Abstract
Background
Recurrent reciprocal 1q21.1 deletions and duplications have been associated with variable phenotypes. Phenotypic features described in association with 1q21.1 microdeletions include developmental delay, craniofacial dysmorphism and congenital anomalies. The 1q21.1 reciprocal duplication has been associated with macrocephaly or relative macrocephaly, frontal bossing, hypertelorism, developmental delay, intellectual disability and autism spectrum disorder.
Methods
Our study describes seven patients, who were referred to us for developmental delay/intellectual disability, dysmorphic features and, in some cases, congenital anomalies, in whom we identified 1q21.1 CNVs by array-CGH.
Results
Our data confirm the extreme phenotypic variability associated with 1q21.1 microdeletion and microduplication. We observed common phenotypic features, described in previous studies, but we also described, for the first time, congenital hypothyroidism in association with 1q21.1 deletion and trigonocephaly associated with 1q21.1 duplication.
Conclusions
The aim of this study is to contribute to the definition of the phenotype associated with reciprocal 1q21.1 deletions and duplications.
Similar content being viewed by others
Background
In recent years, the introduction of new technologies such as comparative genomic hybridization (CGH) allows for the routine detection of submicroscopic deletions and duplications. Several studies of patients with global developmental delay, intellectual disability and/or congenital malformation of unknow cause have led to the identification of new genomic disorders [1,2,3,4].
Recurrent reciprocal 1q21.1 deletions and duplications have been associated with variable phenotypes. Phenotypic features described in association with 1q21.1 microdeletions include developmental delay, craniofacial dysmorphism and congenital anomalies. Developmental delay is usually mild and may involve global or specific areas. Psychiatric and behavioral abnormalities, such as autism spectrum disorders (ASD), schizophrenia and attention deficit hyperactivity disorder (ADHD), are also described in a minority of patients. Dysmorphic features are a common finding; they may include microcephaly (almost 50% of patients), frontal bossing, deep-set eyes, epicanthal folds, large nasal bridge, long philtrum and highly arched palate. Several congenital anomalies may be associated with the deletion: congenital heart disease (CHD), eye abnormalities (microphthalmia, chorioretinal and iris colobomas, strabism, various type of cataracts), skeletal and genitourinary malformations. In some cases, seizures are also described (15%) [5,6,7,8,9]. As it is clear, the phenotype of 1q21.1 microdeletion has a high variability, so it is not possible to define a clinically recognizable syndrome.
Few individuals with 1q21.1 reciprocal duplication have been reported in literature. Most recognizable features are macrocephaly or relative macrocephaly, frontal bossing, hypertelorism, developmental delay, intellectual disability and autism spectrum disorder [6, 7]. Individuals with 1q21.1 copy number variations (CNVs) may also have a normal phenotype.
The 1q21.1 critical region spans approximately 1.35 Mb (from 145 to 146.35 Mb) [6] and includes at least 12 genes, among which PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8, GPR89B. Deletions and duplications can be inherited from a parent in an autosomal dominant manner or occur de novo.
Our study describes seven patients, who were referred to us for developmental delay/intellectual disability, dysmorphic features and, in some cases, congenital anomalies, in whom we identified 1q21.1 CNVs by array-CGH. The aim of this study is to contribute to the definition of the phenotype associated with reciprocal 1q21.1 deletions and duplications.
Methods
Clinical reports
All patients were referred to our Regional Referral Centre for Rare Diseases for the presence of developmental delay, intellectual disability, dysmorphic features and/or congenital anomalies. One patient had a prenatal diagnosis, following the identification of anomalies on the obstetric ultrasound. Written informed consents were obtained from all participants.
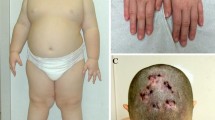
Patient 1 is a 6-year old girl. She is the third child of healthy, non-consanguineous parents. Her family history is positive for intellectual disability (one brother). She has microcephaly and mild dysmorphic features. Neuropsychiatric evaluation revealed intellectual disability, psychomotor and language delay. EEG (electroencephalogram) and brain CT were reported normal.
Patient 2 is a 8-year old boy, the second child of non-consanguineous parents. He was born at term of gestation by cesarean delivery. He was referred to us because of learning disabilities and encopresis. Physical examination showed growth retardation (all growth parameters lower than 3° percentile) and dysmorphic features: dry hair with abnormal implant, hypotelorism, muscular hypotrophy and bilateral clinodactyly of I, II, IV, V fingers. Brain and pituitary MRI was reported normal.
Patient 3 is the only one with a prenatal diagnosis. His family history is positive for intellectual disability and epilepsy. Obstetric ultrasound, performed at 21 weeks of gestation (WG), showed bilateral cysts of the choroid plexus and a reduction in size in the ossification nucleus of the nose. Because of the presence of this anomalies on ultrasound, an amniocentesis was performed. At birth (40,5 WG) the patient was immediately studied: he had no dysmorphic features and his growth parameters were normal. Brain, heart and abdominal ultrasounds were normal.
Patient 4 is a 2-year old boy, the first child of non-consanguineous parents. His family history is positive for intellectual disability, congenital anomalies (first pregnancy: stillbirth with agenesis of the radius and thumb) and chromosomal abnormalities (trisomy 21). He was born at 39,1 WG by cesarean delivery. He was referred to us because of the presence of severe vesicoureteral reflux (VUR), with hydronephrosis detected prenatally, and dysmorphism. Physical examination showed growth retardation (all growth parameters lower than 3° percentile) and dysmorphic features: prominence of the metopic suture, plagiocephaly, hypotelorism, bilateral clinodactyly of IV and V fingers and toes. Brain and heart ultrasounds were reported normal.
Patient 5 is a 8-year old boy. He is the first child of healthy, non-consanguineous parents. His family history is positive for intellectual disability and microcephaly. He was born at 29,2 WG because of premature rupture of membranes (PROM). Because of his prematurity, he was admitted to neonatal intensive care: he had pneumothorax, respiratory distress and retinopathy of prematurity (ROP). When the patient came to our attention, he had intellectual disability, spastic tetraparesis, strabism and craniofacial dysmorphism: microcephaly, protruding ears, prominent nasal bridge, short philtrum, micrognathia and spaced teeth. Brain MRI showed periventricular leukomalacia, polymicrogyria and dilatation of lateral ventricles. Heart and abdominal ultrasounds were reported normal.
Patient 6 is a 21-month old baby, the second child of non-consanguineous parents. His family history is positive for language delay. He was born at 40 WG by cesarean delivery. He was referred to us for the presence of dysmorphic features and psychomotor delay. On physical examination he had trigonocephaly, epicanthus, down-slanting palpebra fissures, large nasal bridge, thin upper lip, large mouth, small and dysplastic ears, thick fingers and broad thumbs and hallux. Heart and abdominal ultrasounds were reported normal, while brain CT and MRI showed areas of periventricular leukomalacia and mild ptosis of the cerebellar tonsils.
Patient 7 is a 2-year old boy, the first child of non-consanguineous parents, born at 36,1 WG by cesarean delivery. His mother has intellectual disability, thyroid hypoplasia and hypothyroidism, reason why she had performed array-CGH, showing a 1q21.1 deletion.
He has dysmorphic features (sloping forehead, prominent occiput, flat nasal bridge, long philtrum, thin upper lip, large mouth, protruding tongue), congenital hypothyroidism and ectopic urethral meatus. The ultrasound study did not document thyroid hypoplasia. Brain and abdominal ultrasounds were reported normal.
Array-CGH analysis
Genomic DNA of the patients (except patient 3) and their parents was extracted from peripheral blood lymphocytes using KingFisher Blood DNA Kit (Thermo Scientific, Vantaa, FI) according to manufacturers’ instructions. Proband and reference DNA (Promega Corporation, Madison, WI, USA) were labeled with Cy5-dUTP and Cy3-dUTP respectively. Whole genome array-CGH was performed using Human Genome CGH Microarray Kit 8x60K (Agilent Technologies, Santa Clara, CA, USA) with an average resolution of 100 kb (Build37: Feb 2009-hg19) according to manufacturers’ instructions. Images of the array were acquired with Agilent scanner G2505B and analyzed with Feature Extraction software v9.5.1 (Agilent Technologies, Santa Clara, CA, USA). Graphical overviews of results were obtained with Genomic Workbench Standard Edition software v5.0.14 (Agilent Technologies, Santa Clara, CA, USA).
Genomic DNA of patient 3 was extracted from amniotic cells. Proband and reference DNA (Promega; G147A) were labeled with Cy5-dUTP and Cy3-dUTP respectively. Whole genome array-CGH was performed using CytoChip Oligo arrays 8x60K (Bluegnome, Cambridge, UK) with an average resolution of 200–250 kb. Images of the array were acquired and analyzed with BlueFuse Multi Software for microarray.
Results
Among our seven patients we detected, using array-CGH, four 1q21.1 deletions, two 1q21.1 duplications and a double rearrangement on the long arm of a chromosome 1, with a 1q21.1 duplication and a 1q21.1-q21.2 deletion.
Before undergoing to array-CGH analysis, a karyotype study was performed in patient 2, 4 and 7: all patients have a normal male karyotype (46,XY).
We detected a 1q21.1 deletion in patient 2, 4, 5 and 7. Patient 2 and 4 have a 1q21.1 deletion that spans approximately 1,2 Mb (146.564.743–147.786.706). Array-CGH analysis in both parents of patient 2 shows that the rearrangement has a paternal origin, while these data are not available for the parents of patient 4. Patient 5 and 7 have a 1q21.1 deletion, of approximately 1,1 Mb (146.641.601–147.786.706)(Fig. 1). The rearrangement is inherited from his mother in patient 7, while it is de novo for what concerns patient 5. This deleted region includes numerous genes: PRKAB2, PDIA3P, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8, GPR89B, PDZK1P1, NBPF11, NBPF24. Patients with a microduplication are patient 1 and 6. Patient 1 has a 1q21.1 duplication of approximately 932 kb (145.632.334–146.564.802), involving the genes: GPR89A, PDZK1, CD160, RNF115, POLR3C, NUDT17, PIAS3, ANKRD35, PEX11B, ITGA10, RBM8A, LIX1L, POLR3GL, HFE2, NBPF10, NOTCH2NL. The origin of this rearrangement is unknown, because array-CGH analysis data concerning parents are not available. Patient 6 has a 1q21.1 duplication that spans approximately 456 kb (145.291.711–145.747.269)(Fig. 2). Array-CGH analysis in both parents shows that the rearrangement has a maternal origin. The duplicated region involves genes such as NBPF20, GPR89A, PDZK1, CD160 and RNF115.
Finally, patient 3 is the only one with a prenatal diagnosis. Fetal karyotype on amniotic cells revealed a chromosomal insertion involving the short arm of a chromosome 3 and the short arm of a chromosome 1: 46, XY, ins(1;3)(p22;p13p23). This rearrangement was confirmed by Fluorescent in-situ Hybridization (FISH): ish ins(1;3)(p22;p13p23)(pVYS218C+;wpc3+). The karyotype study in both parents was normal, this showing that the rearrangement was de novo. In order to better define this chromosomal abnormality, array-CGH analysis was performed. We detected a double rearrangement on the long arm of a chromosome 1: a 1q21.1 duplication extended for 1.2 Mb (from 144.612.681 to 145.799.573) and a 1q21.1-q21.2 deletion extended for 1,7 Mb (from 146.155.983 to 147.824.178). These complex rearrangements were confirmed by Dye-swap. Both rearrangements are de novo. The duplicated region includes many genes: NBPF9, PDE4DIP, NBPF10, HFE2A, TXNIP, RBM8A, PEX11B, ITGA10, PIAS3, CD160, PDZK1, GPR89A. The 1q21.1-q21.2 deletion comprises the 1q21.1 microdeletion syndrome critical region. This region includes genes such as PRKAB2, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8 and GPR89B. Though karyotype and FISH analysis performed prenatally revealed a chromosomal insertion involving the short arm of a chromosome 3 and the short arm of a chromosome 1, array-CGH analysis did not show gain or loss of genetic material of these regions. So we can affirm that it is a balanced rearrangement. At the same time, array-CGH allowed us to highlight a double rearrangement involving the long arm of chromosome 1.
(The main characteristics of the patients are shown in Table 1).
Discussion
Recurrent reciprocal 1q21.1 deletions and duplications have been associated with variable phenotypes. The 1q21.1 microdeletion syndrome is characterized by a high variable clinical phenotype. The most common features, even if not constant, are microcephaly, dysmorphism, developmental delay and mild intellectual disability. Several congenital anomalies may be associated with 1q21.1 deletion: CHD, eye abnormalities, skeletal and genitourinary malformations. Psychiatric and behavioral abnormalities, such as ASD, schizophrenia and ADHD, are also described in a minority of patients.
The 1q21.1 reciprocal duplication have been associated with macrocephaly or relative macrocephaly, dysmorphic features, developmental delay, intellectual disability and autism spectrum disorder. Individuals with 1q21.1 CNVs may also have a normal phenotype.
We describe seven patients, who were referred to us for developmental delay/intellectual disability, dysmorphic features and, in some cases, congenital anomalies, in whom we identified by array-CGH analysis: four 1q21.1 deletions, two 1q21.1 duplications and a double rearrangement on the long arm of a chromosome 1, with a 1q21.1 duplication and a 1q21.1-q21.2 deletion. Duplications detected in patient 3 and 6 involve also the proximal part of 1q21.1 region. 1q21.1 proximal microduplications are associated with variable and not defined phenotypes, including intellectual disabiliy, dysmorphic features and behavior problems.
Our data confirm the extreme phenotypic variability associated with 1q21.1 microdeletion and microduplication. We observed common phenotypic features, described in previous studies, such as microcephaly, mild dysmorphic features, developmental delay and intellectual disability. Three patients also have skeletal anomalies (clinodactyly of fingers and toes, broad thumbs and hallux), while two patients have genitourinary malformation (VUR, ectopic urethral meatus). Furthermore, in our cohort of patients we described, for the first time, congenital hypothyroidism in association with 1q21.1 deletion and trigonocephaly associated with 1q21.1 duplication. None of the patients in our study have CHD.
Disagreeing with the literature, one patient harbouring a duplication (patient 1) has microcephaly instead of relative/absolute macrocephaly.
Only one patient (patient 3), the one with a double rearrangement on the long arm of a chromosome 1 prenatally diagnosed, has no phenotypic manifestations: we suggest a careful follow-up in order to evaluate his psychomotor development and the possible occurence of psychiatric and behavioral abnormalities.
Array-CGH analysis in the parents show a parental origin in three patients: two patients inherited the rearrangement from their mother, while one has a paternal inheritance. Among these, only in one case (patient 7) the mother was affected, having intellectual disability, thyroid hypoplasia and hypothyroidism, while the father of patient 2 and the mother of patient 6 have a normal phenotype. In two of our patients the rearrangement occurs de novo; lastly, in two cases data about parents are not available, so the inheritance remains unknown.
CNVs detected in our patients have different size, ranging from 456 kb to 1,2 Mb (Fig. 3a and b). Many genes are included in the deleted region, but no single gene mutations or haploinsufficiency are known to cause the 1q21.1 microdeletion phenotype. The most studied genes are CHD1L, PRKAB2, GJA5, GJA8 and NBPF genes.
a. Mapping of deletion 1q21.1 in patients 2, 3, 4, 5 and 7. The red box indicates the minimal common deleted region. b. Mapping of duplication 1q21.1 in patients 1, 3 and 6. The red box indicates the minimal common duplicated region. Note that patient 3 is in both Fig. A and B because of his double rearrangement on the long arm of a chromosome 1
CHD1L has been implicated in chromatin remodelling, relaxation and decatenation. Haploinsufficiency or over-expression of CHD1L have been implicated in impaired chromatin remodeling during DNA single strand break repair, suggesting that it has a role in DNA Damage Response [10]. Despite that, the phenotypic consequences of alterations in DNA Damage Response in patients with 1q21.1 CNVs are not clear [11].
PRKAB2 encodes the β2-subunit of AMPK (AMP-activated protein kinase), a regulator of cellular response to a large number of external stimuli, which seems to have an important role in brain function [11].
GJA5 and GJA8 have been identified in subjects with cardiac defect and eye abnormalities, respectively [5, 12, 13].
The NBPF (neuroblastoma breakpoint family) gene family consists of 24 members. Deleted or duplicated regions involved in our patients include: NBPF9, NBPF10, NBPF11, NBPF20 and NBPF24.
This gene family encodes for the DUF1220 protein domains, that seem to be associated with pathological variations in brain-size and neocortex volume. In particular, the loss of DUF1220 copy number has been associated with microcephaly, while the increases in DUF1220 copy number underlie 1q21-associated macrocephaly [14].
In the duplicated region, the most studied gene is PDZK1. Over-expression of this gene has been described in association with an increased risk of ASD and psychiatric diseases [15, 16].
The duplicated region in patient 1 encompasses also the PEX11B (peroxisomal membrane protein 11B) gene. It seems to be involved in the regulation of neuronal differentiation and migration, so it could be responsible for pathological variations in brain-size [17].
Among the genes involved in patient 7 (PRKAB2, PDIA3P, FMO5, CHD1L, BCL9, ACP6, GJA5, GJA8, GPR89B, PDZK1P1, NBPF11, NBPF24), with current knowledge, none seems to be related with congenital hypothyroidism. We believe it is important to report this finding in order to better define this complex phenotype, although we are aware that more studies are needed to explain the relationship between 1q21.1 deletion and hypothyroidism. Likewise, none of the genes duplicated in patient 6 (NBPF20, GPR89A, PDZK1, CD160, RNF115) are known to cause trigonocephaly.
Conclusions
In conclusion, our results confirm the high phenotypic variability of 1q21.1 deletions and duplications, extending the phenotype with the finding of congenital hypothyroidism and trigonocephaly in association with 1q21.1 deletion and duplication, respectively.
Further studies are needed to better define the 1q21.1 microdeletion/microduplication syndrome and to understand how haploinsufficiency or over-expression of genes included in this region can cause this complex phenotype.
Abbreviations
- ADHD:
-
Attention deficit hyperactivity disorder
- ASD:
-
Autism spectrum disorder
- CGH:
-
Comparative genomic hybridization
- CHD:
-
Congenital heart disease
- CNVs:
-
Copy number variations
- EEG:
-
Electroencephalogram
- FISH:
-
Fluorescent in-situ Hybridization
- PROM:
-
Premature rupture of membrans
- ROP:
-
Retinopathy of prematurity
- VUR:
-
Vesicoureteral reflux
- WG:
-
Weeks of gestation
References
Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, Choy CO, Straatman H, van der Vliet W, Huys EH, van Rijk A, Smeets D, van Ravenswaaij-Arts CM, Knoers NV, van der Burgt I, de Jong PJ, Brunner HG, van Kessel AG, Schoenmakers EF, Veltman JA. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003;73:1261–70.
Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, Colleaux L, Bobrow M, Carter NP. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–8.
Friedman JM, Baross A, Delaney AD, Ally A, Arbour L, Asano J, Bailey DK, Barber S, Birch P, Brown-John M, Cao M, Chan S, Charest DL, Farnoud N, Fernandes N, Flibotte S, Go A, Gibson WT, Holt RA, Jones SJM, Kennedy GC, Krzywinski M, Langlois S, Li HI, McGillivray BC, Nayar T, Pugh TJ, Rajcan-Separovic E, Schein JE, Schnerch A, Siddiqui A, Van Allen MI, Wilson G, Yong SL, Zahir F, Eydoux P, Marra MA. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–13.
Koolen DA, Pfundt R, de Leeuw N, Hehir-Kwa JY, Nillesen WM, Neefs I, Scheltinga I, Sistermans E, Smeets D, Brunner HG, van Kessel AG, Veltman JA, de Vries BBA. Genomic microarrays in mental retardation: a practical workflow for diagnostic applications. Hum Mutat. 2009;30:283–92.
Christiansen J, Dyck JD, Elyas BG, Lilley M, Bamforth JS, Hicks M, Sprysak KA, Tomaszewski R, Haase SM, Vicen-Wyhony LM, Somerville MJ. Chromosome 1q21. 1 contiguous gene deletion is associated with congenital heart disease. Circ Res. 2004;94(11):1429–35.
Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, Collins A, Mercer C, Norga K, de Ravel T, Devriendt K, EMHF B, de Leeuw N, Reardon W, Gimelli S, Bena F, Hennekam RC, Male A, Gaunt L, Clayton-Smith J, Simonic I, Park SM, Mehta SG, Nik-Zainal S, Woods CG, Firth HV, Parkin G, Fichera M, Reitano S, Lo Giudice M, Li KE, Casuga I, Broomer A, Conrad B, Schwerzmann M, Räber L, Gallati S, Striano P, Coppola A, Tolmie JL, Tobias ES, Lilley C, Armengol L, Spysschaert Y, Verloo P, De Coene A, Goossens L, Mortier G, Speleman F, van Binsbergen E, Nelen MR, Hochstenbach R, Poot M, Gallagher L, Gill M, McClellan J, King MC, Regan R, Skinner C, Stevenson RE, Antonarakis SE, Chen C, Estivill X, Menten B, Gimelli G, Gribble S, Schwartz S, Sutcliffe JS, Walsh T, SJL K, Sebat J, Romano C, Schwartz CE, Veltman JA, BBA d V, Vermeesch JR, JCK B, Willatt L, Tassabehji M, Eichler EE. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359:1685–99.
Brunetti-Pierri N, Berg JS, Scaglia F, Belmont J, Bacino CA, Sahoo T, Lalani SR, Graham B, Lee B, Shinawi M, Shen J, Kang SHL, Pursley A, Lotze T, Kennedy G, Lansky-Shafer S, Weaver C, Roeder ER, Grebe TA, Arnold GL, Hutchison T, Reimschisel T, Amato S, Geragthy MT, Innis JW, Obersztyn E, Nowakowska B, Rosengren SS, Bader PI, Grange DK, Naqvi S, Garnica AD, Bernes SM, Fong CT, Summers A, Walters WD, Lupski JR, Stankiewicz P, Cheung SW, Patel A. Recurrent reciprocal 1q21. 1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat Genet. 2008;40(12):1466–71.
Brunet A, Armengol L, Heine D, Rosell J, García-Aragonés M, Gabau E, Estivill X, Guitart M. BAC array CGH in patients with Velocardiofacial syndrome-like features reveals genomic aberrations on chromosome region 1q21. 1. BMC medical genetics. 2009;10:144.
Digilio MC, Bernardini L, Consoli F, Lepri FR, Giuffrida MG, Baban A, Surace C, Ferese R, Angioni A, Novelli A. Congenital heart defects in recurrent reciprocal 1q21.1 deletion and duplication syndromes: rare association with pulmunary valve stenosis. Eur J Med Genet. 2013;56(3):144–9.
Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Lackson SP, Owen-Hughes T, Boulton SJ. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodelling enzyme ALC1. Science (New York, NY) 325: 1240-1243.
Harvard C, Strong E, Mercier E, Colnaghi R, Alcantara D, Chow E, Martell S, Tyson C, Hrynchak M, McGillivray B, Hamilton S, Marles S, Mhanni A, Dawson AJ, Pavlidis P, Qiao Y, Holden JJ, Lewis SME, O’Driscoll M, Rajcan-Separovic E. Understanding the impact of 1q21. 1 copy number variant. Orphanet J Rare Dis. 2011;6:54.
Devi RR, Vijayalakshmi P. Novel mutations in GJA8 associated with autosomal dominant congenital cataract and microcornea. Mol Vis. 2006;12(2):190–5.
Arora A, Minogue PJ, Liu X, Addison PK, Russel-Eggitt I, Webster AR, Hunt DM, Ebihara L, Beyer EC, Berthoud VM, Moore AT. A novel connexin50 mutation associated with congenital nuclear pulverulent cataracts. J Med Genet. 2008;45:155–60.
Dumas LJ, O'Bleness MS, Davis JM, Dickens CM, Anderson N, Keeney JG, Jackson J, Sikela M, Raznahan A, Giedd J, Rapoport J, Nagamani SSC, Erez A, Brunetti-Pierri N, Sugalski R, Lupski JR, Fingerlin T, Wai Cheung S, Sikela JM. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am J Hum Genet. 2012;91:444–54.
Casey JP, Magalhaes T, Conroy JM, Regan R, Shah N, Anney R, Shields DC, et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet. 2012;131(4):565–79.
Goodbourn PT, Bosten JM, Bargary G, Hogg RE, Lawrance-Owen AJ, Mollon JD. Variants in the 1q21 risk region are associated with a visual endophenotype of autism and schizophrenia. Genes Brain Behav. 2014;13(2):144–51.
Milone R, Valetto A, Battini R, Bertini V, Valvo G, Cioni G, Sicca F. Focal cortical dysplasia, microcephaly and epilepsy in a boy with 1q21. 1-q21. 3 duplication. Eur J Med Genet. 2016;59(5):278–82.
Acknowledgements
We would like to thank the patients and their family, and all the participants of this study.
Funding
The authors declare no funding for this work.
Availability of data and materials
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors contributed in the same way to collect, analyze and interpret data and write the manuscript; all authors read and approved the final draft.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Written informed consents were obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Busè, M., Cuttaia, H.C., Palazzo, D. et al. Expanding the phenotype of reciprocal 1q21.1 deletions and duplications: a case series. Ital J Pediatr 43, 61 (2017). https://doi.org/10.1186/s13052-017-0380-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13052-017-0380-x