Abstract
Objective
The aim of this research was to evaluate independently the performance of a new isothermal amplification assay for cervical cancer screening compared to two previously validated PCR-based assays and histologic endpoints.
Methods
This is a sub-study from the Chinese multi-center screening trial (CHIMUST). The self-collected and clinician-collected specimens stored in PreservCyt at − 4 °C from 6042 women with complete data were tested with the AmpFire assay. These specimens had been previously tested with Cobas and SeqHPV assays. In the primary study all patients with an abnormal test were referred to colposcopy where all had directed and/or random biopsies plus ECC. No additional patients were called back based on the AmpFire results.
Results
6042/6619 women had complete data (mean age 44.1). There were 57 cases of CIN 2, 35 cases of CIN 3 and 2 cancers. The sensitivity for CIN2+ and CIN3+ were similar among the three assays (both direct and self-collected). For the specificities in all categories (CIN2+/CIN3+ and self and direct collection), isothermal amplification assay was either equal to or more specific than Cobas but consistently less specific than SeqHPV.
Conclusion
The AmpFire HPV assay showed similar sensitivity to Cobas and SeqHPV for CIN2+ and CIN3+ on both self and clinician-collections (P>0.05), with good specificity. The speed, low cost, and simplicity of this assay will make it particularly suited for low and middle resource settings. Its accuracy with self-collection makes it applicable for mass screening programs.
Similar content being viewed by others
Introduction
For over two decades, our academic research group has had a major interest in self-collection technologies for cervical cancer screening [1]. We initially demonstrated how, depending on the assay used, a self-collected vaginal specimen could achieve sensitivity equal to a direct endocervical specimen (clinician-collected) [2]. We have concentrated on lower-resource settings, and initially evaluated faster assay platforms that could accommodate the volume of samples that could be delivered with self-collection [3, 4]. Then we developed a solid media transport card that would be affordable in medically underserved areas to avoid the handling and transport of liquid media [5]. Also, we developed community-based healthcare delivery systems and developed approaches that could reach thousands of women per day [6, 7].
Recently, we learned of the AmpFire assay which presented a unique constellation of characteristics that appeared special among the sea of high-risk human papillomavirus (hrHPV) assays we had explored over the years. The amplification method used was isothermal, making it simple and fast compared to standard thermocycling PCR (polymerase chain reaction). With the simplicity came a significant reduction in per sample cost, as well as the equipment and space required, equal to the least expensive of HPV assays. In addition, there was no deoxyribonucleic acid (DNA) extraction required, since the assay handles raw sample. Therefore, we saw a future opportunity to use a simple dry collection brush with no transport media required (solid or liquid). This would further reduce the cost and facilitate population-based screening in remote communities. The combination of these unique features also represented a real opportunity to integrate clinic-based hrHPV testing into same-day “test-and-treat” cervical screening programs. The platform offers multiplex detection in a single tube of 15 high-risk types with separate detection of types 16/18; or optionally offers full genotyping in 4 tubes. Also, using the same equipment and similar isothermal methodology a panel of STD tests could be done, including SARS-CoV-2 (https://atilabiosystems.com/our-products/). When we first became aware of the AmpFire technology, there were promising analytic data, but we were unable to find clinical validation against histologic outcomes [8]. Therefore, the objective of this study was to compare the AmpFire high-risk HPV assay to two previously validated PCR-based assays, using histologic endpoints.
Methods
Study population
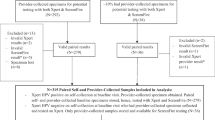
The Chinese Multicenter Screening Trial (CHIMUST) “screening phase” took place between August 2016 and November 2017 [9]. The study was IRB approved by Peking University Shenzhen Hospital (PUSHGYN 2015005) and registered with the WHO designated Chinese Clinical Trial Registry (ChiCTR-EOC-16008456). CHIMUST was designed to compare the standard PCR based assay (Cobas assay, Roche, Pleasanton CA, USA) and a next-gen sequencing assay (SeqHPV assay, BGI Shenzhen, PRC) on both self- and clinician direct collected specimens. It was also designed to study liquid vs solid media sample transport for self-collected specimens. This sub-study of the isothermal amplification assay (AmpFire assay) focuses on 6619 patients from several of the study locations, whose samples were stored at − 4 °C and not frozen [Hebei Province (Pingxiang) – 2035; Hubei Province (Huang Shi) – 1250; Guangdong Province (Chao Zhou) – 1000; Beijing (Men Tao Go) – 988; Inner Mongolia (Xiang Huang Qi) – 1346]. Women aged 30–55 years who had not been screened for cervical cancer in the 3 years prior to entering the study were enrolled. Eligibility required they be non-pregnant, have an intact uterus, and have had no prior pelvic radiation. All the women signed an informed consent for the study, and for the storage and future use of their specimens. The protocol of this trial was approved by the ethics committee of Peking University Shenzhen Hospital (IRB: PUSHGYN 2016001).
Specimen collection
In the primary study (CHIMUST) every woman first took a self-sample, rubbed the brush (with standard 9- μm nylon “Christmas tree head) on the POI solid media transport card [5] and then placed the brush in 6 ml of ThinPrep medium (PreservCyt). Next a speculum was placed, and a physician obtained a direct endocervical specimen also placed in PreservCyt medium (20 cc). Self-collection was processed with the Cobas PCR-based assay and next-gen sequencing assay. The clinician collection was processed for the same two assays, and also for cytology (ThinPrep, Hologic, Marlborough, MA, USA). Some of the patient’s samples in CHIMUST were stored as frozen specimens. There were 6619 patients whose samples (both self and direct collected) in ThinPrep were stored at − 4 °C. These were the specimens tested with the isothermal amplification assay for this study. The specimens had been stored from 1 to 3 years depending on the entry time of the patients into the study at the various study sites.
Isothermal amplification assay (AmpFire HPV assay)
AmpFire is an isothermal amplification real-time fluorescent HPV detection assay, developed in the USA (Atila BioSystems, Inc., Mountain View, CA), which detects HPV directly from clinical samples. No DNA extraction step is required [8] (https://atilabiosystems.com/multiplex-high-risk-hpv-by-fluorescent-detection/). Using a dry brush sample or the isothermal amplification assay transport media total processing time is ≤ 1 h. Using ThinPrep samples requires initial removal of the methanol. There are two isothermal amplification platforms, a multiplex assay: 16, 18, with a thirteen-type high risk pool; and a genotyping assay for 15 specific types. The platforms can be run individually, simultaneously or sequentially. Samples can be processed individually or batched. Formalin fixed paraffin embedded tissue blocks (FFPE) are also acceptable for genotyping studies. The multiplex assay detects 15 “high risk” HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, and 68 in a single tube reaction and simultaneously identifies specifically the presence of types 16 and 18 with real time fluorescent detection [10,11,12]. The human cellular gene beta-globin is used as an internal control to measure sample adequacy. Both the isothermal amplification multiplex assay and genotyping assay were Conformité Européenne (European Community) CE-marked in 2017 and received Chinese Food and Drug Administration CFDA) approval in December 2015.
The standard PCR based assay HPV assay (COBAS HPV assay)
The Cobas 4800 HPV Test was Conformité Européenne (European Community) CE-marked in 2009 and received US Food and Drug Administration approval in April 2011. The Cobas 4800 system platform (Roche Molecular Diagnostics, Pleasanton, CA.), consists of the Cobas × 480 instrument and the Cobas z480 analyzer. It features fully automated nucleic acid extraction in combination with real-time PCR technology plus software that integrates the two components respectively.HPV 16 and 18 are identified separately while 12 other HR-HPV types (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) are detected as a pool. The human cellular gene beta-globin is used as an internal control to measure the sample adequacy and the quality of extraction and amplification.The test was performed according to the manufacturer’s instructions (https://www.hpv16and18.com/labs/lab-efficiencies/COBAS-4800-system.html) [13].
SeqHPV assay
The SeqHPV assay is based on next-generation genomic sequencing. The technology employs a series of unique primers to amplify about 150 base pairs DNA of the L1 gene, and a pair of primers to amplify about 150 base pairs of the human β-globin gene as the internal quality control for identifying the false negatives caused by inadequate DNA or failed PCR. The assay uses multiplex PCR to individually detect a total of 14 HR-HPV genotypes (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68). The assay is approved by the CFDA [3, 4].
Colposcopy and histology examination
Women who tested positive by either the Cobas PCR based assay or next gen sequencing assay on self-collected or clinician-collected specimens, or who had cytology ≥ ASC-US were asked to return for colposcopy. Women having colposcopy were evaluated by our standard research protocol by having a minimum of four small cervical biopsies plus an endocervical curettage [Preventive Oncology International, Inc. (POI) protocol of directed and random biopsies by quadrant] [1]. Histology results from all study sites were independently reassessed by a study gyn pathologist. All histology slides were interpreted as Normal, CIN 1, CIN 2, CIN 3/AIS, or cancer. No additional patients were recalled for a colposcopic examination based on the results of the isothermal amplification assay.
Statistical analysis
HPV type 53 (included only in the isothermal amplification assay) was eliminated from the analysis so a 14- type comparison could be done among the three assays.
Differences in sensitivity and specificity for ≥CIN2,3 were compared using McNemar’s test. P < 0.05 was considered significant. All data were analyzed by SPSS 19.0 [14].
Role of the funding source
This work was supported by a grant from the Shenzhen Health Family Planning Commission, Shenzhen, PR China (Sanming Project of Medicine in Shenzhen, Protocol Number SZSM201412010) and the governmental funds for Shenzhen Leading Gynecological Subject, Shenzhen, PR China,(Science and Education of Shenzhen Health, Protocol Number [2018]61). The two funders had no role whatsoever in the study conduct or analysis.
Results
A total of 6042 women who had all prescribed screening and diagnostic procedures with no missing data are included in the analysis. The mean age of the study population (6042) was 44.1 years. 577 (8.7%) women were dropped from the analysis: 1501 women were asked to return for colposcopy, and 556 (37.04%, 556/1501) did not return to the study doctors. 6 (0.09%, 6/6619) were missing Cobas HPV clinician collection data; 1 (0.02%, 1/6619) was missing SeqHPV clinician data; 10 (0.15%, 10/6619) were missing SeqHPV self-collection results (1 of the these also missing AmpFire self-collection); and 4 (0.06%, 4/6619) had unsatisfactory cytology. The 577 women who were excluded were similar to the total dataset of 6042. 942 women underwent colposcopy and biopsies. Considered as an “intention to treat” analysis, CIN 2 was diagnosed in 0.94% (57/6042) of the total screened population in the analysis, CIN 3 in 0.58% (35/6042) (one of the CIN3 also had AIS), cervical cancer (squamous cell carcinoma) in 0.03% (2/6042). The comparable risk percentages had all women followed protocol, presuming the exclusions were random, would be approximately 1.6 times higher.
The positivity rates of the three assays, and two collection methods, were very similar. Neither assay choice nor clinician versus self-sampling led to important differences in HPV positivity: The positivity rate of the AmpFire HPV assay for clinician collection and self-collection were 10.6% (639/6042) and 11.5% (697/6042), respectively. The positivity rates of Cobas HPV for clinician collection and self-collection were 10.3% (620/6042) and 12.8% (775/6042), respectively. The positivity rates of SeqHPV clinician collection and self-collection were 10.0% (605/6042) and 10.6% (641/6042), respectively. In contrast, the total abnormal cytology (ASC-US or worse) rate was lower than HPV testing, 4.6% (279/6042), with frequencies of ASC-US of 1.9% (114/6042), LSIL of 1.5% (89/6042), ASC-H of 0.45% (27/6042), HSIL of 0.81% (49/6042).
Tables 1 and 2 show the sensitivity and specificity for the three HPV assays for both clinician-collected and self-collected specimens as well as liquid-based cytology.
For ≥CIN2, the sensitivities of the 3 HPV assays for both self and clinician collection were similar. With regard to specificity, for clinician collection, AmpFire was similar to Cobas but less specific than SeqHPV (p < 0.05). For self-collection: Cobas was less specific than AmpFire (p < 0.05), and AmpFire less specific than SeqHPV (p < 0.05) (Table 1).
For ≥CIN3, the sensitivities of the 3 HPV assays for both self and direct collection were similar (p = 1.0). Regarding specificity, for direct collection AmpFire was similar to COBAS (p = 0.09) but less specific than SeqHPV (p = 0.007). For self-collection: Cobas was less specific than AmpFire (p < 0.001), and AmpFire less specific than SeqHPV (p < 0.001) (Table 2).
Table 3 provides a comparison of AmpFire assay to Cobas for type detection; agreement was very good to excellent based on the kappa statistic. The histologic outcomes for the discrepant results were examined, and did not reveal important differences; specifically, there was only 1 case of ≥CIN3 with discrepant typing results, of the many thousands of typing results.
Discussion
The CHIMUST dataset is highly validated due to liberal patient referral to colposcopy, and a minimum of 5 biopsies obtained from all colposcopy patients (POI protocol of directed and random biopsies) [1]. The AmpFire assay, for both self and direct collected samples, generated similar numbers of positive results and had similar accuracy compared with Cobas and SeqHPV with reference to histologic CIN2 or CIN3. Sensitivity was the same, while AmpFire was equal or more specific than COBAS but generally less specific than SeqHPV.
The comparable accuracy of AmpFire, in our opinion, tells only part of the story. Importantly, AmpFire does not require the complexities and costs of DNA extraction. A loss of viral copies, which can average about 40% with extraction, is reduced to a minimum by eliminating that step [14]. Dry brush samples can now be easily self-collected and delivered to simple non-specialized table-top laboratories [15]. Procedures are easy to learn because only basic pipetting skills are required. Product is not wasted since specimens can be processed individually, in small numbers, or batched.
The same equipment and methodology also being directly applicable for all routine sexually transmitted disease testing, and for SARS-CoV-2. Finally, this assay can function very effectively in a hospital laboratory or large clinic, but can be moved to less developed settings too.
In the future, the AmpFire assay might be redesigned to be a 14-type or even 13-type assay (removing type 53 and possibly type 66), although we had limited data to address the change. There were only 10 cases in our study cohort that were single type HPV 53 by AmpFire assay whom we discovered had colposcopy during the trial (5 were positive for other types by COBAS and/or SeqHPV), and all 10 had negative colposcopy and cytology <LSIL.
Conclusion
We believe the data presented in this study demonstrates the AmpFire assay compares favorably with the Cobas and the SeqHPV assays. In consideration of the aforementioned characteristics, this assay may be a useful choice for primary screening and triage in low- and middle-income regions of the world, especially applicable to self-collection.
Abbreviations
- PCR:
-
polymerase chain reaction
- CHIMUST:
-
Chinese multi-center screening tria
- HPV:
-
human papillomavirus
- ECC:
-
endocervical curettage
- CIN:
-
cervical intraepithelial neoplasia
- CIN1:
-
cervical intraepithelial neoplasia grade 1
- CIN2:
-
cervical intraepithelial neoplasia grade 2
- CIN3:
-
cervical intraepithelial neoplasia grade 3
- AIS:
-
Adenocarcinoma in situ
- DNA:
-
deoxyribonucleic acid
- IRB:
-
Institutional Review Board
- POI:
-
Preventive Oncology International, Inc.
- FFPE:
-
Formalin fixed paraffin embedded
- CFDA:
-
Chinese Food and Drug Administration
- ASCUS:
-
atypical squamous cells of undetermined significance
- LSIL:
-
low-grade grade squamous intraepithelial lesions
- HSIL:
-
high-grade squamous intraepithelial lesions
- ASC-H:
-
atypical squamous cells cannot rule out HSIL
References
Belinson J, Qiao YL, Pretorius R, et al. Shanxi Province cervical Cancer screening study: a cross-sectional comparative trial of multiple techniques to detect cervical neoplasia. Gynecol Oncol. 2001;83(2):439–44.
Belinson JL, Hu S, Niyazi M. Prevalence of type-specific human papillomavirus in endocervical, upper and lower vaginal, perineal, and vaginal self-collected specimens; implications for vaginal self-collection. Int J Gynecol Cancer. 2010;127(5):1151–7.
Yi X, Zou J, Xu J, et al. Development and Validation of a New HPV Genotyping Assay Based on Next Generation Sequencing. Am J Clin Pathol. 2014;141:796–804. https://doi.org/10.1309/AJCP9P2KJSXEKCJB Cuzick J, Clavel C, Petry KU, et al. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int J Cancer. 2006;119(5):1095–101.
Du H, Yi J, Wu R, et al. A New PCR based Mass Spectrometry System for High-Risk HPV Part II – Clinical Trial. Am J Clin Pathol. 2011;136(6):920–3 Mayrand MH D-FE, Rodrigues I,Walter SD, et al.cervical cancer screening trial study group. Human Papillomavirus DNA versus Papanicolaou screening tests for cervical cancer. N Engl J Med 2007;357(16):1579–88.
Kathryn Maurer MD, Hongxue Luo MD, Zhiyong Shen PD, et al. The development and evaluation of a new solid media specimen transport card for population based cervical Cancer prevention. J Clin Virol. 2016;76:14–9. https://doi.org/10.1016/j.jcv.2015.12.010 Epub 2016 Jan 2.
Belinson JL, Wang GX, Qu XF, et al. The development and evaluation of a community based model for cervical Cancer screening based on self-sampling. Gynecol Oncol. 2014;132(3):636–42. https://doi.org/10.1016/j.ygyno.2014.01.006 Epub 2014 Jan 14.
Wu R, Xinfeng Q, Hui D, et al. A Pilot Study to Evaluate an Internet-Based Cervical Cancer Screening Model Based on Self-Sampling. Health. 2016;8:672–67. https://doi.org/10.4236/health.2016.87071.
Wang L, Jiang M, Chao XF, et.al. Evaluation the Performance of An Isothermal HPV Amplification Method for Cervical Cancer Screening. Chinese J Virol. 2018;34(4):498–504.
Wu R, Du H, Belinson JL, et al. Evaluation of COBAS HPV and SeqHPV NGS assays in the Chinese multi-center screening trial (CHIMUST). Atlanta USA: ASCCP; 2019.
Kleeman M, Chen X, Reuter C, et al. Evaluation of a novel isothermal amplification assay for rapid HPV detection and genotyping: Eurogin Meeting Abstracts. 2018. p. 00489.
Tang YW, Lozano L, Chen X, et al. An Isothermal, Multiplex Amplification Assay for Detection and Genotyping of Human Papillomaviruses in Formalin-Fixed, Paraffin-Embedded Tissues. J Mol Diagnostics. 2020;22(3):419–28.
Paytubi S, Taberna M, Mena M, et al. Assessment of isothermal amplification AmpFire assay for detection and genotyping of hpv in Formalinfixed paraffin-embedded head and neck cancer samples: Eurogin Meeting Abstracts. 2018.
Stoler MH, Wright TC, Sharma A, et al. High-risk human papillomavirus testing in women with ASC-US cytology: results from the ATHENA HPV study. Am J Clin Pathol. 2011;135(3):468–75.
Kemp BM, Winters M, Monroe C, Barta JL. How much DNA is lost? Measuring DNA loss of short-tandem-repeat length fragments targeted by the PowerPlex 16® system using the Qiagen MinElute purification kit. Hum Biol. 2014;86(4):313–29.
Sun J, Wu S, Hu L, Shang H, Yang Y, Pretorius R, Huang Y, Yang X, Wu X, Belinson J. Evaluation of liquid vs. dry specimen transport with a newly validated isothermal amplification HR-HPV assay. J Lower Genital Tract Dis. 2020;24(3):243-6.
Acknowledgements
None.
Authors’ contributions
All authors except for MS helped conduct the project and/or the testing of specimens for HPV and/or data analysis. MS and JLB finalized the data analysis and composition of the manuscript. Of note, MS is the corresponding author for submission of the manuscript but the corresponding authors responsible for the mansuscript as listed: The author(s) read and approved the final manuscript.
Funding
Supported by Shenzhen Health Family Planning Commission, Shenzhen, PR China (No. SZLY2017005) Sanming Project of Medicine in Shenzhen (SZSM201412010). The governmental funds were for Shenzhen Leading Gynecological Subject(Science and Education of Shenzhen Health [2018]61).
Availability of data and materials
Data available upon reasonable request from corresponding authors. Registered: Chinese Clinical Trials Registry – Chinese Multicenter Cervical Cancer Screening Trial (CHIMUST) ChiCTR-EOC-16008456.
Ethics approval and consent to participate
The study was approved by the IRBs of the Peking University Shenzhen Hospital, Shenzhen, China.
Consent for publication
The authors agree to publication.
Competing interests
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, W., Du, H., Huang, X. et al. Evaluation of an isothermal amplification HPV detection assay for primary cervical cancer screening. Infect Agents Cancer 15, 65 (2020). https://doi.org/10.1186/s13027-020-00328-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13027-020-00328-1