Abstract
Background
The FOXG1 gene plays a vital role in mammalian brain differentiation and development. Intra- and intergenic mutations resulting in loss of function or altered expression of the FOXG1 gene cause FOXG1 syndrome. The hallmarks of this syndrome are severe developmental delay with absent verbal language, post-natal growth restriction, post-natal microcephaly, and a recognizable movement disorder characterized by chorea and dystonia.
Case presentation
Here we describe a case of a 7-year-old male patient found to have a de novo balanced translocation between chromosome 3 at band 3q14.1 and chromosome 14 at band 14q12 via G-banding chromosome and Fluorescence In Situ Hybridization (FISH) analyses. This rearrangement disrupts the proximity of FOXG1 to a previously described smallest region of deletion overlap (SRO), likely resulting in haploinsufficiency.
Conclusions
This case adds to the growing body of literature implicating chromosomal structural variants in the manifestation of this disorder and highlights the vital role of cis-acting regulatory elements in the normal expression of this gene. Finally, we propose a protocol for reflex FISH analysis to improve diagnostic efficiency for patients with suspected FOXG1 syndrome.
Similar content being viewed by others
Introduction
The Forkhead Box G1 (FOXG1) gene [OMIM: 164874], located on chromosome 14q12, encodes the protein forkhead box protein G1 (FOXG1). It belongs to a class of winged-helix transcriptional regulators and contains a highly conserved fork head DNA-binding domain. This protein plays an important role in mammalian brain development, with high levels of expression in the developing fetal telencephalon [1,2,3,4]. Specifically, it is expressed in the rostral forebrain prior to differentiation into the telencephalon and diencephalon, indicating its role in early differentiation between these structures [5]. It exerts its effects via DNA binding-dependent and -independent mechanisms to encourage neocortical progenitor proliferation and prevent precocious differentiation [6]. In addition to regulating neocortical progenitor cell populations, it also plays an important role in controlling post-mitotic pyramidal cortical neuron migration and post-migration dorsal–ventral patterning to establish normal cortical laminar structure and the corpus callosum [7].
Both intra- or inter-genic mutations resulting in altered FOXG1 expression or protein function causes FOXG1 syndrome. The syndrome is characterized by post-natal growth deficiency, postnatal microcephaly, intellectual disability, restricted language development, autism-like social deficits, stereotypies and dyskinesias, epilepsy, poor sleep, irritability, excessive crying episodes, recurrent aspiration, and gastroesophageal reflux [8]. Characteristic findings on imaging include: frontal predominant simplified gyral patterning, reduced white matter volume, and callosal hypogenesis [8]. The course of epilepsy in patients with FOXG1 syndrome varies based on the underlying genetic mutation. Those with deletions and intragenic mutations tend to have a wide variety of seizure types, ranging from complex partial to generalized tonic–clonic, while those with duplications frequently exhibit infantile spasms [9]. Clinically, the developmental encephalopathy index (DEI) has proven useful in delineating MECP2 and FOXG1 syndrome, showing that those with FOXG1 syndrome had greater impairment overall, with significantly worse function in the domains of fine motor skills, receptive language, reciprocity and ability to walk [10].
Different mutation mechanisms, including intragenic point or indel (insertions and deletions) mutations, microdeletions, or balanced chromosomal rearrangements can lead to FOXG1 haploinsufficiency resulting in FOXG1 syndrome. This makes efficient genetic diagnosis of the disease quite challenging [11,12,13,14]. Here we report a case of FOXG1 syndrome in a 7-year-old Puerto Rican male patient found to have a de novo balanced chromosomal rearrangement with the breakpoint in an intergenic region between FOXG1 and a nearby smallest critical enhancer region (SRO). This report shows that chromosomal rearrangements disrupting a distant regulatory enhancer of FOXG1 is a recurrent event, and that a follow-up FISH analysis is important to reach a definitive genetic diagnosis for patients with balanced rearrangements involving 14q11-q13 with FOXG1 syndrome in the differential diagnosis.
Clinical report
The patient is a 7-year-old male born in Puerto Rico at 33-week gestation to an 18-year-old gravida 1 para 0 female of Puerto Rican descent. No prenatal complications or exposures are reported in the available medical record. Birth weight was 1871 g (35th percentile; standard deviations (SD) − 0.38) and length was 48 cm (93rd percentile; SD 1.82). Head circumference at birth is not available in the medical record. Newborn screening was reportedly normal. The patient was admitted to the neonatal intensive care unit for poor weight gain and jaundice requiring phototherapy and discharged after 8 days.
At approximately 4 months of age the patient’s parents noted developmental motor delays with an inability to raise his head, push himself up, and limited overall movement as well as microcephaly. An initial work-up at the time was notable for: metabolic screening showed non-specific elevations in glutamate and mild abnormalities in excretion of 4-hydroxyphenylacetate, mildly elevated NH3, and elevated lactate at two to three times the upper reference limit. Magnetic resonance imaging (MRI, unavailable for review) was reportedly unremarkable. Cytogenetic studies showed a balanced (3;14) translocation.
The patient had no speech development at 1 year of age. Seizures began at 18 months and were characterized by staring episodes which would progress to full-body limpness, stiffening, followed by jerking movements. The patient had five seizures prior to control with levetiracetam.
At 3 years of age the patient’s mother moved with him to the United States where evaluation showed a severely underweight child (12.08 kg; 5th percentile; SD − 1.61) with microcephaly (head circumference 42.9 cm; SD − 4.0), congenital esotropia, reduced muscle bulk, decreased axial and increased appendicular tone with antigravity strength throughout. The patient remained non-verbal but exhibited social smiling, and the ability to track objects. The patient had poor weight gain due to severe oropharyngeal dysphagia with silent aspiration on pharyngogram, eventually necessitating G-tube placement.
EEG performed at 4 years of age showed a disordered background with slower than expected posterior dominant rhythm consistent with a mild diffuse encephalopathy. There were no focal or epileptiform changes noted.
At present the patient continues to have severe global developmental delay, with no speech development, but he attempts to make sounds. He is able to roll over and grab onto objects. He continues to exhibit upper and lower extremity spasticity with grossly ataxic movements, and excessive purposeless arm movements. His seizures continue to be well-controlled. He has recently displayed episodes of inappropriate laughing and crying.
Methods
G-banding chromosome and FISH analysis
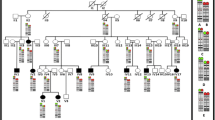
Peripheral blood samples were cultured using standard cytogenetic methods for 72 h with phytohemagglutinin (PHA) stimulation. Chromosomes were analyzed by G-banding using trypsin digestion and Wright’s staining (GTW). Twenty metaphase spreads were analyzed. The karyotypes were described according to An International System for Human Cytogenetic Nomenclature (ISCN 2016). Fluorescent In Situ Hybridization (FISH) analyses were performed with standard techniques using bacterial artificial chromosome (BAC) probes (Empire Genomics, NY; listed in Fig. 1d). All genomic coordinates are based on the Human GRCh37/hg19 Genome Assembly.
Cytogenetic Analysis. a Chromosome G-banding analysis identified an apparently balanced de novo translocation involving chromosome 3 and chromosome 14 in the proband. b Selected BAC clones on chromosome 14 were fluorescent-dye labeled as indicated. Five probes within the interval of chr14:29,000,000-30,500,000 are scaled in terms of their size and location, while four peripheral ones are not. c and d Fluorescent in situ hybridization (FISH) analysis of metaphase chromosomes reveals the breakpoint indicated by a blue line at approximately chr14:29,689,627-29,728,030 between the FOXG1 gene (chr14:29,236,278-29,239,483) and the previous defined smallest region of deletion overlap(SRO, chr14:29,875,672-30,173,942). c, inverted DAPI staining; d, DAPI staining. SG, spectrum green; SO, spectrum orange. dn, de novo
Comparative genomic hybridization (CGH)
DNA was extracted from the patient’s peripheral blood using QIAamp® DNA Blood Mini Kit (Qiagen, CA). A Nanodrop ND-1000 spectrometer (Thermo Scientific, DE) was used for determination of DNA concentrations. Microarray experiments were performed using the SurePrint G3 Human CGH Microarray 4 × 180 K platform (Agilent Technologies, CA). Data were analyzed and visualized using the Agilent CytoGenomics 4.0 software (Agilent Technologies, CA). Commercially available pooled male DNA (Promega, WI) was used as control DNA. Briefly, after the initial DNA denaturation step, the patient’s DNA and control DNA (500 ng) were labeled with dyes Cyanine-5dUTP and Cyanine-3dUTP, respectively, by using Agilent’s Universal Linkage System (ULS™) technology as per the manufacturer’s recommendations. The hybridization and subsequent steps were performed as per the manufacturer’s recommendations (Agilent Technologies, CA). The slide was scanned in Agilent’s high-resolution Model #G2505C scanner at 3 µm resolution. The scanned image file was directly imported to the Agilent CytogGenomics 4.0 software for the visualization and analysis.
Results
Chromosome analysis identified a balanced reciprocal translocation involving breakpoints on the short arm of chromosome 3 at band 3p13 and the long arm of chromosome 14 at band 14q11.2 (Fig. 1a). The karyotype was defined as 46,XY,t(3;14)(p13;q11.2). Chromosome analysis on peripheral blood of both parents revealed normal karyotypes, indicating that the identified translocation in the proband is de novo. Chromosomal rearrangement has been shown to cause disruption or dysregulation of a gene (or genes) near the breakpoint, so detailed breakpoint characterization has been beneficial to identify and annotate genes important in human development [15]. In addition, such chromosomal rearrangements may be more complex when analyzed at a higher level of resolution, and are associated with deletions around breakpoints in 37% cases [15]. Microarray analysis revealed this proband carries no clinically significant genomic imbalances and no identifiable aberrations in the proximity of breakpoints (data not shown). We hypothesize that the de novo balanced (3;14) translocation is likely pathogenic and elected to map the breakpoints using FISH analysis.
FISH analysis using a series of bacteria artificial chromosome (BAC) probes (RP11-905F6, RP11-689N19, RP11-844M13 on chromosome 3, and RP11-91A10, RP11-1001O21, RP11-1005D8 and RP11-661M22 on chromosome 14) defined the breakpoint within a 2.7 Mb interval of chr3:66,441,621-69,183,895 at band 3p14.1, and the breakpoint within a 5.9 Mb interval of chr14:24,994,466-30,870,173 within band 14q12 (data not shown). The karyotype was therefore refined as 46,XY,t(3;14)(p14.1;q12)dn. Genes within these regions were investigated in detail by literature search and genotype–phenotype comparison. There are 10 protein-coding genes, including EOGT (OMIM#614789) and LMOD3 (OMIM#616112) within the chr3 breakpoint interval and 7 refseq protein-coding genes, including FOXG1 (OMIM#164874) and PRKD1 (OMIM#605435) within the chr14 breakpoint interval. EOGT is associated with autosomal recessive Adams-Oliver syndrome 4, and LMOD3 with autosomal recessive Nemaline myopathy 10. FOXG1 is associated with autosomal dominant FOXG1 syndrome, and PRKD1 with autosomal dominant congenital heart defects and ectodermal dysplasia. Haploinsufficiency of FOXG1 caused by long-range position effects of intergenic structural variants is one underlying molecular mechanism causing FOXG1 syndrome [11]. As the proband presented a clinical phenotype similar to FOXG1 syndrome (discussed below), we focused on the fine mapping of the breakpoint on chromosome 14. BAC probe (RP11-829F20), which covers the entire FOXG1 gene, was selected and co-hybridized along with the RP11-1001O21 BAC probe, and revealed the breakpoint is distal to the FOXG1 gene (the gene itself is likely not disrupted), and within a 1.6 Mb interval of chr14:29,303,506-30,870,173. In order to further define the breakpoint relative to the FOXG1 gene, we selected four additional BAC probes (Fig. 1b). The RP11-829F20 and RP11-880C14 probes label the derivative chromosome 14, while the RP11-58H4, RP11-845I7, and RP11-840N18 probes stain the derivative chromosome 3 (Fig. 1c, d). These findings indicate the breakpoint occurred in an intergenic ~ 38 Kb region (approximately around chr14:29,689,627- 29,728,030) between the FOXG1 gene (~ 450 Kb proximal) and the smallest region of deletion overlap previously defined (SRO, ~ 148 Kb distal) [11]. In conclusion, metaphase FISH analysis using two BAC probes of RP11-829F20 (FOXG1) and RP11-840N18 (SRO) defined the breakpoint for this case, and can in principle diagnose 16/16 reported FOXG1 syndrome cases which present structural variants involving chromosome 14 [11].
Discussion
FOXG1 syndrome results from intragenic or intergenic mutations resulting in haploinsufficiency of FOXG1. FOXG1 syndrome has a well-defined clinical phenotype characterized in numerous case reports and cohort studies as postnatal growth restriction, post-natal microcephaly, global developmental delay with absence of language, movement disorder characterized by chorea and dystonia, deficient social reciprocity, variable forms of epilepsy, poor sleep patterns, paroxysmal laughter/crying, recurrent aspiration, with common neuroimaging findings of simplified gyral patterning reduced white matter frontal lobe volume, hypogenesis of the corpus callosum, and delayed myelination [8, 16,17,18].
Here we report a case of developmental encephalopathy in a 7 year-old-male patient with microcephaly, global developmental delay, seizures, appendicular spasticity with decreased axial tone, stereotypies, and inappropriate laughing/screaming spells. From the available medical record, microcephaly was not noted until approximately 4 months of age, though a final determination of whether it is truly post-natal microcephaly is not possible. An MRI was performed and reportedly was normal, without agenesis or malformation of the corpus callosum, but the images were not available for our review. Cytogenetic analysis using chromosome, microarray, and FISH demonstrated a de novo balanced translocation between chromosome 3 at band 3q14.1 and chromosome 14 at band 14q12. The breakpoint on chromosome 14 involves an intergenic region approximately 450 kb distal to the FOXG1 gene.
Overall the patient’s phenotype as described above is consistent with a developmental encephalopathy, and cytogenetic analysis confirms a diagnosis of FOXG1 syndrome [10]. It should be noted, however, that medical records are not available for the patients first 3 years of life, and much of the history from that time period was obtained via the patient’s family. This represents a potential weakness of this paper, and highlights the difficulty of using diagnostic criteria reliant on regular healthcare access and detailed neonatal and pediatric records.
This case is notable due to its unique cytogenetics. The translocation described does not disrupt the FOXG1 gene itself, but rather its proximity to an enhancer element. This structural variant likely disrupts the normal cis-acting regulatory elements typically responsible for controlling FOXG1 expression, resulting in effective haploinsufficiency. Chromosomal structural variants resulting in a “congenital variant of Rett syndrome” have been previously described, as well as cases of deletions resulting in the disruption of FOXG1 regulatory elements [19,20,21]. Other reports have found examples of chromosomal structural variants disrupting FOXG1 regulatory elements [8, 13, 22,23,24]. Mehrjouy et al. published one of the more comprehensive investigations into regulatory variants in FOXG1 syndrome, examining the role of topologically associated domains and long range positional effects on FOXG1 expression [11]. They defined a smallest region of deletion overlap (SRO) of approximately 430 kb located over 600 kb 3′ to FOXG1. They conclude that structural variants resulting in disruption of regulatory elements within the SRO results in the FOXG1 phenotype. In our case, the breakpoint event (~ 450 Kb distal to FOXG1) also occurs between FOXG1 and the SRO region, presumably resulting in loss of FOXG1 expression and a disease phenotype. This supports the idea that such disease-causing chromosomal rearrangements disrupting a distant regulatory enhancer are recurrent in FOXG1 syndrome. A unique genomic/chromatin characteristic at this locus may predispose it to breakage and rearrangement, warranting further molecular investigation.
A genotype–phenotype correlation study has also been performed in a cohort of 83 patients with FOXG1 intragenic variants and revealed high phenotypic variability. Mild cases were associated with missense variants in the forkhead conserved site 1, severe phenotypes with truncating variants, and the most severe cases with the N-terminal truncating mutations [17]. We summarized the clinical features associated with different FOXG1 variants, including FOXG1 intragenic mutations [17], balanced translocations and microdeletions resulting in enhancer disruption [11], and microduplications associated with increased gene dosage [11] (Table 1). For intragenic mutations, we listed the most severe group with N-terminal truncating variants and the mildest group with missense variants within the forkhead conserved site 1 [17]. It is interesting that structural variants with a presumable enhancer disruption appeared to show higher severity than the truncating group, while the microduplication group showed milder phenotypes than the missense group (Table 1). The phenotype observed in this study is consistent with the severe phenotype associated with an enhancer disruption, despite our ability to accurately assess anomalies of the corpus collosum (Table 1). This suggests that the enhancer disruption may decrease gene expression of the FOXG1 gene during development to a level of a complete gene deletion, resulting in a null allele and a severe haploinsufficient phenotype. We suspect that the defined enhancer may act as a super enhancer and regulate more genes within its proximity. Haploinsufficiency of adjacent genes could contribute to the phenotypic severity observed in this group. More detailed molecular characterization of the enhancer disruption will help better characterize this genotype–phenotype association.
This case further supports many of the features common to the FOXG1 syndrome phenotype. In addition, it confirms that such a phenotype occurs in a case involving a chromosomal structural variant resulting in haploinsufficiency via disruption of FOXG1 regulatory elements located within a previously defined SRO. It also shows that for patients with a similar phenotype and karyotype consistent with balanced structural variants involving 14q11-q13, reflex metaphase FISH analysis using BAC probes of RP11-829F20 (FOXG1) and RP11-840N18 (SRO)(Fig. 1d) provides an efficient option for making a final diagnosis. By examining the breakpoints of previously reported FOXG1 syndrome cases (reviewed by Mehrjouy et al. [11]), we are able to conclude that the proposed FISH analysis would help make a genetic diagnosis for 12/12 cases with balanced translocations and for 4/4 cases with submicroscopic microdeletions. It also suggests that submicroscopic balanced rearrangements, such as subtle inversions or insertions, may cause the disease, but would remain undiagnosed without the reflex FISH. Taken together, FISH and FOXG1 gene sequencing would identify a majority of disease-causing mutation types (Table 1), including translocation, microdeletion, single nucleotide and small indels, and are therefore recommended as a diagnostic algorithm for FOXG1 syndrome.
Availability of data and materials
The raw data of CGH and FISH were available upon request.
References
Online Mendalian Inheritance in Man, OMIM. Baltimore: Johns Hopkins University, No Title.
Bredenkamp N, Seoighe C, Illing N. Comparative evolutionary analysis of the FoxG1 transcription factor from diverse vertebrates identifies conserved recognition sites for microRNA regulation. Dev Genes Evol. 2007;217(3):227–33. https://doi.org/10.1007/s00427-006-0128-x.
Murphy DB, Wise S, Burfeind P, et al. Human grain factor 1, a new member of the fork head gene family. Genomics. 1993;21:551–7.
Li J, Chang HW, Lai E, Parker EJ, Vogt PK. The oncogene qin codes for a transcriptional repressor. Cancer Res. 1995;55:5540–5.
Tao W, Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8(5):957–66. https://doi.org/10.1016/0896-6273(92)90210-5.
Hanashima C, Shen L, Li SC, Lai E. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J Neurosci. 2018;22(15):6526–36. https://doi.org/10.1523/jneurosci.22-15-06526.2002.
Cargnin F, Kwon JS, Katzman S, Chen B, Lee JW, Lee SK. FOXG1 orchestrates neocortical organization and cortico-cortical connections. Neuron. 2018;100(5):1083-1096.e5. https://doi.org/10.1016/j.neuron.2018.10.016.
Kortüm F, Das S, Flindt M, et al. The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet. 2011;48(6):396–406. https://doi.org/10.1136/jmg.2010.087528.
Seltzer LE, Ma M, Ahmed S, et al. Epilepsy and outcome in FOXG1-related disorders. Epilepsia. 2014;55(8):1292–300. https://doi.org/10.1111/epi.12648.
Ma M, Adams HR, Seltzer LE, Dobyns WB, Paciorkowski AR. Phenotype differentiation of FOXG1 and MECP2 disorders: a new method for characterization of developmental encephalopathies. J Pediatr. 2016;178(233–240):e10. https://doi.org/10.1016/j.jpeds.2016.08.032.
Mehrjouy MM, Fonseca ACS, Ehmke N, et al. Regulatory variants of FOXG1 in the context of its topological domain organisation/631/208/200/631/208/1516 article. Eur J Hum Genet. 2018;26(2):186–96. https://doi.org/10.1038/s41431-017-0011-4.
Papa FT, Mencarelli MA, Caselli R, et al. A 3 Mb deletion in 14q12 causes severe mental retardation, mild facial dysmorphisms and Rett-like features. Am J Med Genet A. 2008;146A(15):1994–8. https://doi.org/10.1002/ajmg.a.32413.
Shoichet SA, Kunde SA, Viertel P, et al. Haploinsufficiency of novel FOXG1B variants in a patient with severe mental retardation, brain malformations and microcephaly. Hum Genet. 2005;117(6):536–44. https://doi.org/10.1007/s00439-005-1310-3.
Ariani F, Hayek G, Rondinella D, et al. FOXG1 is responsible for the congenital variant of rett syndrome. Am J Hum Genet. 2008;83(1):89–93. https://doi.org/10.1016/j.ajhg.2008.05.015.
Higgins AW, Alkuraya FS, Bosco AF, et al. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. Am J Hum Genet. 2008;82(3):712–22. https://doi.org/10.1016/j.ajhg.2008.01.011.
Zhang Q, Wang J, Li J, et al. Novel FOXG1 mutations in Chinese patients with Rett syndrome or Rett-like mental retardation. BMC Med Genet. 2017;18(1):96. https://doi.org/10.1186/s12881-017-0455-y.
Mitter D, Pringsheim M, Kaulisch M, et al. FOXG1 syndrome: genotype–phenotype association in 83 patients with FOXG1 variants. Genet Med. 2018;20(1):98–108. https://doi.org/10.1038/gim.2017.75.
Vegas N, Cavallin M, Maillard C, et al. Delineating FOXG1 syndrome: From congenital microcephaly to hyperkinetic encephalopathy. Neurol Genet. 2018. https://doi.org/10.1212/nxg.0000000000000281.
Takagi M, Sasaki G, Mitsui T, Honda M, Tanaka Y, Hasegawa T. A 2.0 Mb microdeletion in proximal chromosome 14q12, involving regulatory elements of FOXG1, with the coding region of FOXG1 being unaffected, results in severe developmental delay, microcephaly, and hypoplasia of the corpus callosum. Eur J Med Genet. 2013;56(9):526–8. https://doi.org/10.1016/j.ejmg.2013.05.012.
Caporali C, Signorini S, De Giorgis V, Pichiecchio A, Zuffardi O, Orcesi S. Early-onset movement disorder as diagnostic marker in genetic syndromes: Three cases of FOXG1-related syndrome. Eur J Paediatr Neurol. 2018;22(2):336–9. https://doi.org/10.1016/j.ejpn.2018.01.007.
Ellaway CJ, Ho G, Bettella E, et al. 14q12 microdeletions excluding FOXG1 give rise to a congenital variant Rett syndrome-like phenotype. Eur J Hum Genet. 2013;21(5):522–7. https://doi.org/10.1038/ejhg.2012.208.
Perche O, Haddad G, Menuet A, et al. Dysregulation of FOXG1 pathway in a 14q12 microdeletion case. Am J Med Genet Part A. 2013;161(12):3072–7. https://doi.org/10.1002/ajmg.a.36170.
Goubau C, Devriendt K, Van Der Aa N, et al. Platelet defects in congenital variant of Rett syndrome patients with FOXG1 mutations or reduced expression due to a position effect at 14q12. Eur J Hum Genet. 2013;21(12):1349–55. https://doi.org/10.1038/ejhg.2013.86.
Alosi D, Klitten LL, Bak M, Hjalgrim H, Møller RS, Tommerup N. Dysregulation of FOXG1 by ring chromosome 14. Mol Cytogenet. 2015. https://doi.org/10.1186/s13039-015-0129-4.
Acknowledgements
We thank the family and patient for their participation in this study.
Funding
This work was supported by the Department of Pathology and Laboratory Medicine at University of Rochester Medical Center. No other funding was received.
Author information
Authors and Affiliations
Contributions
CPC and BZ conceived, designed, and performed the experiments. CPC, EC, CF, MAI, ARP, and BZ wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Informed consent to participate and for publication was obtained from the study participant.
Competing interests
The authors have declared that no conflict of interest exists.
Consent for publication
Informed consent to participate and for publication was obtained from the study participant.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Craig, C.P., Calamaro, E., Fong, CT. et al. Diagnosis of FOXG1 syndrome caused by recurrent balanced chromosomal rearrangements: case study and literature review. Mol Cytogenet 13, 40 (2020). https://doi.org/10.1186/s13039-020-00506-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-020-00506-1