Abstract
We aimed to explore role of lncRNA PAX-interacting protein 1-antisense RNA1 (PAXIP1-AS1) in ovarian cancer (OC). RT-qPCR analysis identified upregulation of PAXIP1-AS1 in OC cell lines. Functionally, PAXIP1-AS1 knockdown inhibited cell proliferation, accelerated cell apoptosis, and suppressed cell migration and epithelial-mesenchymal transition (EMT) process. Upregulation of PAXIP1-AS1 was induced by CBP-mediated H3K27 acetylation (H3K27ac) via bioinformatic analysis and ChIP assay. Furthermore, PAXIP1-AS1 served as a competing endogenous RNA (ceRNA) to regulate PCBP2 expression by sponging microRNA-6744-5p (miR-6744-5p). Restoration experiments showed that overexpressed PCBP2 rescued effects of silenced PAXIP1-AS1 on cell proliferation, apoptosis, migration and EMT. Overall, lncRNA PAXIP1-AS1 activated by H3K27ac functioned as a tumor promoter in OC via mediating miR-6744-5p/PCBP2 axis, which provided promising insight into exploration on OC therapy.
Similar content being viewed by others
Introduction
As a female gynecologic malignancy, ovarian cancer (OC) is one of the most common type of lethal tumors with higher mortality rate worldwide [1]. Each year, there is a continuous increase in the newly-diagnosed OC cases and in OC-related deaths [2]. In recent decades, therapeutic approaches, such as surgery, chemotherapy and radiotherapy, have achieved great progress. However, the long-term survival is still poor due to the imperceptible symptoms at early stage and the distant metastasis at advanced stage [3, 4]. Thus, it is quite imperative to explore the underlying mechanisms and develop novel methods for the treatment of OC.
Long noncoding RNAs (lncRNAs), a new group of noncoding RNAs with over 200 nucleotides in length, are recently identified [5]. Although no potential ability in protein-coding, lncRNAs have been found to exert important effect on multiple biological processes [6, 7]. Extensive studies have suggested that lncRNAs could modulate survival, cell proliferation, stemness, differentiation, and epithelial-mesenchymal transition (EMT) [8,9,10]. For example, lncRNA CRNDE shows a high level in tongue squamous cell carcinoma tissues and inhibits miR-384 to facilitate cell proliferation and metastasis [11]. In addition, high-expressed LINC01296 is revealed to predict poor prognosis in lung cancer patients and enhances tumor growth via modulating miR-598/Twist1 pathway [12]. Importantly, mounting reports indicate that lncRNAs regulate gene expression to mediate cancer progression through multiple ways, such as histone modification, transcriptional and post-transcriptional regulation [13]. Among which, lncRNAs were widely reported as competing endogenous RNAs (ceRNAs) that involved in lncRNA/miRNA/mRNA network [14]. This is a new model which proposes that lncRNAs regulate expression of target genes via miRNA response elements [15]. Because of the importance of lncRNAs in cancers, much more research should be conducted to depict their functions in OC.
LncRNA PAX-interacting protein 1-antisense RNA1 (PAXIP1-AS1) has been reported as a tumor promoter in glioma by aggravating cell invasion and angiogenesis [16]. However, its functional role in other cancers, especially in OC remains obscure. This study focused on the biological role and underlying mechanism of PAXIP1-AS1 in OC. The results demonstrated that PAXIP1-AS1 induced by H3K27ac was upregulated in OC cells and promoted cellular process in OC by sponging miR-6744-5p and targeting PCBP2, providing a new molecular mechanism implicated in OC cellular development.
Materials and methods
Cell lines
Human ovarian epithelial cell line (HOSEpiC) and OC cell lines (SKOV3, A2780, OVCAR3 and COV362) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The four OC cell lines were identified for study since functional experiments and examination of gene expression were successfully conducted using these cell lines in previous studies [17,18,19,20]. All above cell lines were cultured at 37 °C in RPMI-1640 medium with a supplement of 100 U/ml penicillin, 100 mg/ml streptomycin and 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA) in a humidified atmosphere containing 5% CO2. C646, an inhibitor of acetylation, was commercially provided by Sigma Chemical (St. Louis, MO).
Cell transfection
The miR-6744-5p mimics/inhibitor and their matched negative control (NC mimics/inhibitor), short hairpin RNAs (shRNAs) against PAXIP1-AS1 or CBP (sh-PAXIP1-AS1#1/2 or sh-CBP#1/2) and their negative control (sh-NC), and overexpressing plasmids pcDNA3.1/PCBP2 (PCBP2) and its negative control pcDNA3.1 (Vector) were synthesized by GenePharma (Shanghai, China). Using Lipofectamine 2000 (Invitrogen), cell transfection was conducted with manufacturer’s instructions. After 48 h of cell culture, RT-qPCR was conducted to validate the transfection efficiency.
RT-qPCR
First, TRIzol reagent (Invitrogen) was used to isolate total RNAs from OC cell lines. Next, the reverse transcription of miRNA or lncRNA/mRNA was performed by the miRNA First-Stand cDNA Synthesis Kit (GeneCopoeia) or the cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). Afterwards, RT-qPCR was performed on Applied Biosystems 7500 Real-time PCR Systems (Thermo Fisher Scientific). Gene expression was quantified by 2−ΔΔCt method and normalized to U6 or GAPDH.
Colony formation assay
Cells at a density of 600 cells/well were grown in 6-well plates with RPMI-1640 medium containing 10% FBS. After 2 weeks of cell culture, methanol was used for fixing colonies for 15 min at room temperature, and then the cells were stained with 0.1% crystal violet (Invitrogen) for 15 min. Finally, the number of visible colonies was manually counted.
EdU assay
After transfection, SKOV3 or OVCAR3 cells were collected and plated to 96-well plates at the density of 1 × 104 cells each well. Later, the plates were added with EdU assay kit (Ribobio) at 37 °C for 2 h. Cell nuclei was stained by DAPI solution. Finally, a fluorescence microscope (Olympus, Tokyo, Japan) was applied to observe proliferative cells.
Flow cytometry analysis
SKOV3 or OVCAR3 cells were seeded in 6-well plate. After centrifugalization, the binding buffer, resuspended with residue, was added with 5-μL Annexin V-FITC and 5-μL propidium (PI). Next, a flow cytometer (BD, Franklin Lake, NJ, USA) was used to measure cell apoptotic rate, and results were analyzed by the software WinMDI 2.9 (Invitrogen).
Western blot analysis
Cell lysis was conducted in the RIPA lysis buffer, and then the lysate was separated by 10% SDS-PAGE and transferred to PVDF membranes (Millipore). Blocked with 5% nonfat milk, the membranes were incubated with primary antibodies overnight at 4 °C. After an incubation with secondary antibody at 37 °C for 1 h, the enhanced chemiluminescence (ECL) Plus kit (Beyotime, Shanghai, China) was used to visualize the protein bands.
Transwell assay
The migratory ability of SKOV3 and OVCAR3 cells was assessed using transwell chambers without matrigel. In brief, the cells (1 × 105) were seeded to upper chambers containing 200 ul serum-free media. The lower chambers were added with 600 ul of 20% FBS media. Incubated for 48 h, the cells without invasion in the upper chambers were wiped away by cotton wool. Fixed with methanol, the cells were stained with 0.5% crystal violet in the lower chamber. Finally, five random fields were photographed under an inverted microscope.
ChIP assay
ChIP assays were carried out by ChIP Assay Kits (Thermo Fisher Scientific) with SKOV3 or OVCAR3 cells. For DNA-protein cross-links, SKOV3 or OVCAR3 cells were incubated with 1% formaldehyde for 10 min, and then an ultrasound machine was utilized to break the cross-linked chromatin DNAs into segments sized 200 to 1000 bp. The chromatin lysate was precipitated by anti-H3K27ac (Abcam), anti-CBP (Abcam) or anti-IgG (Abcam). Finally, RT-qPCR was performed to analyze the ChIP samples.
Subcellular fractionation assay
First, the nuclear and cytoplasmic fractions of SKOV3 and OVCAR3 cells were obtained. Next, with the Cytoplasmic & Nuclear RNA Purification Kit (Norgen), the fractions were separated and purified as per the manual. At last, RT-qPCR was performed to analyze the isolated RNA (GADPH, U6, PAXIP1-AS1). GAPDH was the internal control for cytoplasm, and the distribution of PAXIP1-AS1 in nucleus was normalized to U6.
Luciferase reporter assay
To conduct the luciferase reporter assay, pmirGLO vector (Promega, Madison, WI) was used. Briefly, the wild-type (WT) PAXIP1-AS1/PCBP2 or mutant (Mut) PAXIP1-AS1/PCBP2 binding element in the sequence of miR-6744-5p was cloned into pmirGLO vector for the construction of PAXIP1-AS1-WT/Mut or PCBP2-WT/Mut. Then, the reporters were separately co-transfected with miR-6744-5p mimics in SKOV3 or OVCAR3 cells. After 48 h, the relative luciferase activity was testified by the dual-luciferase reporter assay system (Promega).
RNA pulldown assay
NC-miRNA and miR-6744-5p-WT/Mut were labeled with biotin and then transfected into SKOV3 and OVCAR3 cells. Streptavidin magnetic beads were incubated with the cell lysates for 4 h at 4 °C. Using precooled lysis buffer and salt buffer, the beads were rinsed. After that, PAXIP1-AS1 or PCBP2 level was detected following the extraction of pull-down RNAs.
RIP assay
For RNA immunoprecipitation, the EZMagna RIP kit (Millipore) was applied. Cells were lysed in RIP lysis buffer after being harvested. Then, the cell lysate was incubated with magnetic beads absorbed anti-IgG (Millipore) or anti-Ago2 antibody (Millipore). Finally, RT-qPCR analyzed the purified RNA.
Statistical analysis
Three biological repeats were applied to all experimental procedures. Shown as the mean ± SD, data were statistically analyzed through GraphPad Prism 6 (GraphPad). Differences between two groups were analyzed by Student’s t test or one-way ANOVA for multiple groups with p < 0.05 as a cut-off value.
Results
Upregulated PAXIP1-AS1 in OC enhanced cell proliferation, migration and inhibited cell apoptosis
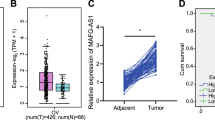
First, PAXIP1-AS1 expression pattern in OC cells was assessed by RT-qPCR for the investigation of its biological role. Compared with HOSEpiC cell line, high expression of PAXIP1-AS1 was observed in OC cell lines (SKOV3, A2780, OVCAR3 and COV362) (Fig. 1a). Then, SKOV3 and OVCAR3 cells presenting higher PAXIP1-AS1 expression were chose to probe functional role of PAXIP1-AS1 in OC. Accordingly, we designed and conducted loss-of-function assays by transfecting sh-PAXIP1-AS1 (sh-PAXIP1-AS1#1/2) into SKOV3 and OVCAR3 cells. The results of RT-qPCR analysis confirmed that PAXIP1-AS1 expression was apparently downregulated after sh-PAXIP1-AS1 transfection (Fig. 1b). Through colony formation assay, we found silenced PAXIP1-AS1 significantly decreased colonies of SKOV3 and OVCAR3 cells (Fig. 1c). EdU assay further confirmed the inhibitory effect of PAXIP1-AS1 deficiency on cell proliferation (Fig. 1d). In addition, flow cytometry analysis suggested that cell apoptosis was remarkably promoted by sh-PAXIP1-AS1 transfection (Fig. 1e). To further confirm this, levels of apoptosis-related proteins after PAXIP1-AS1 knockdown were tested by western blot analysis. The results showed that silenced PAXIP1-AS1 reduced Bcl-2 protein level and lifted Bax, caspase 3 and caspase 9 levels (Fig. 1f). Transwell assay denoted that PAXIP1-AS1 knockdown obviously lessened migrated cells (Fig. 1g). Moreover, expression levels of migration-related proteins (MMP2, MMP9) and EMT-relevant proteins (E-cadherin, N-cadherin) with sh-PAXIP1-AS1 transfection were testified. As we observed, MMP2, MMP9 and N-cadherin protein levels were decreased while E-cadherin protein level was induced by PAXIP1-AS1 silencing (Fig. 1h). Overall, PAXIP1-AS1 was upregulated in OC and exhibited an oncogenic role by facilitating cell proliferation, migration, EMT and suppressing cell apoptosis.
Expression pattern and functional role of PAXIP1-AS1 in OC cells. a RT-qPCR data of PAXIP1-AS1 expression in HOSEpiC cell line and OC cell lines. b Knockdown of PAXIP1-AS1 in SKOV3 and OVCAR3 cells validated by RT-qPCR. c-d Proliferation of SKOV3 and OVCAR3 cells upon PAXIP1-AS1 silencing was evaluated via colony formation assay and EdU assay. e Apoptosis of SKOV3 and OVCAR3 cells after PAXIP1-AS1 silencing was assessed through flow cytometry analysis. f Protein levels of Bax, Bcl-2, caspase-3 and caspase-9 under sh-PAXIP1-AS1 transfection were detected by western blot. g Migration of SKOV3 and OVCAR3 cells transfected with sh-PAXIP1-AS1 was confirmed by Transwell assay. h MMP2, MMP9, E-cadherin and N-cadherin protein levels were testified with western blot upon PAXIP1-AS1 knockdown. *p < 0.05
PAXIP1-AS1 was transcriptionally activated by CBP-mediated H3K27ac
Thereafter, we explored the cause of PAXIP1-AS1 upregulation in OC. Existing reports highlighted that lncRNAs could be activated through H3K27ac at transcriptional level [21, 22]. Afterwards, high density of H3K27ac enrichment was predicted in PAXIP1-AS1 promoter region through genome bioinformatics analysis (Fig. 2a). To further validate this, ChIP assay was performed. As demonstrated in Fig. 2b, PAXIP1-AS1 promoter region was enriched with H3K27ac in both OC cells and HOSEpiC cell line. Importantly, H3K27ac enrichment level was significantly increased in SKOV3 and OVCAR3 cell lines compared with that in HOSEpiC cell line. Interestingly, we found that PAXIP1-AS1 expression was significantly downregulated when OC cells were treated with C646, an inhibitor of histone acetyltransferase (HAT) (Fig. 2c). Thus, we wondered whether there were other vital enzymes involved in H3K27ac process. Previous studies have confirmed that CBP is crucial for chromatin acetylation and responsible for the promotive acetylation [23]. To test this, RT-qPCR was used to evaluate CBP expression in OC cells. As expected, compared with CBP in HOSEpiC cell line, CBP was upregulated in OC cells (Fig. 2d). Subsequently, results of ChIP assay uncovered that CBP precipitates was obviously enriched by PAXIP1-AS1 promoter (Fig. 2e). Then, CBP was stably silenced by sh-CBP and the transfection efficiency was validated by RT-qPCR (Fig. 2f). For assessing the effect of CBP on H3K27ac enrichment, ChIP assay was conducted. As expected, CBP knockdown significantly decreased the enrichment of H3K27ac at PAXIP1-AS1 promoter (Fig. 2g). Furthermore, CBP knockdown also led to an obvious decrease on PAXIP1-AS1 expression in OC cells (Fig. 2h). In conclusion, PAXIP1-AS1 upregulation in OC was caused by CBP-mediated H3K27ac at its promoter region.
Transcriptional upregulation of PAXIP1-AS1 in OC via CBP-modulated K3K27ac. a UCSC predicted H3K27ac enrichment at PAXIP1-AS1 promoter. b H3K27ac level on PAXIP1-AS1 promoter in HOSEpiC, SKOV3 and OVCAR3 cells was determined by ChIP assay. c PAXIP1-AS1 expression in SKOV3 and OVCAR3 cells treated with DMSO or C646 was confirmed by RT-qPCR. d RT-qPCR results of CBP expression in OC cell lines and HOSEpiC cell line. e Binding of CBP on PAXIP1-AS1 promoter was detected via ChIP assay. f RT-qPCR analysis of CBP silencing in SKOV3 and OVCAR3 cells. g ChIP assay determined the effect of CBP knockdown on H3K27ac at PAXIP1-AS1 promoter. h PAXIP1-AS1 expression upon CBP deficiency in SKOV3 and OVCAR3 cells was assessed by RT-qPCR. *p < 0.05
PAXIP1-AS1 sponged miR-6744-5p in OC
Next, downstream molecular mechanism of PAXIP1-AS1 in OC was explored. The subcellular fractionation assay implied that PAXIP1-AS1 was mainly localized in the cytoplasm of OC cells (Fig. 3a). Increasing reports indicated that cytoplasmic lncRNAs regulated cancer progression by sequestering miRNAs [24]. Hence, we aimed to find the potential miRNAs for PAXIP1-AS1. Through DIANA tool, five miRNAs (binding score > 0.9) were predicted with binding site to PAXIP1-AS1 (Fig. 3b). Through RT-qPCR analysis, miR-6744-5p demonstrated a low level in OC cells, while miR-3942-3p, miR-6505-5p, miR-6796-5p and miR-1976 failed to show expression differences (Fig. 3c). Later, we obtained the binding sequence of PAXIP1-AS1 on miR-6744-5p and mutated the site to conduct luciferase reporter assay (Fig. 3d). Meanwhile, miR-6744-5p was overexpressed in SKOV3 and OVCAR3 cells with transfection of miR-6744-5p mimics (Fig. 3e). As observed, the luciferase activity of PAXIP1-AS1-WT, but not PAXIP1-AS1-Mut, exhibited an overt reduction upon miR-6744-5p overexpression (Fig. 3f). Furthermore, RNA pulldown assay revealed a great enrichment of PAXIP1-AS1 in wide type miR-6744-5p pellets (Fig. 3g). Data above highlighted the interaction between PAXIP1-AS1 and miR-6744-5p.
PAXIP1-AS1 was a sponge of miR-6744-5p in OC cells. a Distribution of PAXIP1-AS1 in SKOV3 and OVCAR3 cells was validated by the subcellular fractionation assay. b Predicted miRNAs for PAXIP1-AS1 by DIANA tool. c RT-qPCR analysis of miR-3942-3p, miR-6744-5p, miR-6505-5p, miR-6796-5p and miR-1976 in OC cells. d Binding site of miR-6744-5p on PAXIP1-AS1 sequence. e Overexpression efficiency of miR-6744-5p mimics was tested by RT-qPCR. f Binding of PAXIP1-AS1 to miR-6744-5p was assessed through luciferase reporter assay. g The reaction between PAXIP1-AS1 and Bio-miR-6744-5p-WT or Bio-miR-6744-5p-Mut was evaluated via RNA pulldown assay. *p < 0.05
PAXIP1-AS1 positively regulated PCBP2 expression through miR-6744-5p
It was commonly recognized that lncRNAs release downstream genes by competitively combining with miRNAs [25]. Therefore, we explored the potential downstream targets of miR-6744-5p. Combining the prediction results of miRDB, miRTarBase and TargetScan (three bioinformatics websites), nine candidate genes were found (Fig. 4a). Considering that CCND2, CKS2, RNF187 and YWHAZ have been investigated in OC, the rest five mRNAs (PCBP2, YBX1, EPHA4, FAM102A and NMNAT2) were selected for further study. Through RT-qPCR analysis, we found only PCBP2 was apparently downregulated by miR-6744-5p overexpression (Fig. 4b). Then, miR-6744-5p site on PCBP2 sequence was identified through TargetScan website, and the mutation was designed (Fig. 4c). Through the luciferase reporter assay, miR-6744-5p overexpression attenuated the luciferase activity of PCBP2-WT reporter rather than PCBP2-Mut reporter (Fig. 4d). Later, biotinylated miR-6744-5p-WT was revealed to show significant enrichment of PCBP2 by RNA pulldown assay (Fig. 4e). An RIP assay validated the enrichments of PAXIP1-AS1, miR-6744-5p, and PCBP2 in the precipitates of anti-Ago2 (Fig. 4f). We then inhibited the expression of miR-6744-5p in SKOV3 and OVCAR3 cells by using miR-6744-5p inhibitor (Fig. 4g). Subsequently, we confirmed that miR-6744-5p inhibition counteracted the inhibitory role of silenced PAXIP1-AS1 in PCBP2 mRNA expression and protein levels (Fig. 4h). Collectively, PAXIP1-AS1 sequestered miR-6744-5p to positively regulated PCBP2 expression.
PAXIP1-AS1 regulated PCBP2 expression via miR-6744-5p. a Potential targets of miR-6744-5p combined from results of miRTarBase, TargetScan and miRDB online tools. b Effect of miR-6744-5p mimics on the expressions of PCBP2, YBX1, EPHA4, FAM102A and NMNAT2. c The binding sequence between PCBP2 and miR-6744-5p. d Luciferase activity of PCBP2-WT/Mut reporter under the transfection of miR-6744-5p mimics. e The RNA pulldown assay was performed to assess the interaction between miR-6744-5p and PCBP2. f Enrichments of PAXIP1-AS1, miR-6744-5p and PCBP2 in the beads conjugated with anti-Ago2 or anti-IgG was tested by the RIP assay. g MiR-6744-5p expression in cells with transfection of miR-6744-5p inhibitor. h PCBP2 mRNA and protein levels were tested in each group by RT-qPCR and western blot. *p < 0.05
PAXIP1-AS1 regulated OC cellular process by upregulating PCBP2 expression
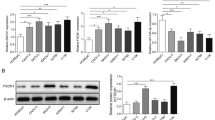
At last, we probed whether PCBP2 was necessary for the regulation of PAXIP1-AS1 on cellular process in OC. First, we transfected pcDNA3.1/PCBP2 into SKOV3 and OVCAR3 cells to overexpress PCBP2 (Fig. 5a). Colony formation and EdU assays depicted that cell proliferation hampered by PAXIP1-AS1 knockdown was recovered by PCBP2 overexpression (Fig. 5b-c). Moreover, the apoptosis of OC cells was promoted by PAXIP1-AS1 silencing, and such promotion was rescued by PCBP2 upregulation (Fig. 5d). Furthermore, overexpressed PCBP2 also reserved the effect of PAXIP1-AS1 knockdown on levels of apoptosis-relevant proteins (Fig. 5e). Transwell assay demonstrated that cell migration inhibited by silenced PAXIP1-AS1 was counteracted by pcDNA3.1/PCBP2 transfection (Fig. 5f). Additionally, the role of PAXIP1-AS1 downregulation in levels of proteins associated with migration and EMT process was countervailed via overexpressing PCBP2 (Fig. 5g). In conclusion, PAXIP1-AS1 accelerated OC cellular processes through regulating PCBP2 expression.
PCBP2 was a target of PAXIP1-AS1 in regulating OC cellular process. a Expression of PCBP2 in cells transfected with pcDNA3.1/PCBP2. b-c Cell proliferation with indicated transfection was tested by colony formation and EdU assays. d-e Apoptotic rate and levels of apoptosis-relevant proteins were respectively determined by flow cytometry analysis and western blot. F Cell migration in each group was measured through Transwell assay. g Levels of migration-related proteins and EMT-associated proteins in cells transfected with appointed plasmids were evaluated using western blot. *p < 0.05
Discussion
Over the past decades, significant attention has been paid on the effect of dysregulated lncRNAs in the progression of cancers, including OC. Previous studies have identified numerous lncRNAs as tumor facilitator or suppressor in OC. For example, lncRNA FLVCR1-AS1 enhances cell migration and EMT process in OC through mediating miR-513/YAP1 axis [26]. LncRNA WDFY3-AS2 acts as a tumor suppressor to inhibit tumor growth in OC via delaying miR-18a [27]. LncRNA HAND2-AS1 represents anti-oncogenic property in OC by targeting BCL2L11 [28]. This study was to explore the function of PAXIP1-AS1 in OC. Previously, it has been stated that PAXIP1-AS1 was upregulated and served as an oncogenic lncRNA in glioma [16]. Herein, we found high expression level of PAXIP1-AS1 in OC cell lines. Functional assays revealed that PAXIP1-AS1 accelerated proliferation, restrained apoptosis, and promoted migration and EMT process in OC cells. This suggested that PAXIP1-AS1 played a carcinogenic role in OC.
Histone H3 on lysine 27 acetylation (H3K27ac) has been known as a common type of histone posttranslational regulation, associating with active enhancer modulatory elements to transcriptionally activate gene expression [29, 30]. Previous researchers have suggested that H3K27ac at promoter regions led to the overexpression of some carcinogenic lncRNAs, such as PLAC2 and lnc-SLC4A1–1 [31, 32], thus facilitating tumor development. Herein, we discovered high enrichment of H3K27ac at PAXIP1-AS1 promoter region via UCSC genome browser. Then, H3K27ac high level on the promoter region of PAXIP1-AS1 was validated through ChIP assay. The decreased PAXIP1-AS1 expression by HAT inhibitor (C646) further verified that PAXIP1-AS1 upregulation was attributed to H3K27ac modification. Previously, increasing evidence has implied that CBP is an essential regulator on histone acetylation and gene transcription, including lncRNA [23, 31]. Thus, we first validated that CBP interacted with PAXIP1-AS1 promoter to trigger H3K27ac and upregulated PAXIP1-AS1 expression.
Mechanistically, it was widely accepted that lncRNAs play the role of ceRNAs by which lncRNAs released mRNAs from posttranscriptional silence through sponging miRNAs [24, 33]. Our study first found that miR-6744-5p could be potentially targeted by PAXIP1-AS1. Moreover, we conducted luciferase reporter assay and RNA pulldown assay to confirm the strongest affinity of miR-6744-5p with PAXIP1-AS1. Previously, miR-6744-5p was reported to accelerate anoikis by directly targeting NAT1 enzyme in breast cancer [34]. Furthermore, we identified that PCBP2 was the target gene for miR-6744-5p. Former studies have showed the oncogenic role of PCBP2 in cancers. As reported, PCBP2 is involved in cell proliferation and migration in bladder cancer [35]. PCBP2 enhances cell viability through regulating CDK2 in gastric cancer [36]. PCBP2 is overexpressed in glioblastoma and associated with unfavorable prognosis [37]. In our study, we first found that miR-6744-5p combined with PCBP2 to repress its expression, and that PAXIP1-AS1 upregulated PCBP2 expression through miR-6744-5p. Rescue assays delineated that PAXIP1-AS1 aggravated cell growth and migration by targeting PCBP2 in OC.
In conclusion, our study first depicted that H3K27ac-induced lncRNA PAXIP1-AS1 promoted cellular process through miR-6744-5p/PCBP2 axis, suggesting PAXIP1-AS1 as an underlying novel biomarker for improving research on OC molecular therapy.
Availability of data and materials
None.
Change history
20 September 2022
A Correction to this paper has been published: https://doi.org/10.1186/s13048-022-01038-5
19 December 2023
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1186/s13048-023-01327-7
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29.
Song X, et al. LncRNA SPRY4-IT1 regulates breast cancer cell stemness through competitively binding miR-6882-3p with TCF7L2. J Cell Mol Med. 2019;24(1):772.
Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. https://doi.org/10.1016/j.bpobgyn.2016.08.006.
La Vecchia C. Ovarian cancer: epidemiology and risk factors. Eur J Cancer Prev. 2017;26(1):55–62. https://doi.org/10.1097/CEJ.0000000000000217.
Lan H, Zeng J, Chen G, Huang H. Survival prediction of kidney renal papillary cell carcinoma by comprehensive LncRNA characterization. Oncotarget. 2017;8(67):110811–29. https://doi.org/10.18632/oncotarget.22732.
Ye B, et al. LncKdm2b controls self-renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J. 2018;37:8.
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B, et al. LncBRM initiates YAP1 signalling activation to drive self-renewal of liver cancer stem cells. Nat Commun. 2016;7(1):13608. https://doi.org/10.1038/ncomms13608.
Xu Q, Deng F, Qin Y, Zhao Z, Wu Z, Xing Z, et al. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell Death Dis. 2016;7(6):e2254. https://doi.org/10.1038/cddis.2016.149.
Yong W, Yu D, Jun Z, Yachen D, Weiwei W, Midie X, et al. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018;9(9):861. https://doi.org/10.1038/s41419-018-0908-z.
Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J, et al. Lnc-β-Catm elicits EZH2-dependent β-catenin stabilization and sustains liver CSC self-renewal. Nat Struct Mol Biol. 2016;23(7):631–9. https://doi.org/10.1038/nsmb.3235.
Ren Y, He W, Chen W, Ma C, Li Y, Zhao Z, et al. CRNDE promotes cell tongue squamous cell carcinoma cell growth and invasion through suppressing miR-384. J Cell Biochem. 2019;120(1):155–63. https://doi.org/10.1002/jcb.27206.
Xu L, Wei B, Hui H, Sun Y, Liu Y, Yu X, et al. Positive feedback loop of lncRNA LINC01296/miR-598/Twist1 promotes non-small cell lung cancer tumorigenesis. J Cell Physiol. 2019;234(4):4563–71. https://doi.org/10.1002/jcp.27235.
Xiong G, Feng M, Yang G, Zheng S, Song X, Cao Z, et al. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett. 2017;397:94–102. https://doi.org/10.1016/j.canlet.2017.02.020.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–8. https://doi.org/10.1016/j.cell.2011.07.014.
Barabási AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5(2):101–13. https://doi.org/10.1038/nrg1272.
Xu H, Zhao G, Zhang Y, Jiang H, Wang W, Zhao D, et al. Long non-coding RNA PAXIP1-AS1 facilitates cell invasion and angiogenesis of glioma by recruiting transcription factor ETS1 to upregulate KIF14 expression. J Exp Clin Cancer Res. 2019;38(1):486. https://doi.org/10.1186/s13046-019-1474-7.
Hao T, Huang S, Han F. LINC-PINT suppresses tumour cell proliferation, migration and invasion through targeting miR-374a-5p in ovarian cancer. Cell Biochem Funct. 2020;38(8):1089–99. https://doi.org/10.1002/cbf.3565.
Wang X, Yang B, She Y, Ye Y. The lncRNA TP73-AS1 promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. J Cell Biochem. 2018;119(9):7790–9. https://doi.org/10.1002/jcb.27158.
Liu HY, Zhang YY, Zhu BL, Feng FZ, Yan H, Zhang HY, et al. miR-21 regulates the proliferation and apoptosis of ovarian cancer cells through PTEN/PI3K/AKT. Eur Rev Med Pharmacol Sci. 2019;23(10):4149–55. https://doi.org/10.26355/eurrev_201905_17917.
Hofmann NA, Yang J, Trauger SA, Nakayama H, Huang L, Strunk D, et al. The GPR 55 agonist, L-α-lysophosphatidylinositol, mediates ovarian carcinoma cell-induced angiogenesis. Br J Pharmacol. 2015;172(16):4107–18. https://doi.org/10.1111/bph.13196.
Ding G, Li W, Liu J, Zeng Y, Mao C, Kang Y, et al. LncRNA GHET1 activated by H3K27 acetylation promotes cell tumorigenesis through regulating ATF1 in hepatocellular carcinoma. Biomed Pharmacother. 2017;94:326–31. https://doi.org/10.1016/j.biopha.2017.07.046.
Zhang E, Han L, Yin D, He X, Hong L, Si X, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45(6):3086–101. https://doi.org/10.1093/nar/gkw1247.
Bose DA, et al. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell. 2017;168(1–2):135–149.e22.
Liu K, et al. Functional role of a long non-coding RNA LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to pohotodynamic therapy. Biochim Biophys Acta Mol basis Dis. 2018;1864(9 Pt B):2871–80.
Chen X, Zeng K, Xu M, Hu X, Liu X, Xu T, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9(10):982. https://doi.org/10.1038/s41419-018-0962-6.
Yan H, Li H, Silva MA, Guan Y, Yang L, Zhu L, et al. LncRNA FLVCR1-AS1 mediates miR-513/YAP1 signaling to promote cell progression, migration, invasion and EMT process in ovarian cancer. J Exp Clin Cancer Res. 2019;38(1):356. https://doi.org/10.1186/s13046-019-1356-z.
Li W, Ma S, Bai X, Pan W, Ai L, Tan W. Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting AS a competing endogenous RNA of microRNA-18a in ovarian cancer. J Cell Physiol. 2020;235(2):1141–54. https://doi.org/10.1002/jcp.29028.
Chen J, Lin Y, Jia Y, Xu T, Wu F, Jin Y. LncRNA HAND2-AS1 exerts anti-oncogenic effects on ovarian cancer via restoration of BCL2L11 AS a sponge of microRNA-340-5p. J Cell Physiol. 2019;234(12):23421–36. https://doi.org/10.1002/jcp.28911.
Wang S, Zang C, Xiao T, Fan J, Mei S, Qin Q, et al. Modeling cis-regulation with a compendium of genome-wide histone H3K27ac profiles. Genome Res. 2016;26(10):1417–29. https://doi.org/10.1101/gr.201574.115.
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–6. https://doi.org/10.1073/pnas.1016071107.
Chen F, Qi S, Zhang X, Wu J, Yang X, Wang R. lncRNA PLAC2 activated by H3K27 acetylation promotes cell proliferation and invasion via the activation of Wnt/β-catenin pathway in oral squamous cell carcinoma. Int J Oncol. 2019;54(4):1183–94. https://doi.org/10.3892/ijo.2019.4707.
Yi T, Zhou X, Sang K, Huang X, Zhou J, Ge L. Activation of lncRNA lnc-SLC4A1-1 induced by H3K27 acetylation promotes the development of breast cancer via activating CXCL8 and NF-kB pathway. Artif Cells Nanomed Biotechnol. 2019;47(1):3765–73. https://doi.org/10.1080/21691401.2019.1664559.
Cheng JL, Li DJ, Lv MY, Pei YJ, Zhang XJ, Li L, et al. LncRNA KCNQ1OT1 regulates the invasion and migration of hepatocellular carcinoma by acting on S1PR1 through miR-149. Cancer Gene Ther. 2020. https://doi.org/10.1038/s41417-020-0203-x.
Malagobadan S, Ho CS, Nagoor NH. MicroRNA-6744-5p promotes anoikis in breast cancer and directly targets NAT1 enzyme. Cancer Biol Med. 2020;17(1):101–11. https://doi.org/10.20892/j.issn.2095-3941.2019.0010.
Wang J, Zhang H, Situ J, Li M, Sun H. KCNQ1OT1 aggravates cell proliferation and migration in bladder cancer through modulating miR-145-5p/PCBP2 axis. Cancer Cell Int. 2019;19(1):325. https://doi.org/10.1186/s12935-019-1039-z.
Chen C, Lei J, Zheng Q, Tan S, Ding K, Yu C. Poly (rC) binding protein 2 (PCBP2) promotes the viability of human gastric cancer cells by regulating CDK2. FEBS Open Bio. 2018;8(5):764–73. https://doi.org/10.1002/2211-5463.12408.
Luo K, Zhuang K. High expression of PCBP2 is associated with progression and poor prognosis in patients with glioblastoma. Biomed Pharmacother. 2017;94:659–65. https://doi.org/10.1016/j.biopha.2017.07.103.
Acknowledgments
We appreciate all participants who provide help for this study.
Funding
None.
Author information
Authors and Affiliations
Contributions
Both two co-authors participated in the literature search, analysis and interpretation of the data, and the writing of the manuscript. All authors saw and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1186/s13048-023-01327-7
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ma, Y., Zheng, W. RETRACTED ARTICLE: H3K27ac-induced lncRNA PAXIP1-AS1 promotes cell proliferation, migration, EMT and apoptosis in ovarian cancer by targeting miR-6744-5p/PCBP2 axis. J Ovarian Res 14, 76 (2021). https://doi.org/10.1186/s13048-021-00822-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13048-021-00822-z