Abstract
Background
The effect of competing endogenous RNA (ceRNA) can regulate gene expression by competitively binding microRNAs. Fascin-1 (FSCN1) plays an important role in the regulation of cellular migration and invasion during tumor progression, but how its regulatory mechanism works through the ceRNA effect is still unclear in bladder cancer (BLCA).
Methods
The role of fascin-1, miR-200b, and ZEB1-AS1 in BLCA was investigated in vitro and in vivo. The interaction between fascin-1, miR-200b, and ZEB1-AS1 was identified using bioinformatics analysis, luciferase activity assays, RNA-binding protein immunoprecipitation (RIP), quantitative PCR, and western blotting. Loss (or gain)-of-function experiments were performed to investigate the biological roles of miR-200b and ZEB1-AS1 on migration, invasion, proliferation, cell apoptosis, and cell cycle.
Results
ZEB1-AS1 functions as a competing endogenous RNA in BLCA to regulate the expression of fascin-1 through miR-200b. Moreover, the oncogenic long non-coding RNA ZEB1-AS1 was highly expressed in BLCA and positively correlated with high tumor grade, high TNM stage, and reduced survival of patients with BLCA. Moreover, ZEB1-AS1 downregulated the expression of miR-200b, promoted migration, invasion, and proliferation, and inhibited apoptosis in BLCA. Furthermore, we found TGF-β1 induced migration and invasion in BLCA by regulating the ZEB1-AS1/miR-200b/FSCN1 axis.
Conclusion
The observations in this study identify an important regulatory mechanism of fascin-1 in BLCA, and the TGF-β1/ZEB1-AS1/miR-200b/FSCN1 axis may serve as a potential target for cancer therapeutic purposes.
Similar content being viewed by others
Background
Bladder cancer (BLCA) is the most common urinary cancer worldwide [1]. Several genes have been associated with BLCA tumorigenesis. However, the molecular mechanisms underlying the progression of BLCA are still unclear.
Fascin-1 (FSCN1) is a globular actin-bundling protein [2]. There are three isoforms of fascin in vertebrate cells. Among them, FSCN1 is expressed in mesenchymal tissues and the nervous system, FSCN2 is expressed in the retinal photoreceptors, and FSCN3 is a testis-specific protein [3]. FSCN1 is located at the membrane ruffles, micro spikes, and stress fibers and induces membrane protuberance. The main function of FSCN1 is the formation of parallel actin bundles, which support filopodial and lamellipodial cell protrusions that are key cellular structures related to environmental guidance and cell movement, migration, and adhesion [4,5,6].
Recent studies have shown that FSCN1 levels are significantly increased in transformed epithelial cells as well as in various types of carcinomas [7,8,9], including BLCA [10, 11]. Our previous studies have found that FSCN1 promoted migration and invasion of BLCA [12, 13].
In the present study, through bioinformatic analysis, we found that the microRNA (miRNA) miR-200b may regulate FSCN1 expression. miR-200b has been shown to be a tumor suppressor in multiple cancer types, including BLCA. However, the expression pattern of miR-200b in BLCA is intriguing, in that it is higher in BLCA tissues than in normal bladder tissues, but lower in high grade tumors than in low grade tumors [14].
Long non-coding RNAs (lncRNAs) have been the focus of numerous studies in recent years. It has been suggested that lncRNAs act as “sponges” for microRNAs, reducing their effect on mRNAs and therefore regulating several biological processes. In the present study, we found that the lncRNA ZEB1-AS1 may regulate miR-200b. Zinc Finger E-Box Binding Homeobox 1 (ZEB1) is a promoter of epithelial-to-mesenchymal transition (EMT) and a target of miR-200b. Additionally, lncRNA ZEB1-AS1, which is the antisense of ZEB1, has been shown to be an oncogenic lncRNA in many kinds of cancers [15].
Transforming growth factor (TGF)-β has been shown to be an oncogene that can affect several signaling pathways in cancer cells [16, 17]. It has been previously reported that TGF-β1 downregulates the expression of miR-200 s [18] and upregulates the expression of FSCN1 [19]. However, the molecular details underlying this process are still unclear. In the present study, we found that ZEB1-AS1 is a downstream target of TGF-β1 and is involved in its regulatory mechanism on cell migration and invasion by affecting miR-200b/FSCN1 axis in BLCA cells. Our data might add insight on the diagnosis and treatment of BLCA.
Methods
Human samples
Ten paired BLCA and corresponding noncancerous tissues (located > 3 cm from the tumor), 60 BLCA tissue samples, and 23 normal tissue samples were obtained from patients at the Department of Urology, The First Hospital of China Medical University, between 2012 and 2017. The protocols used in the study were approved by the Hospital’s Protection of Human Subjects Committee.
Cell culture and transfection
The human bladder carcinoma cell line RT4 was cultured in McCoy5A medium (HyClone, Logan, UT, USA), while the cell lines SW780, BIU, 5637, J82, T24, TCC-SUP, UM-UC3 were cultured in RPMI 1640 (HyClone). McCoy5A and RPMI 1640 were supplemented with 10% fetal bovine serum (FBS; HyClone), and the cells were cultured in a humidified atmosphere with 5% CO2 at 37 °C. Lipofectamine™ 3000 (Invitrogen, Carlsbad, CA, USA) was used for transfections, according to the manufacturer’s recommendations.
Plasmids and short interfering RNA (siRNA)
Human pcDNA3.1-ZEB1-AS1 plasmid, pcDNA3.1-negative control (NC), siRNA against ZEB1-AS1 (siZEB1-AS1), siRNA against FSCN1 (siFSCN1), hsa-mir-200b-3p mimics (miR-200b), mimics NC (miR-NC), hsa-mir-200b-3p inhibitor (ant miR-200b), inhibitor NC (ant miR-NC), and the pmirGLO luciferase reporter plasmid were synthesized by and purchased from GenePharm (Shanghai, China). RNAi sequences are shown in Additional file 1: Table S1.
Dual luciferase reporter assay
Cells were seeded (4 × 104 cells/well) in triplicate in 24-well plates and cultured for 24 h. RNA/DNA was transfected according to the experimental purpose. Luciferase and Renilla signals were measured 48 h after treatment using a Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
RNA extraction and quantitative PCR (qPCR)
Total RNA (including miRNA) from cells and bladder tissues was extracted using the miRNeasy™ Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s recommendations. Nuclear RNA from cells was extracted with the miRNeasy™ Mini Kit after nuclear extraction with a Nuclear Extraction Kit (Solarbio, Beijing, China).
cDNA (except for cDNA from miRNA) was synthesized with the PrimeScript™ RT Master Mix (Takara, Beijing, China). cDNA of miRNA was synthesized using the Mir-X™ miRNA First-Strand Synthesis Kit (Clontech Laboratories). qPCR was performed using the SYBR Premix EX Taq™ (Takara). The 2-ΔΔCT method was used to calculate the relative expression level. Primer pairs used for qPCR are shown in Additional file 1: Table S2.
Western blotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer. Protein concentrations were detected using a bicinchoninic acid (BCA) assay kit. Equal amounts of protein samples were separated by 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and then transferred to polyvinylidene fluoride membranes. The membranes were blocked with 5% skim milk in Tris-buffered saline with 1% Tween 20 (TBS-T) for 1 h and then incubated with the appropriate primary antibodies at 4 °C overnight. After washing with TBS-T, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies at 37 °C for 1 h. The membranes were then washed and the enhanced chemiluminescence method was used for protein detection according to the manufacturer’s instructions. Antibodies against FSCN1, E-cadherin and N-cadherin were purchased from Abcam (Cambridge, MA, USA). The antibody against vimentin was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). The antibody against glyceraldehyde 3-phosphate dehydrogenase (GAPDH; loading control) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Transwell assays
Cell invasion and migration were measured using transwell chambers with 8-μm pores in 24-well tissue culture plates (Corning Costar, Corning, NY, USA). Transwell chambers coated with Matrigel (BD, San Diego, CA, USA) were used to determine cell invasiveness, while transwell chambers without Matrigel were used to measure cell motility. After transfection of DNA/RNA for 48 h, the cells were re-suspended in RPMI 1640 containing 1% FBS, and 0.2 mL cell suspension (1 × 104 cells per well for cell migration, 2 × 104 cells per well for cell invasion) was seeded into the top chamber, whereas the lower chamber was filled with 0.6 ml per well of RPMI 1640 containing 10% FBS, used as a chemoattractant.
After 24 h of incubation at 37 °C, the cells that remained on the upper side of the filter were removed using cotton swabs, and those that had migrated to the lower side were fixed and stained with 1.0% crystal violet. Images were captured (100 ×) by microscope, and cells were counted using the ImageJ 1.48v software (National institutes of health, Bethesda, Maryland, USA).
Cell cycle analysis
Cells were trypsinized and washed in ice-cold phosphate-buffered saline (PBS) and then fixed in ice-cold 75% ethanol in PBS. A propidium iodide (PI)/RNase staining buffer (BD) was added, and the cells were incubated at 4 °C for 30 min. Cell cycle profiles were analyzed using a FACS Calibur flow cytometer (BD).
Cell apoptosis assays
Cells were detached using trypsin without ethylenediaminetetraacetic acid (EDTA). An Annexin V- fluorescein isothiocyanate (FITC)/PI (BD) solution was added, and the cells were incubated for 15 min at room temperature. Cell apoptosis were analyzed using the FACS Calibur.
5-Ethynyl-2′-deoxyuridine (EdU)
T24 and RT4 cells were seeded in 96-well plates at a density of 5000 cells per well and cultured for 24 h. The solution of an EdU Kit (Ribobio, Guangzhou, China) was diluted in proportion to 1: 1000 in cell medium. Cells were incubated with the EdU solution for 2 h and were stained following the manufacturer’s instructions. Images were acquired using a fluorescence microscope.
Real time cell analysis (RTCA)
T24 and RT4 cells were seeded in E-plates at a density of 5000 cells per well and incubated in culture medium at 37 °C in a 5% CO2 atmosphere for 72 h. Cell growth curves were automatically recorded by the RTCA xCELLigence S16.
Fluorescence in situ hybridization (FISH)
The probe of ZEB1-AS1 was designed and purchased from GenePharm (Shanghai, China).
Sequences of the ZEB1-AS1 probe are shown in Additional file 1: Table S3. T24 and RT4 cells were seeded in 48-well plates at a density of 10,000 cells per well and incubated in culture medium at 37 °C in a 5% CO2 atmosphere overnight. Next, the cells were washed twice with PBS, fixed with 4% paraformaldehyde for 15 min. As per the manufacturer’s instructions, the plate was placed at 73 °C for 5 min and incubated at 37 °C for 12~16 h in an incubator after adding the mixture of 100 μl (10 μM) probes. Then, the cells were stained with 200 μl (500 ng/ml) of 4′, 6-diamidino-2-phenylindole (DAPI; Beyotime, Shanghai, China) per well in the dark for 1 min after being washed following the manufacturer’s instructions. Images were acquired using a fluorescence microscope.
RNA-binding protein immunoprecipitation (RIP)
The Magna RIP kit (Millipore) was used for RIP assays with the anti-Ago2 antibody (Abcam), according to the manufacturer’s recommendations.
Phalloidin staining
T24 cells were seeded in 24-well plates. When the cell density reached 50%, the cells were washed twice with PBS, fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton-X-100. Next, the cells were incubated with 200 μl (5 μg/ml) Rhodamine Phalloidin (YEASEN, Shanghai, China) per well in the dark for 30 min and then strained with 200 μl (500 ng/ml) of 4′, 6-diamidino-2-phenylindole (DAPI; Beyotime, Shanghai, China) per well in the dark for 1 min. Images were acquired using a fluorescence microscope.
Xenograft tumor model
BALB/c nude mice (4–6 weeks old, 14–16 g) were purchased from Beijing Vital River Experimental Animal Technology Co, Ltd. (Beijing, China). The mice were housed in barrier facilities on a 12-h light/dark cycle. The Institutional Animal Care and Use Committee of China Medical University approved all the experimental procedures. Twelve mice were inoculated subcutaneously with UM-UC3 cells (5 × 106) in the right dorsal flanks. The mice were randomly divided into two groups (n = 6/group), injected with UM-UC3 cells transfected with pcDNA3.1-ZEB1-AS1 or pcDNA3.1-NC. Tumors were examined twice weekly; their length, width, and thickness were measured with calipers, and tumor volumes were calculated using the equation (Length× Width2)/2. On day 60, the animals were euthanized, and the tumors were excised, weighed, and paraffin-embedded. Serial 6.0-μm sections were cut and subjected to immunohistochemistry (IHC). The Ki67 and FSCN1 antibodies were used in these assays.
TGF-β treatment
TGF-β (R&D Systems) was stored at − 20 °C after reconstitution in sterile ddH2O. When needed, TGF-β was diluted in serum-free medium to a concentration of 10 ng/ml and added to the cells.
Statistical analysis
All data were expressed as the mean ± standard deviation (SD), and represented as the average of at least three experiments, with each experiment performed in triplicate. Statistical significance was determined using the Student’s t-test (two-tailed) or Mann-Whitney U test. The correlation between expression of different genes was evaluated by Spearman correlation analysis. The correlation between patients’ clinical pathological characteristics and ZEB1-AS1 expression were analyzed by the Chi-square test. In bar graphs, * and ** indicate P < 0.05 and P < 0.01, respectively. A P value < 0.05 was considered to indicate statistical significance.
Results
The expression of FSCN1 is negatively correlated with that of miR-200b in BLCA tissues and cells
Firstly, we searched for miRNAs that could potentially regulate FSCN1 expression using the Targetscan server (http://www.targetscan.org) [20] and found that miR-200b was a valid candidate on the basis of the potential miR-200b binding site on FSCN1 mRNA.
According to OncoLnc (www.oncolnc.org; a TCGA data portal) [21], high levels of FSCN1 indicated poor prognosis (Fig. 1a); conversely, high miR-200b expression indicated good prognosis (Fig. 1b). Additionally, FSCN1 expression was negatively correlated with miR-200b in BLCA (Additional file 2: Figure S1A). Next, we detected FSCN1 and miR-200b levels in 23 normal bladder samples and 60 BLCA tissues. We found that FSCN1 was upregulated (Fig. 1c) in cancer tissues, but there was no significant difference in miR-200b expression between BLCA tissues and normal bladder tissues (Fig. 1d). However, in the BLCA tissues, the expression of FSCN1 was negatively correlated with that of miR-200b (Fig. 1e). We then detected the mRNA levels of FSCN1 and miR-200b in BLCA cell lines and found that FSCN1 was highly expressed in some high grade BLCA cell lines (Tccsup, T24, UM-UC3), in which the expression level of miR-200b was low (Fig. 1f and g).
Differential expression of fascin-1 (FSCN1) and miR-200b in bladder cancer (BLCA) tissues and cells. a and b Kaplan–Meier analysis of overall survival based on FSCN1 (a) and miR-200b (b) expression levels in patients with BLCA from the TCGA database. Patients with BLCA were divided into FSCN1 (or miR-200b) high expression group (33% of the total, with the highest FSCN1 (or miR-200b) expression) and low expression group (33%, with the lowest FSCN1 (or miR-200b) expression). c and d The mRNA level of FSCN1 (c) and miR-200b (d) in 23 normal bladder tissues and 60 BLCA tissues was measured by qPCR. Data were analyzed by Mann-Whitney U test. e Correlation between FSCN1 and miR-200b of 60 BLCA tissues. Data were analyzed by Spearman correlation analysis. f and g The mRNA level of FSCN1 (f) and miR-200b (g) in BLCA cell lines was measured by quantitative PCR (qPCR) and normalized by RT4 cell line. Data were analyzed by T-test. Data are presented as the mean ± standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
These results suggest that the expression of FSCN1 is negatively correlated with that of miR-200b in BLCA tissues and cells.
miR-200b is a potential suppressor for FSCN1
To verify the direct binding of miR-200b to FSCN1, we performed luciferase reporter assays. We constructed a pmirGLO luciferase reporter plasmid, containing the putative (wild-type or mutated) miR-200b binding site sequence of FSCN1. We found that miR-200b significantly reduced the luciferase activity of the cells (T24, RT4 or 293 T) transfected with the construct containing the wild-type FSCN1 sequence; however, miR200b did not have any effect on the construct with the mutated FSCN1 sequence (Fig. 2a). Additionally, in T24 and RT4 cells, FSCN1 was found to be downregulated by miR-200b and upregulated by miR-200b inhibitors by qPCR (Fig. 2b).
miR-200b inhibits fascin-1 (FSCN1) expression, migration, invasion. a Luciferase assays were performed after T24, RT4 and 293 T cells were co-transfected for 24 h with miR-negative control (miR-NC) or miR-200b and a plasmid containing wild-type or mutant-type FSCN1 3′ untranslated region (UTR) upstream the luciferase gene. The firefly luciferase activity of each sample was normalized by Renilla luciferase activity. Data were analyzed by T-test. b The mRNA level of FSCN1 in T24 and RT4 cells was measured by quantitative PCR (qPCR) after miR-200b was silenced or overexpressed. Data were analyzed by T-test. c The protein level of E-cadherin, N-cadherin, vimentin and FSCN1 in T24 and RT4 cells was measured by western blot after miR-200b was silenced or overexpressed. Data were analyzed by T-test. d The protein level of FSCN1 in T24 cells was measured by western blot after the cells were co-transfected with ant miR-NC or ant miR-200b and siNC or siFSCN1. Data were analyzed by T-test. e and f The migration (e) and invasion (f) abilities of T24 and RT4 cells were detected in transwell assays (without or with Matrigel) after miR-200b was silenced or overexpressed. g and h The cell migration (g) and invasion (h) abilities of T24 and RT4 were detected in transwell assays (without or with Matrigel) after co-transfection with miR-NC or miR-200b and siNC or siFSCN1. Data were analyzed by T-test. All images were taken at 100× magnification. Data are presented as the mean ± standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
We then examined the expression of FSCN1 and EMT markers by western blot and observed that the overexpression of miR-200b increased the expression of E-cadherin and inhibited that of N-cadherin, vimentin and FSCN1, whereas the inhibition of miR-200b had opposite effect (Fig. 2c). In a rescue experiment, in T24 cells, western blot assays showed that the inhibition of miR-200b reversed the effect of FSCN1 silencing (Fig. 2d). We then performed transwell assays to explore the effect of miR-200b on the migration and invasion abilities of BLCA cells. The overexpression of miR-200b in T24 and RT4 cells inhibited cell migration and invasion, whereas the inhibition of miR-200b had opposite effect (Fig. 2e and f). In rescue experiments in T24 and RT4 cells, the inhibition of miR-200b reversed the effect of FSCN1 silencing on cell migration and invasion (Fig. 2g and h). Next, we explored the effect of miR-200b on cell cycle and proliferation of BLCA cells. Flow cytometry assays showed that the percentage of cells in G0/G1 phase significantly increased upon miR-200b overexpression and decreased upon miR-200b knockdown (Additional file 2: Figure S1B). Additionally, in EdU assays and RTCA on T24 and RT4 cells, we found that the proliferation of cells significantly decreased upon miR-200b overexpression, while miR-200b inhibition had the opposite effect (Additional file 2: Figure S1C and D).
These results suggest that miR-200b downregulates the expression of FSCN1, which leads to the inhibition of migration and invasion, and inhibits cell cycle progression and proliferation of BLCA cells.
ZEB1-AS1 is a potential regulator on the expression of miR-200b and FSCN1
By downregulating miRNA, lncRNA can regulate their function: this is the so-called “sponge-effect.” To find potential lncRNA that could regulate miR-200b, we used the DIANA-LncBase tool (www.microrna.gr/LncBase) [22] and found that the lncRNA ZEB1-AS1 might regulate the expression of miR-200b. We used the GEPIA server (http://gepia.cancer-pku.cn; a TCGA data portal) [23] to investigate the relation between ZEB1-AS1 expression and the survival of patients with BLCA. We found that there was no significant correlation between ZEB1-AS1 levels and the patient’s overall survival (Fig. 3a). On the other hand, ZEB1-AS1 levels were positively correlated with disease free survival (Fig. 3b), indicating that high levels of ZEB1-AS1 might result in recurrence of BLCA. As shown in Table 1, in 60 BLCA tissues, the levels of ZEB1-AS1 were higher in high-grade tumors than in low-grade tumors. Next, we examined the correlation between ZEB1-AS1 and miR-200b or ZEB1-AS1 and FSCN1 and found that the expression of ZEB1-AS1 was negatively correlated with that of miR-200b (Additional file 2: Figure S2A) and positively correlated with that of FSCN1 (Additional file 2: Figure S2B) according to the database.
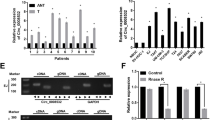
ZEB1-AS1 is upregulated in BLCA tissues and correlates with miR-200b and fascin-1 (FSCN1). (A and B) Kaplan–Meier analysis of overall (a) and disease free (b) survival based on ZEB1-AS1 expression levels in patients with BLCA from the TCGA database. Patients with BLCA were divided into ZEB1-AS1 high expression group (21% of the total, with the highest ZEB1-AS1 expression) and low expression group (21% of the total, with the lowest ZEB1-AS1). c ZEB1-AS1 expression levels were detected in 10 pairs of BLCA and adjacent normal mucosa tissue samples by qPCR. Data were analyzed by T-test. d The mRNA level of ZEB1-AS1 in 23 normal bladder tissues and 60 BLCA tissues was measured by qPCR. Data were analyzed by Mann-Whitney U test. e and f The correlation between ZEB1-AS1 and miR-200b (e), ZEB1-AS1 and FSCN1 (f) of 60 BLCA tissues. Data were analyzed by Spearman correlation analysis. g The mRNA level of ZEB1-AS1 in BLCA cell lines was measured by quantitative (qPCR). Data were analyzed by T-test. h Fluorescence in situ hybridization (FISH) assay in T24 and RT4 cells showing ZEB1-AS1 (red); nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI; blue). i The mRNA level of ZEB1-AS1 in the nuclear and cytoplasmic fraction of T24 and RT4 cells by qPCR. Data are presented as mean ± standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
To verify this finding, we detected the expression of ZEB1-AS1 in ten pairs of BLCA tissues and adjacent normal bladder mucosa and found significant upregulation of ZEB1-AS1 in BLCA samples compared to normal controls (Fig. 3c). Similar results were obtained in 23 normal bladder tissues and 60 BLCA tissues (Fig. 3d). Additionally, in the BLCA samples, we found a negative correlation between ZEB1-AS1 and miR-200b (Fig. 3e), and a positive correlation between ZEB1-AS1 and FSCN1 (Fig. 3f). We then detected the mRNA level of ZEB1-AS1 (Fig. 3g) in BLCA cell lines and found that ZEB1-AS1 expression was high in some high grade BLCA cell lines (Tccsup, T24, and UM-UC3), in which the expression of miR-200b was low, while the expression of FSCN1 was high. To act as a sponge for miRNAs, lncRNAs need be located in the cytoplasm. Fluorescence in situ hybridization (FISH) showed that ZEB1-AS1 was located in both the nucleus and the cytoplasm in T24 and RT4 cells (Fig. 3h). Similar results were obtained in qPCR assays in nuclear and cytoplasmic RNA fractions (Fig. 3i).
These results suggest that ZEB1-AS1 may be a regulator on the expression of miR-200b and FSCN1.
ZEB1-AS1 induces migration and invasion potentially through regulating the expression of miR-200b and FSCN1
To verify the potential interaction between miR-200b and ZEB1-AS1, we performed luciferase reporter assays. We constructed a pmirGLO luciferase reporter plasmid containing the putative miR-200b binding sequence (wild-type or mutated) of ZEB1-AS1. Luciferase assays showed that miR-200b significantly reduced the luciferase activity of cells (T24, RT4 or 293 T) transfected with the construct containing the wild-type ZEB1-AS1 sequence (Fig. 4a). Additionally, RIP assays confirmed the interaction between ZEB1-AS1 and miR-200b in T24 and RT4 cells through AGO2 (Fig. 4b).
ZEB1-AS1 downregulates miR-200b, upregulates fascin-1 (FSCN1), and promotes cell migration and invasion. a Luciferase assays were performed in T24, RT4, and 293 T cells co-transfected for 24 h with miR-negative control (NC) or miR-200b and a plasmid containing wild-type or mutant-type ZEB1-AS1 3′untranslated region (UTR) upstream the luciferase gene. Firefly luciferase activity of each sample was normalized by Renilla luciferase activity. Data were analyzed by T-test. b RNA-binding protein immunoprecipitation (RIP) assays with anti-AGO2 antibodies were performed in T24 and RT4 cells transiently transfected with miR-200b; ZEB1-AS1 levels were detected by quantitative PCR (qPCR); 10% input was used as positive control and RIP with anti-IgG antibodies served as negative control. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the internal control. Data were analyzed by T-test. c Transfection efficiency of ZEB1-AS1 knockdown (si1, si2) and overexpression (OE) in T24 and RT4 cells was detected by qPCR. Data were analyzed by T-test. d and e The levels of miR-200b (d) and FSCN1 (e) were measured by qPCR after ZEB1-AS1 knocked down or overexpressed in T24 and RT4 cells. Data were analyzed by T-test. f E-cadherin, N-cadherin, vimentin, and FSCN1 protein expression in T24 and RT4 cells in which ZEB1-AS1 had been knocked down or overexpressed. Data were analyzed by T-test. (g) FSCN1 levels in T24 cells co-transfected with miR-NC or miR-200b and with an empty vector (EV) or a plasmid overexpressing ZEB1-AS1. Data were analyzed by T-test. h and i Transwell assays (without or with Matrigel) to detect cell migration (h) and invasion (i) of T24 and RT4 after ZEB1-AS1 silencing or overexpression. j and k Transwell assays (without or with Matrigel) to detect cell migration (j) and invasion (k) of T24 and RT4 co-transfected with miR-NC or miR-200b and with an EV or a plasmid overexpressing ZEB1-AS1. Data were analyzed by T-test. Data are presented as the mean ± standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
Next, we transfected T24 and RT4 cells with siRNA against ZEB1-AS1 (called si1 or si2), ZEB1-AS1 overexpression vector (OE) and their corresponding controls; Fig. 4c shows the transfection efficiency of these constructs. The expression of miR-200b was decreased and that of FSCN1 was increased after inhibiting the expression of ZEB1-AS1 by siRNA, while overexpression of ZEB1-AS1 had opposite effect (Fig. 4d and e). We also evaluated the expression of FSCN1 and EMT markers by western blot. We observed that the inhibition of ZEB1-AS1 expression increased the expression of E-cadherin and decreased the expression of N-cadherin, vimentin, and FSCN1, whereas the overexpression of ZEB1-AS1 was associated with opposite results (Fig. 4f). In rescue experiments in T24 cells, western blot assays showed that the overexpression of miR-200b counteracted the overexpression of ZEB1-AS1 (Fig. 4g).
We then performed transwell assays to explore the effect of ZEB1-AS1 on migration and invasion of BLCA cells. Decreasing the expression of ZEB1-AS1 by siRNA in T24 and RT4 cells inhibited cell migration and invasion, whereas overexpressing ZEB1-AS1 had the opposite effect (Fig. 4h and i). In rescue experiments in T24 and RT4 cells, we found that the overexpression of miR-200b reversed the effect of the overexpression of ZEB1-AS1 on cell migration and invasion (Fig. 4j and k). Therefore, these results suggest that ZEB1-AS1 affects migration and invasion likely through regulating miR-200b and FSCN1 in BLCA cells.
Furthermore, cellular morphology images (Additional file 2: Figure S3A) and cytoskeleton staining (Additional file 2: Figure S3B) indicated that ZEB1-AS1 might affect the cytoskeleton of BLCA cells and therefore impact cell migration.
ZEB1-AS1 inhibits apoptosis, and promotes cell cycle progression and proliferation
To further explore the function of ZEB1-AS1 in BLCA cells, we investigated the effect of ZEB1-AS1 on several cellular processes. Apoptosis assays in T24 and RT4 cells showed that ZEB1-AS1 silencing was associated with a significant increase of apoptosis, and ZEB1-AS1 overexpression had the opposite effect (Fig. 5a). In cell cycle analysis, we found that the percentage of cells in the G0/G1 phase was increased upon ZEB1-AS1 silencing and decreased when ZEB1-AS1 was overexpressed (Fig. 5b). Additionally, EdU assays and RTCA showed that the proliferation of cells was decreased when ZEB1-AS1 was silenced and increased upon ZEB1-AS1 overexpression (Fig. 5c and d).
ZEB1-AS1 inhibits apoptosis and promotes cell cycle and proliferation. a-b Apoptosis (a) and cell cycle (b) were detected by flow cytometry in T24 and RT4 cells after ZEB1-AS1 silencing or overexpression. Data were analyzed by T-test. c 5-Ethynyl-2′-deoxyuridine (EdU) assays on T24 and RT4 after ZEB1-AS1 silencing or overexpression. Data were analyzed by T-test. All images were taken at 200× magnification. d The proliferation of ZEB1-AS1-silenced or -overexpressing T24 and RT4 were detected by real time cell analysis (RTCA). Data were analyzed by T-test. e-h Xenograft model in nude mice. e Tumor growth was measured at the indicated days. f Nude mice at the end of the experiment. g Images of the excised tumors from each group. h Staining of Ki-67 and FSCN1 in the excised tumors. Data are presented as the mean ± standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
We extended this result in xenograft models. First, we generated UM-UC3 cells stably overexpressing ZEB1-AS1. Then, we injected mice with these cells (or control UM-UC3) and observed the formation of tumors. The tumors derived from ZEB1-AS1 overexpressing cells grew faster than control cells (Fig. 5e-g). The IHC assay showed a high expression level of Ki67 and FSCN1 in tumors derived from ZEB1-AS1 overexpressing cells (Fig. 5h).
These results suggest that ZEB1-AS1 inhibits apoptosis and enhances tumor growth both in vitro and in vivo.
ZEB1-AS1 enhances cell migration and invasion that is induced by TGF-β1
It is well known that TGF-β1 induces cell migration and invasion and downregulates the expression of miR-200b in many kinds of cancers. In our previous studies, we found that TGF-β1 induced cell migration and invasion by upregulating the expression of FSCN1 in BLCA and kidney renal clear cell carcinoma (KIRC). Therefore, we asked whether TGF-β1 regulated the ZEB1-AS1/miR-200b/FSCN1 axis. Interestingly, we found that TGF-β1 treatment in T24 and RT4 cells was associated with upregulation of ZEB1-AS1 and FSCN1 and downregulation of miR-200b (Fig. 6a and b).
Transforming growth factor (TGF)-β1 inhibits miR-200b and promotes ZEB1-AS1 and fascin-1 (FSCN1) expression. a The mRNA level of ZEB1-AS1, miR-200b and FSCN1 in T24 and RT4 after treated with TGF-β1. Data were analyzed by T-test. b FSCN1 protein expression in T24 and RT4 cells treated with TGF-β1. Data were analyzed by T-test. c FSCN1 protein expression in T24 cells treated with TGF-β1 and transfected with siNC or siZEB1-AS1; untreated and not-transfected cells were used as control. Data were analyzed by T-test. d FSCN1 protein levels in T24 cells treated with TGF-β1 and transfected with miR-negative control (miR-NC) or miR-200b; untreated and not-transfected cells were used as control. Data were analyzed by T-test. e and f Transwell assays (without or with Matrigel) to detect migration (e) and invasion (f) of T24 and RT4 treated with TGF-β1 and transfected with siNC or siZEB1-AS1; untreated and not-transfected cells were used as control. g and h Transwell assays (without or with Matrigel) to detect the migration (g) and invasion (h) of T24 and RT4 treated with TGF-β1 and transfected with miR-NC or miR-200b untreated and not-transfected cells were used as control. Data were analyzed by T-test. i Hypothetical model illustrating the TGF-β1/ZEB1-AS1/miR-200b/FSCN1 axis. Data are presented as the mean ± standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant
To further verify this observation, we performed rescue experiments. Western blot assays showed that ZEB1-AS1 silencing reversed the upregulation of FSCN1 induced by TGF-β1 treatment in T24 cells (Fig. 6c). At the same time, miR-200b overexpression had a similar effect (Fig. 6d) on the regulation of FSCN1 by TGF-β1. Similar results were obtained in cell migration (Fig. 6e and g) and invasion (Fig. 6f and h) assays in T24 and RT4 cells.
Therefore, we concluded that, in BLCA, ZEB1-AS1 is likely to be involved in TGF-β1 regulation on cell migration and invasion potentially through impacting the expression of miR-200b and FSCN1 (Fig. 6i).
Discussion
In our previous study, we found that FSCN1 is a biomarker of BLCA [12, 13]. In this study, we investigated the regulatory mechanisms through which FSCN1 exerts its effect in BLCA.
miRNAs are non-coding RNA that regulate the expression of genes through the RNA-induced silencing complex (RISC) [24, 25]. Through bioinformatics analysis, we found that miR-200b might regulate the expression of FSCN1 and verified this hypothesis by performing experiments on BLCA cells. Even though the expression of miR-200b in BLCA samples and normal tissues was not significantly different probably due to the small sample size in our study, miR-200b was still correlated with good prognosis (overall survival).
miR-200b inhibits cell migration and invasion, negatively affects cell cycle progression and inhibits cell proliferation, and is therefore considered a tumor suppressor in BLCA. miR-200b exerts a similar function in other types of cancer [26, 27]. Some studies showed that the levels of miR-200 family were increased in BLCA samples compared with normal tissues but decreased in high grade and stage tumors in both non-muscle invasive bladder cancer (NMIBC) and muscle invasive bladder cancer (MIBC) compared with low grade and stage tumor, still acting as a tumor suppressor [14, 28,29,30]. These observations are in agreement with our results. How miR-200b causes these effects in BLCA needs to be further investigated.
The “sponge effect” of lncRNAs is the classic mechanism through which they regulate miRNAs [31,32,33,34]. Bioinformatics analysis indicated that ZEB1-AS1 might regulate the function of miR-200b. ZEB1-AS1 is an oncogenic lncRNA correlated with higher histopathological grade in many kinds of cancer [35,36,37,38], and recent studies have suggested that ZEB1-AS1 regulates the miR-200c in prostate cancer [37], and miR-200 family in osteosarcoma [39]. However, whether there is a regulatory relation between ZEB1-AS1 and miR-200b in BLCA is still unknown.
Our bioinformatics analysis suggested ZEB1-AS1 is more expressed in BLCA tissues than in normal ones and correlates with disease-free survival and not with overall survival in patients with BLCA. Therefore, ZEB1-AS1 might promote tumor progression by increasing the recurrence of BLCA. Further experiments demonstrated that ZEB1-AS1 acted as a competing endogenous RNA (ceRNA) in BLCA by interacting with miR-200b and indirectly upregulating FSCN1. Furthermore, in functional experiments, we found that ZEB1-AS1 decreased cell apoptosis and G0/G1 arrest and induced BLCA cell proliferation, migration, and invasion, thereby exerting a tumor promoting role via the miR-200b/FSCN1 axis.
TGF-β1 affects many biological processes [16] such as migration, invasion, and proliferation. In our previous studies, we found that TGF-β1 upregulated the expression of FSCN1 in BLCA [19] and KIRC [40]. Some studies have shown that TGF-β1 downregulates the expression of miR-200b [18, 41, 42]. Therefore, we investigated if, in our system, TGF-β1 acted through the ZEB1-AS1/miR-200b/FSCN1 axis and found that TGF-β1 upregulated the expression of ZEB1-AS1. These results suggest that TGF-β1 induces migration and invasion in BLCA by regulating the ZEB1-AS1/miR-200b/FSCN1 axis.
In the current study, we focused on the effect of the ZEB1-AS1/miR-200b/FSCN1 axis on cell migration and invasion. This axis might affect other cell functions; however, their mechanism has not been investigated in this study and might be the focus of future research.
Some studies have shown that TGF-β1 increases FSCN1 expression via the extracellular-signal-regulated kinase (ERK) and c-Jun N-terminal kinase (JNK) signaling pathways [40, 43]. Meanwhile, some studies reported that ZEB1-AS1 could affect cell function by regulating the Wnt/β-catenin [44] and p38MAPK signaling pathways, [45] which were also regulated by TGF-β1 [46,47,48,49]. Therefore, we hypothesize that some signaling network must participate in the regulation of the TGF-β1/ZEB1-AS1/miR-200b/FSCN1 axis. Future studies might investigate the conditions or cases in which the axes are activated.
Conclusions
Our study suggests that the lncRNA ZEB1-AS1 is upregulated in BLCA, correlates with high tumor grade and high TNM stage and indicates poor prognosis of patients. Functional experiments showed that ZEB1-AS1 induces cell migration, invasion, and proliferation in BLCA. TGF-β1 might induce migration and invasion, at least in part, by regulating the ZEB1-AS1/miR-200b/FSCN1 axis in BLCA. Thus, the TGF-β1/ZEB1-AS1/miR-200b/FSCN1 axis may be a potential therapeutic target for BLCA.
Abbreviations
- BLCA:
-
Bladder cancer
- ceRNA:
-
Competing endogenous RNA
- DAPI:
-
4′, 6-diamidino-2-phenylindole
- EDTA:
-
Ethylenediaminetetraacetic acid
- EdU:
-
5-Ethynyl-2′- deoxyuridine
- EMT:
-
Epithelial-to-mesenchymal transition
- ERK:
-
Extracellular-signal-regulated kinase
- EV:
-
Empty vector
- FBS:
-
Fetal bovine serum
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- FSCN1:
-
Fascin-1
- GAPDH:
-
Glyceraldehyde 3-phosphate dehydrogenase
- IHC:
-
Immunohistochemistry
- JNK:
-
c-Jun N-terminal kinase
- lncRNA:
-
Long non-coding RNA
- MIBC:
-
Muscle invasive bladder cancer
- miRNA:
-
microRNA
- NC:
-
Negative control
- NMIBC:
-
Non-muscle invasive bladder cancer
- OE:
-
Overexpression vector
- PBS:
-
Phosphate-buffered saline
- PI:
-
Propidium iodide
- qPCR:
-
Quantitative PCR
- RIP:
-
RNA-binding Protein Immunoprecipitation
- RISC:
-
RNA-induced silencing complex
- RTCA:
-
Real time cell analysis
- SD:
-
Standard deviation
- siRNA:
-
Short interfering RNA
- TBS-T:
-
Tris-buffered saline with 1% Tween 20
- TCGA:
-
The Cancer Genome Atlas
- TGF:
-
Transforming growth factor
- UTR:
-
Untranslated region
- ZEB1:
-
Zinc Finger E-Box Binding Homeobox 1
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Adams JC. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol. 2004;16(5):590–6.
Kureishy N, Sapountzi V, Prag S, Anilkumar N, Adams JC. Fascins, and their roles in cell structure and function. Bioessays. 2002;24(4):350–61.
Anilkumar N, Parsons M, Monk R, Ng T, Adams JC. Interaction of fascin and protein kinase Calpha: a novel intersection in cell adhesion and motility. EMBO J. 2003;22(20):5390–402.
Takikita M, Hu N, Shou JZ, et al. Fascin and CK4 as biomarkers for esophageal squamous cell carcinoma. Anticancer Res. 2011;31(3):945–52.
Buda A, Pignatelli M. E-cadherin and the cytoskeletal network in colorectal cancer development and metastasis. Cell Commun Adhes. 2011;18(6):133–43.
Durmaz A, Kurt B, Ongoru O, Karahatay S, Gerek M, Yalcin S. Significance of fascin expression in laryngeal squamous cell carcinoma. J Laryngol Otol. 2010;124(2):194–8.
Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adhes Migr. 2011;5(5):421–30.
Park SH, Song JY, Kim YK, et al. Fascin1 expression in high-grade serous ovarian carcinoma is a prognostic marker and knockdown of fascin1 suppresses the proliferation of ovarian cancer cells. Int J Oncol. 2014;44(3):637–46.
Karasavvidou F, Barbanis S, Pappa D, et al. Fascin determination in urothelial carcinomas of the urinary bladder: a marker of invasiveness. Arch Pathol Lab Med. 2008;132(12):1912–5.
Soukup V, Babjuk M, Dusková J, et al. Does the expression of fascin-1 and tumor subclassification help to assess the risk of recurrence and progression in t1 urothelial urinary bladder carcinoma. Urol Int. 2008;80(4):413–8.
Bi JB, Zhu Y, Chen XL, et al. The role of fascin in migration and invasion of urothelial carcinoma of the bladder. Urol Int. 2013;91(2):227–35.
Bi J, Chen X, Zhang Y, et al. Fascin is a predictor for invasiveness and recurrence of urothelial carcinoma of bladder. Urol Oncol. 2012;30(5):688–94.
Martínez-Fernández M, Dueñas M, Feber A, et al. A Polycomb-mir200 loop regulates clinical outcome in bladder cancer. Oncotarget. 2015;6(39):42258–75.
Li J, Li Z, Leng K, et al. ZEB1-AS1: a crucial cancer-related long non-coding RNA. Cell Prolif. 2018;51(1).
Kajdaniuk D, Marek B, Borgiel-Marek H, Kos-Kudła B. Transforming growth factor β1 (TGFβ1) in physiology and pathology. Endokrynol Pol. 2013;64(5):384–96.
Liu S, Chen S, Zeng J. TGF-β signaling: a complex role in tumorigenesis (review). Mol Med Rep. 2018;17(1):699–704.
Gregory PA, Bracken CP, Smith E, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22(10):1686–98.
Zhang N, Bi X, Zeng Y, et al. TGF-β1 promotes the migration and invasion of bladder carcinoma cells by increasing fascin1 expression. Oncol Rep. 2016;36(2):977–83.
Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4.
Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2:e67. https://doi.org/10.7717/peerj-cs.67.
Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–8.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102.
Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression. Nat Rev Genet. 2014;15(9):599–612.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Tang Q, Li M, Chen L, Bi F, Xia H. miR-200b/c targets the expression of RhoE and inhibits the proliferation and invasion of non-small cell lung cancer cells. Int J Oncol. 2018;53(4):1732–42.
Li J, Yuan J, Yuan X, et al. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int J Oncol. 2016;48(2):541–50.
Lee H, Jun SY, Lee YS, Lee HJ, Lee WS, Park CS. Expression of miRNAs and ZEB1 and ZEB2 correlates with histopathological grade in papillary urothelial tumors of the urinary bladder. Virchows Arch. 2014;464(2):213–20.
Adam L, Zhong M, Choi W, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15(16):5060–72.
Santos M, Martínez-Fernández M, Dueñas M, et al. In vivo disruption of an Rb-E2F-Ezh2 signaling loop causes bladder cancer. Cancer Res. 2014;74(22):6565–77.
Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20(19):R858–61.
Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363.
Calin GA, Liu CG, Ferracin M, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–29.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language. Cell. 2011;146(3):353–8.
Li T, Xie J, Shen C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35(12):1575–84.
Gong H, Wen H, Zhu X, et al. High expression of long non-coding RNA ZEB1-AS1 promotes colorectal cancer cell proliferation partially by suppressing p15 expression. Tumour Biol. 2017;39(6):1010428317705336.
Su W, Xu M, Chen X, et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16(1):142.
Lin J, Zhan Y, Liu Y, et al. Increased expression of ZEB1-AS1 correlates with higher histopathological grade and promotes tumorigenesis in bladder cancer. Oncotarget. 2017;8(15):24202–12.
Liu C, Pan C, Cai Y, Wang H. Interplay between long noncoding RNA ZEB1-AS1 and miR-200s regulates osteosarcoma cell proliferation and migration. J Cell Biochem. 2017;118(8):2250–60.
Yang J, Zhang N, Gao R, et al. TGF-β1 induced fascin1 expression facilitates the migration and invasion of kidney carcinoma cells through ERK and JNK signaling pathways. Biochem Biophys Res Commun. 2018;501(4):913–9.
Xiong M, Jiang L, Zhou Y, et al. The miR-200 family regulates TGF-β1-induced renal tubular epithelial to mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol. 2012;302(3):F369.
Izumchenko E, Chang X, Michailidi C, et al. The TGFβ-miR200-MIG6 pathway orchestrates the EMT-associated kinase switch that induces resistance to EGFR inhibitors. Cancer Res. 2014;74(14):3995–4005.
Fu H, Hu Z, Wen J, Wang K, Liu Y. TGF-beta promotes invasion and metastasis of gastric cancer cells by increasing fascin1 expression via ERK and JNK signal pathways. Acta Biochim Biophys Sin Shanghai. 2009;41(8):648–56.
Lv SY, Shan TD, Pan XT, et al. The lncRNA ZEB1-AS1 sponges miR-181a-5p to promote colorectal cancer cell proliferation by regulating Wnt/β-catenin signaling. Cell Cycle. 2018;17(10):1245–54.
Gan L, Chen Y, Liu H, Ju WH. Long non-coding RNA ZEB1-antisense 1 affects cell migration and invasion of cervical Cancer by regulating epithelial-mesenchymal transition via the p38MAPK signaling pathway. Gynecol Obstet Investig. 2018:1–9.
Gomes LR, Terra LF, Wailemann RA, Labriola L, Sogayar MC. TGF-β1 modulates the homeostasis between MMPs and MMP inhibitors through p38 MAPK and ERK1/2 in highly invasive breast cancer cells. BMC Cancer. 2012;12:26.
Hill KS, Erdogan E, Khoor A, et al. Protein kinase Cα suppresses Kras-mediated lung tumor formation through activation of a p38 MAPK-TGFβ signaling axis. Oncogene. 2014;33(16):2134–44.
Wang JL, Qi Z, Li YH, Zhao HM, Chen YG, Fu W. TGFβ induced factor homeobox 1 promotes colorectal cancer development through activating Wnt/β-catenin signaling. Oncotarget. 2017;8(41):70214–25.
Bae WJ, Lee SH, Rho YS, Koo BS, Lim YC. Transforming growth factor β1 enhances stemness of head and neck squamous cell carcinoma cells through activation of Wnt signaling. Oncol Lett. 2016;12(6):5315–20.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (81372722).
Availability of data and materials
The dataset(s) supporting the findings of this study are included within the article.
Author information
Authors and Affiliations
Contributions
BJ and KC contributed to the experiment design, manuscript draft, and data analysis. GR contributed to the experiment implementation, manuscript draft and data analysis. ZN, YJ, ZY and ZZ contributed to the experiment implementation. WJ and XX contributed to data analysis. LZ, LX, LZ and LJ collected clinical samples. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The protocols used in the study were approved by the Hospital’s Protection of Human Subjects Committee. The Institutional Animal Care and Use Committee of China Medical University approved all the experimental procedures of animal experiments.
Consent for publication
Consent for publication was obtained from all participants.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Sequences of RNAi for transfection. Table S2. Sequences of primer pairs for qPCR. Table S3. Sequences of ZEB1-AS1 probe for FISH. (DOCX 13 kb)
Additional file 2:
Figure S1. miR-200b is correlated with FSCN1 and inhibits cell cycle and proliferation (A) Correlation between FSCN1 and miR-200b in patients with BLCA from the TCGA database. Data were analyzed by Spearman correlation analysis. (B) The cell cycle of T24 and RT4 cells was detected by flow cytometry after miR-200b was silenced or overexpressed. Data were analyzed by T-test. (C) 5-Ethynyl-2′-deoxyuridine (EdU) assays were performed on T24 and RT4 cells after miR-200b was silenced or overexpressed. Data were analyzed by T-test. All images were taken at 200× magnification. (D) The proliferation of T24 and RT4 cells was detected by real time cell analysis (RTCA) for 72 h after miR-200b was silenced or overexpressed. Data were analyzed by T-test. Data are presented as the mean ± standard deviation (SD). *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Figure S2. ZEB1-AS1 is correlated with miR-200b and FSCN1 according to TCGA database (A and B) Correlation between ZEB1-AS1 and miR-200b (A) and ZEB1-AS1 and FSCN1 (B) in patients with BLCA from the TCGA database. Data were analyzed by Spearman correlation analysis. Figure S3. The cellular morphological changes after ZEB1-AS1 has been overexpressed. (A) Cellular morphology images of T24 and RT4 cells in which ZEB1-AS1 had been overexpressed. (B) Cytoskeleton staining images of T24 cells in which ZEB1-AS1 had been overexpressed. (TIF 3996 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Gao, R., Zhang, N., Yang, J. et al. Long non-coding RNA ZEB1-AS1 regulates miR-200b/FSCN1 signaling and enhances migration and invasion induced by TGF-β1 in bladder cancer cells. J Exp Clin Cancer Res 38, 111 (2019). https://doi.org/10.1186/s13046-019-1102-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13046-019-1102-6