whether the genomic region around rs10865710 has enhancer activity, as predicted through bioinformatics analysis. First, we generated plasmids and conducted luciferase assays to examine whether allelic differences alter the efficiency of PPARG transcription in HEK293T cells. The allele-specific constructs containing the sepsis risk allele (G) ofrs10865710 showed lower transcriptional enhancer activity than did the other construct containing the C allele ofrs10865710 (Figs. 2, 0.068 ± 0.002 vs. 0.096 ± 0.002, P = 0.0005). These experiments were performed three times with similar results. Overall, the results demonstrate that rs10865710 variants significantly influence enhancer activity and that the risk allele rs10865710G has lower enhancer activity compared to the non-risk allele rs10865710C.
Rs10865710 decreased PPARG transcriptional enhancer activity. a Plasmid constructs used for transfection. b Transcriptional enhancer activities of rs10865710 measured by luciferase (luc) activity 48 h after transfection. Values of relative luciferase activity are shown as means ± SDs from three independent experiments. *P = 0.005, Student t test. C, C allele; G, G allele
Identification of CREB-2 as the transacting factor binding to the PPARG enhancer region
As rs10865710C/G may play a role in traumatic sepsis susceptibility and transcriptional regulation of PPARG mRNA, we next focused on determining which transacting factor binds to rs10865710C/G. In silico searches using TFSEARCH revealed that the genomic region containing the rs10865710 G allele has lost a consensus sequence corresponding to the putative binding element of CREB2. We examined allelic differences with regard to the binding of nuclear proteins using EMSA. As shown in Fig. 3a (arrow), the signal intensity of the DNA-protein complex derived with the G allele was lower than that with the C allele in the presence of THP-1 nuclear extract, suggesting differential transcription factor binding dependent on the rs10865710 allele. Self-competition with a 200-fold excess of unlabeled “C”-allele probe led to complete abrogation of this band (lane 4). Furthermore, the addition of an anti-CREB2 antibody resulted in a super-shift with the rs10865710C-allele probe (lanes 3 and 4), which indicates that CREB2 is a component of this complex (Fig. 3b).
The rs10865710 risk allele disrupts transcription factor CREB2 binding. a EMSA with biotin-labeled probes containing either the C or the G allele of rs10865710, incubated with THP-1 cell nuclear extract. The arrow indicates an allele-specific band that interacts with nuclear protein. 10× and 200× indicate 10- and 200-fold unlabeled probes excess over labeled probes. “+” and “−” mean added and unadded, respectively. b Super-shift EMSA of rs10865710. Two independent experiments were performed with similar results. Ab, antibody; C, C allele; G, G allele
Discussion
Our current two-stage cohort genetic association studies integrating biological experiments confirmed that the G allele of rs10865710 was significantly associated with an increased risk of traumatic sepsis, and different alleles (C/G) of rs10865710 might affect enhancer activity through differential transcription factor binding, as revealed by dual luciferase reporter assays and EMSAs. Therefore, integrating the results of e-QTL analysis and the important role that rs10865710 plays in regulating expression of PPARG, we propose that rs10865710 contributes to traumatic sepsis susceptibility by impacting transcription and further affecting expression of PPARG.
PPARG is one of the three subtypes of PPARs and is by far the most widely studied. PPARG forms heterodimers with retinoid X receptors (RXRs), influencing transcription of many target genes. The PPARG gene is located at chromosome3 p25 and contains eight exons; the expressed protein contains 479 amino acids. A common polymorphism (rs10865710) in PPARG is a C → G substitution in intron 1. The MAF of this polymorphism is similar between different populations (fluctuating between 21 and 36%) according to the 1000 Genomes Project. In our study, the frequency of the G allele was approximately 34%, similar to that of the CHB population reported by the 1000 Genomes Project. Moreover, previous studies have suggested a potential correlation of this SNP with systemic sclerosis [19], obesity [20], asthma [21, 22], coronary artery disease [23], and lipid metabolism [24, 25], with most suggesting that variation in rs10865710 is a disease risk factor. In addition, our previous study [12] analyzing correlation between PPAR polymorphisms and sepsis risk found that rs10865710G allele carriers had higher sepsis morbidity, MOD scores, and LPS-induced TNF-α production, which was confirmed in the present study. Functional studies have shown that the C → G substitution results in a decreased binding capacity of transcription factor CREB2 to the enhancer region of PPARG, reducing transcriptional activity, as shown by dual luciferase reporter assays and EMSAs. Furthermore, the results of e-QTL and western blotting revealed that individuals carrying rs10865710CG/GG genotypes express lower levels of PPARγ than do individuals carrying the CC genotype, which indicates that the C to G transition may downregulate PPARγ expression, which may contribute to increase TNF-α production by peripheral blood leukocytes. One previous study [8] reported that PPARγ exerts anti-inflammatory effects through transrepression of the inflammatory activities of nuclear factor κB (NF-κB), signal transducers and activators of transcription 1 (STAT-1), and activator protein-1 (AP-1) via physical interaction, thereby influencing the occurrence and progression of sepsis.
To the best of our knowledge, this is the first study to comprehensively explore the potential regulation of thers10865710 polymorphism in traumatic sepsis. Nevertheless, several limitations of our study should be acknowledged. First, our clinical study was restricted to Han Chinese patients, and whether the findings can be generalized to other ethnic groups needs further evaluation. Second, the relatively small sample size of the second stage resulted in limited statistical power (68% at a significance of 0.05). Additional large studies are needed for validating the clinical relevance of this polymorphism. Finally, as we were unable to obtain additional blood samples to determine PPARG mRNA levels, we could only make our inference based on e-QTL data from the present database, which was further confirmed through protein expression analysis.
Conclusion
Our two-stage cohort genetic association study integrating biological experiments provided solid evidence that rs10865710 may decrease transcriptional activity and PPARG expression, thus conferring susceptibility to traumatic sepsis. Our findings provide additional insight into rs10865710 as a novel predictive biomarker to improve the early identification of high risk for traumatic sepsis or MODS. Further studies with larger sample sizes and indifferent ethnic populations are warranted to validate the findings, and more biological experiments are also needed to explore the role of rs10865710 in the pathogenesis of traumatic sepsis.
Availability of data and materials
The datasets used for analysis during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AP-1:
-
Activatorprotein-1
- CI:
-
Confidence interval
- CREB:
-
cAMP-response element binding protein
- DNA:
-
Deoxyribonucleic acid
- EMSA:
-
Electrophoretic mobility shift assay
- e-QTL:
-
Expression quantitative trait locus
- HWE:
-
Hardy-Weinberg equilibrium
- ICU:
-
Intensive care unit
- IL:
-
Interleukin
- iMLDR:
-
Improved multiplex ligation detection reaction
- ISS:
-
Injury Severity Score
- LPS:
-
Lipopolysaccharide
- MAF:
-
Minor allele frequency
- MOD:
-
Multiple organ dysfunction
- NC:
-
Nitrocellulose
- NF-κB:
-
Nuclear factor κB
- OR:
-
Odds ratios
- PPARγ:
-
Peroxisome proliferator-activated receptor gamma
- RXR:
-
Retinoid X receptor
- SD:
-
Standard deviation
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SNP:
-
Single nucleotide polymorphism
- SOFA:
-
Sepsis-related Organ Failure Assessment
- STAT-1:
-
Signal transducers and activators of transcription 1
- TNF:
-
Tumor necrosis factor
References
Jin H, Liu Z, Xiao Y, Fan X, Yan J, Liang H. Prediction of sepsis in trauma patients. Burns Trauma. 2014;2(3):106–13.
Vincent JL. The clinical challenge of sepsis identification and monitoring. PLoS Med. 2016;13(5):e1002022.
Chousterman BG, Swirski FK, Weber GF. Cytokine storm and sepsis disease pathogenesis. Semin Immunopathol. 2017;39(5):517–28.
Reddy AT, Lakshmi SP, Reddy RC. PPARgamma in bacterial infections: a friend or foe? PPAR research. 2016;2016:7963540.
Almeida PE, Carneiro AB, Silva AR, Bozza PT. PPARgamma expression and function in mycobacterial infection: roles in lipid metabolism, immunity, and bacterial killing. PPAR Res. 2012;2012:383829.
Croasdell A, Duffney PF, Kim N, Lacy SH, Sime PJ, Phipps RP. PPARgamma and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015;2015:549691.
Wang D, Shi L, Xin W, Xu J, Li Q, Xu Z, Wang J, Wang G, Yao W, He B, et al. Activation of PPARgamma inhibits pro-inflammatory cytokines production by upregulation of miR-124 in vitro and in vivo. Biochem Biophys Res Commun. 2017;486(3):726–31.
Zingarelli B, Sheehan M, Hake PW, O'Connor M, Denenberg A, Cook JA. Peroxisome proliferator activator receptor-gamma ligands, 15-deoxy-Delta (12,14)-prostaglandin J2 and ciglitazone, reduce systemic inflammation in polymicrobial sepsis by modulation of signal transduction pathways. J Immunol. 2003;171(12):6827–37.
Araujo CV, Campbell C, Goncalves-de-Albuquerque CF, Molinaro R, Cody MJ, Yost CC, Bozza PT, Zimmerman GA, Weyrich AS, Castro-Faria-Neto HC, et al. A PPARgamma agonist enhances bacterial clearance through neutrophil extracellular trap formation and improves survival in sepsis. Shock. 2016;45(4):393–403.
Wang G, Liu L, Zhang Y, Han D, Liu J, Xu J, Xie X, Wu Y, Zhang D, Ke R, et al. Activation of PPARgamma attenuates LPS-induced acute lung injury by inhibition of HMGB1-RAGE levels. Eur J Pharmacol. 2014;726:27–32.
Villapol S. Roles of peroxisome proliferator-activated receptor gamma on brain and peripheral inflammation. Cell Mol Neurobiol. 2018;38(1):121–32.
Gao JW, Zeng L, Zhang AQ, Wang X, Pan W, Du DY, Zhang LY, Gu W, Jiang JX. Identification of haplotype tag single-nucleotide polymorphisms within the PPAR family genes and their clinical relevance in patients with major trauma. Int J Environ Res Public Health. 2016;13(4):374.
Zhang AQ, Gu W, Zeng L, Zhang LY, Du DY, Zhang M, Hao J, Yue CL, Jiang J. Genetic variants of microRNA sequences and susceptibility to sepsis in patients with major blunt trauma. Ann Surg. 2015;261(1):189–96.
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):801–10.
Marshall JC, Cook DJ, Christou NV, Bernard GR, Sprung CL, Sibbald WJ. Multiple organ dysfunction score: a reliable descriptor of a complex clinical outcome. Crit Care Med. 1995;23(10):1638–52.
Grossi V, Forte G, Sanese P, Peserico A, Tezil T, Lepore Signorile M, Fasano C, Lovaglio R, Bagnulo R, Loconte DC, et al. The longevity SNP rs2802292 uncovered: HSF1 activates stress-dependent expression of FOXO3 through an intronic enhancer. Nucleic Acids Res. 2018;46(11):5587–600.
Zhang AQ, Zeng L, Gu W, Zhang LY, Zhou J, Jiang DP, Du DY, Hu P, Yang C, Yan J, et al. Clinical relevance of single nucleotide polymorphisms within the entire NLRP3 gene in patients with major blunt trauma. Crit Care. 2011;15(6):R280.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
Marangoni RG, Korman BD, Allanore Y, Dieude P, Armstrong LL, Rzhetskaya M, Hinchcliff M, Carns M, Podlusky S, Shah SJ, et al. A candidate gene study reveals association between a variant of the Peroxisome Proliferator-Activated Receptor Gamma (PPAR-gamma) gene and systemic sclerosis. Arthritis research & therapy. 2015;17:128.
Bai X, Xu C, Wen D, Chen Y, Li H, Wang X, Zhou L, Huang M, Jin J. Polymorphisms of peroxisome proliferator-activated receptor gamma (PPARgamma) and cluster of differentiation 36 (CD36) associated with valproate-induced obesity in epileptic patients. Psychopharmacology. 2018;235(9):2665–73.
Zhang DD, Wang WT, Xiong J, Xie XM, Cui SS, Zhao ZG, Li MJ, Zhang ZQ, Hao DL, Zhao X, et al. Long noncoding RNA LINC00305 promotes inflammation by activating the AHRR-NF-kappaB pathway in human monocytes. Sci Rep. 2017;7:46204.
Li W, Dai W, Sun J, Zhang W, Jiang Y, Ma C, Wang C, He J. Association of peroxisome proliferator-activated receptor-gamma gene polymorphisms and gene-gene interaction with asthma risk in a Chinese adults population. Int J Clin Exp Med. 2015;8(10):19346–52.
Zhang X, Lv S, Guo C, Shi C, Chi Y, Zhao L, Wang G, Wang Z. Gene-gene interaction between PPARG and CYP1A1 gene on coronary artery disease in the Chinese Han population. Oncotarget. 2017;8(21):34398–404.
Fan W, Shen C, Wu M, Zhou ZY, Guo ZR. Association and interaction of PPARalpha, delta, and gamma gene polymorphisms with low-density lipoprotein-cholesterol in a Chinese Han population. Genet Test Mol Biomarkers. 2015;19(7):379–86.
Cao CY, Li YY, Zhou YJ, Nie YQ, Wan YJ. The C-681G polymorphism of the PPAR-gamma gene is associated with susceptibility to non-alcoholic fatty liver disease. Tohoku J Exp Med. 2012;227(4):253–62.
Acknowledgements
We acknowledge MM. Xu Wang for his technical help and Dr. Dong-po Jiang, Army Medical University, and Dr. JunYang, Chongqing Emergency Medical Center, for the collection of the blood samples.
Funding
This work was supported by National Natural Science Foundation of China (81601677 and 81571892), Science and Technology Innovation Project for Academicians of Chongqing (cstc2017jcyj-yszxX0007), and Medical Research Funding of PLA (17QNP005 and AWS14C003).
Author information
Authors and Affiliations
Contributions
H-XL and D-LW was the main researcher for this study. L-YZ, D-YD, and JD were involved in the collecting of blood samples and clinical data. J-HS did the technical work. J-XJ and A-QZ planned the study, wrote the protocol, and was involved in the genetic and clinical aspects of data analyses and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Ethical and Protocol Review Committee of the Army Medical University (No. TMMU2012009). Informed consent was obtained from the patients or their next of kin.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1.
The flow chart of trauma sample screening for genetic association and expression analysis of rs10865710.
Additional file 2: Table S1.
Overall characteristics of patients with major trauma.
Additional file 3: Table S2.
Distribution of the rs10865710C/G in the PPARG among trauma patients.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lu, H., Wen, D., Sun, J. et al. Enhancer polymorphism rs10865710 associated with traumatic sepsis is a regulator of PPARG gene expression. Crit Care 23, 430 (2019). https://doi.org/10.1186/s13054-019-2707-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13054-019-2707-z