Abstract
Background
Inaccurate meniscus allograft size is still an important problem of the currently used sizing methods. The purpose of this study was to evaluate a new three-dimensional (3D) meniscus-sizing method to increase the accuracy of the selected allografts.
Methods
3D triangular surface models were generated from 280 menisci based on 50 bilateral and 40 unilateral knee joint magnetic resonance imaging (MRI) scans. These models served as an imaginary meniscus allograft tissue bank. Meniscus sizing and allograft selection was simulated for all 50 bilateral knee joints by (1) the closest mean surface distance (MeSD) (3D-MRI sizing with contralateral meniscus), (2) the smallest meniscal width/length difference in MRI (2D-MRI sizing with contralateral meniscus), and (3) conventional radiography as proposed by Pollard (2D-radiograph (RX) sizing with ipsilateral tibia plateau). 3D shape and meniscal width, length, and height were compared between the original meniscus and the selected meniscus using the three sizing methods.
Results
Allograft selection by MeSD (3D MRI) was superior for all measurement parameters. In particular, the 3D shape was significantly improved (p < 0.001), while the mean differences in meniscal width, length, and height were only slightly better than the allograft selected by the other methods. Outliers were reduced by up to 55% (vs. 2D MRI) and 83% (vs. 2D RX) for the medial meniscus and 39% (vs. 2D MRI) and 56% (vs. 2D RX) for the lateral meniscus.
Conclusion
3D-MRI sizing by MeSD using the contralateral meniscus as a reconstruction template can significantly improve meniscus allograft selection. Sizing using conventional radiography should probably not be recommended.
Trial registration
Kantonale Ethikkommission Zürich had given the approval for the study (BASEC-No. 2018-00856).
Similar content being viewed by others
Background
The meniscus plays an important role in the kinematics of the knee joint, reduces contact pressure [1], and improves joint stability [2, 3]. These functions disappear after subtotal or total meniscectomy with resultant early osteoarthritis [4].
Meniscus allograft transplantation seems to be a valuable option for pain reduction and improvement of function in patients with (chronic) postmeniscectomy syndrome [5]. The first meniscus allograft transplantation was performed in 1989 [6] and has been widely used with encouraging results. Several studies demonstrated good or excellent short- to medium-term results [6,7,8,9,10,11,12]. However, while midterm survivorship is reported to be 85–90%, long-term survivorship decreases to 50–70% [13].
Reconstruction of a geometrically similar meniscus seems to be crucial for physiological joint pressure distribution [14,15,16,17,18,19,20,21] and good clinical results [22,23,24,25]. Undersized grafts could lead to excessive loads due to poor congruity with the femoral condyle, while oversized grafts lose their function by extrusion from the compartment [14]. A mismatch of a few millimeters is supposed to result in poorer biomechanical outcomes [17, 19, 26,27,28,29] and increased degenerative changes [18, 30]. Therefore, sizing should be as close to the native meniscus as possible.
Different methods have been described to determine the size of a meniscus [4, 31,32,33,34,35]. Today, meniscus sizing is most commonly performed by conventional radiography according to the Pollard method [31] or by magnetic resonance imaging (MRI) scans [32, 33], and the latter appears to be superior [33, 36]. Unfortunately, inaccuracy of sizing is still a relevant problem in allograft surgery by these methods and has to be improved [17, 25, 31, 36,37,38,39].
3D meniscus sizing was recently proposed as a solution to increase the accuracy and precision of meniscus allograft selection [40]. The contralateral meniscus can be used as a very precise reconstruction template. However, advantages of 3D-MRI sizing with the contralateral meniscus as a reconstruction template compared with existing sizing methods have not been shown to date. We hypothesized that 3D-MRI sizing with the contralateral side is clearly superior compared with the currently used sizing methods.
Methods
The following description of the approaches used for the validation of the different sizing methods is divided into three parts. Part 1 focuses on describing the basic material, imaging, and measurement methods. Part 2 is dedicated to explaining the three different sizing methods in detail. Finally, part 3 contains the sizing validation methods.
Part 1: Material/imaging/measurements
Material
Fifty patients with bilateral and 40 patients with unilateral complete imaging (34 men, 56 women; mean age 26.7 years (range 15–50)) were retrospectively included in this study. All patients had a patellofemoral disorder. The inclusion criteria were patients with available MRI scan and conventional radiography, mature skeletal age, healthy contralateral meniscus, and no tibio-femoral osteoarthritis (Kellgren and Lawrence grade 0) [41]. The presence of a completely closed growth plate at the distal femur and the proximal tibia in the MRI images was used to determine mature skeletal age. Meniscus integrity was assessed by a fellowship-trained musculoskeletal radiologist, and the presence of meniscus tears, degeneration, or extrusion led to exclusion.
Imaging
All radiographs (RX) were performed in a standard fashion, with a plain anteroposterior (AP) and lateral (LAT) view with the ampoule placed 1 m distant from the knee using calibrators for the correction of magnification (Optimus 50 X-ray Generator; Philips, USA).
All MRI scans were performed in our institution on a 3.0-T scanner (Skyra-fit, Siemens Healthineers, Erlangen, Germany) with a send/receive knee coil, and the patient was examined in the supine position. All MRI examinations consisted of sagittal, coronal, and axial sequences as part of the standard MRI protocol [40]: (1) coronal short tau inversion recovery (STIR) sequence (repetition time 4200 ms; echo time 34 ms; inversion time 210 ms; slice thickness 3 mm; number of slices 23; bandwidth 245 Hz/pixel; flip angle 150°; matrix 384 × 384; field of view 16 cm); (2) sagittal intermediate-weighted sequence with the Dixon technique (repetition time 4200 ms; echo time 39 ms; slice thickness 3 mm; number of slices 30; bandwidth 250 Hz/pixel; flip angle 150; matrix 448 × 448; field of view 16 cm), with in-phase image, and fat-suppressed water image; and (3) axial intermediate-weighted fat-suppressed sequence (repetition time 4990 ms; echo time 40 ms; slice thickness 2.5 mm; number of slices 39; bandwidth 150 Hz/pixel; flip angle 180; matrix 384 × 384; field of view 15 cm).
Measurement methods
The complete dataset was further analyzed based on the following models and measurement methods:
-
1)
3D surface model of medial and lateral meniscus;
-
2)
3D-calculated meniscal width, length, and height;
-
3)
2D-calculated meniscal width and length in MRI; and
-
4)
2D-derived meniscal width and length in RX.
-
1)
3D surface model of medial and lateral meniscus
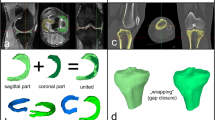
3D surface models were created, as described in a previous paper [40], using the Materialise Interactive Medical Control System (MIMICS) 18.0 3D reconstruction software program (Materialise, Leuven, Belgium). The segmentation of the medial and lateral meniscus was manually performed by two trained orthopedic surgeons in sagittal and coronal slides (biplanar). Finally, all 3D surface models were smoothed (gap closing distance 0.0 mm, smallest detail 1.0 mm), and all left menisci were mirrored to the right surface models.
-
2)
3D-calculated meniscal width, length, and height (Fig. 1a)
Measurement of meniscal width/length/height. a Width/length/height: meniscal dimensions are measured using an oriented bounding box. The box was aligned to the meniscus roots, and the size was adjusted as long as the entire meniscus body was enclosed. The dimensions of the meniscus correspond to the length of the sides of the box: y-axis (green arrow) = meniscal width; x-axis (red arrow) = meniscal length; z-axis (blue arrow) = meniscal height. b 2D-MRI sizing based on MRI of the contralateral side. c 2D-RX sizing according to Pollard on ipsilateral radiography
The 3D measurement of meniscal width, length, and height was performed in the same way as described in a previous paper [40] with an oriented bounding box (= minimal-volume rectangular box that fully encloses the meniscal model) using the in-house developed planning software CASPA (Computed Assisted Surgery Planning Application, Balgrist CARD AG). The box was initially automatically generated around the 3D models. Then, the box was aligned parallel to a line from the anterior to the posterior meniscus root by rotation around the z-axis. Finally, the box dimensions were adjusted as long as the entire meniscus body was enclosed. Meniscus width (y-axis), length (x-axis), and height (z-axis) were now represented by the dimensions of the bounding box.
-
3)
2D-calculated width and length in MRI (Fig. 1b)
For the measurement of the 2D meniscal width and length in MRI, we used the following method. The measurements were performed on axial slides with reference to sagittal slides. Anterior and posterior meniscus roots were identified and connected by a first line. Parallel to this, a second line was placed adjacent to the outer contour of the meniscal base. Two additional lines, perpendicular to the first two lines, were set on the outer contours of the meniscus. Thereby, a 2D bounding box was created. The width and length of the meniscus corresponded to the dimensions of the box and could be measured.
-
4)
2D-derived width and length in RX (Fig. 1c)
The tibial plateau width was measured in the AP view perpendicular to the joint line as the distance between the margin of the tibial metaphysis to the medial lateral tibial eminence. According to Pollard, meniscal width is supposed to be equal to the tibia plateau width for medial and lateral menisci [31]. Tibia plateau length was measured in the LAT view perpendicular to the joint line as the distance between the anterior surface of the tibia above the tuberosity to the posterior margin of the tibia plateau. According to Pollard, the medial and lateral meniscal lengths can be calculated as 80% and 70%, respectively, of the tibia plateau length (medial meniscal length = 0.8 × tibia plateau length; lateral meniscal length = 0.7 × tibia plateau length).
Part 2: Sizing methods
For the 3D-MRI meniscus-sizing method and for the 2D-MRI meniscus-sizing method, we used the contralateral meniscus as a template [40]. Therefore, both sizing methods are direct sizing methods, i.e., they were compared with the size of the meniscus on the contralateral side. For the 2D-RX sizing method, sizing was performed using an indirect calculation based on the bony anatomy of the same side [31].
-
1)
3D-MRI meniscus sizing (Fig. 2)
-
Principle: Direct meniscus sizing, based on the mean surface distance (MeSD) of the contralateral meniscus.
-
Requirements: MRI scan of the healthy contralateral knee and 3D surface models of the allografts.
-
Surface distance: The similarity of two objects can be represented by the closest surface point distance as previously described [40]. To this end, the objects were automatically superimposed by using the iterative closest point (ICP) algorithm so that the distances of all surface points from one object to the other (and vice versa) were as small as possible. The maximum surface distance (MaSD) is the widest measured distance, and the MeSD is the mean of all surface distances. For this analysis, the Hausdorff distance, the highest distance value between the two objects, was used (Fig. 3).
-
Allograft selection: The best fitting allografts were selected based on the closest MeSD values between the 3D template and all available 3D allograft surface models.
-
2)
2D-MRI meniscus sizing
-
Principle: Direct meniscus sizing, based on calculated meniscal width and length of the contralateral meniscus.
-
Requirements: MRI scan of the healthy contralateral knee, and the measured width and length of the allografts.
-
Allograft selection: The best fitting allografts were selected by the smallest differences in width and length between the calculated sizes and the available allografts.
-
3)
2D-RX meniscus sizing
-
Principle: Indirect meniscus sizing, based on derived meniscal width and length calculated from the measurement of the ipsilateral tibia plateau according to Pollard [31].
-
Requirements: Conventional radiography of AP and mediolateral view of the ipsilateral side with calibrators.
-
Allograft selection: The best fitting allografts were selected by the smallest differences in width and length between the calculated sizes and the available allografts.
3D meniscus sizing (3D-MRI sizing). a Right knee with missing medial meniscus. b MRI scan of the contralateral side. c Meniscus segmentation and mirroring (see also Fig. 1). d Meniscus matching by all menisci in the tissue bank by mean and maximum surface distances (see also Fig. 3). e Selection of best fitting meniscus for meniscus allograft surgery
Surface distance (SD). The SD between two 3D models can be calculated after automatic superimposition by using the iterative closest point algorithm so that the distances of all surface points from one model to the other (and vice versa) are as small as possible. Maximum SD (MaSD) is calculated as the widest distance between “meniscus A” and “meniscus B.” The mean SD (MeSD) is the mean value of all surface points between “meniscus A” and “meniscus B.” For this analysis, the Hausdorff distance, i.e., the highest values, is used
Part 3: Sizing validation
First, different terms must be clearly defined for better understanding. The missing meniscus—i.e., after meniscectomy—is hereby called the “original meniscus.” The 3D meniscus surface model for meniscus sizing is hereby called the “template meniscus.” The best fitting meniscus that was selected by a particular sizing method is hereby called the “selected meniscus.” In other words, we used different sizing methods of the template meniscus to choose the selected meniscus and then compared the selected meniscus with the original meniscus.
Because sizing with surface distances does not allow a simple comparison of two values (i.e., width and length) of the original and selected meniscus, as was done in previous studies, sizing validation had to be performed by a more complex simulation (see Fig. 4). Therefore, the 50 patients with bilateral imaging served as the “validation group,” resulting in 100 (= 50 right and 50 left) different medial and lateral menisci. An imaginary tissue bank (“allograft pool”) was composed of all 100 menisci, together with the menisci from the unilateral imaging of another 40 patients, resulting in 140 different menisci. The menisci of each tested patient in the validation group were excluded from the potential menisci that could be selected for them, which finally resulted in an allograft pool of 138 different medial and lateral menisci. Therefore, for the validation group, we needed bilateral MRI scans to compare the previously described sizing methods. For the additional patients, this was not necessary, and we aimed to have as many allografts as possible in our tissue bank.
Validation. The validation was repeated for each of the 100 menisci from the 50 patients as follows (“validation group”): meniscus No. 1 of patient No. 1 was removed from the available pool. The sizing was performed by the “template meniscus.” The 3D surface model of the ipsilateral side (“original meniscus”) served as the best possible allograft (gold standard). The 3D surface model of the contralateral side corresponded to the 3D-MRI sizing. The meniscal width/length of the contralateral side in MRI corresponded to the 2D-MRI sizing. The derived meniscal width/length by the ipsilateral tibia plateau in RX corresponded to the 2D-RX sizing method. The best match (“selected meniscus”) out of the “allograft pool” was selected based on the different “template menisci.” Finally, the “selected meniscus” was compared with the “original meniscus.” This procedure was repeated for all 100 menisci of the “validation group”
The simulation was then performed for each meniscus (meniscus No. 1 to meniscus No. 100) of the validation group. An in-house developed computer program automatically calculated the closest MeSD and MaSD between all 100 original (validation group) and 138 different allografts (allograft pool). Overall, there were 13,800 possibilities for allograft selection by MeSD (= 100 originals × 138 allografts), and the 100 closest menisci served as the best possible allografts, which served as the gold standard. In a second step, the same validation was now performed by the contralateral meniscus surface model as the template, which corresponds to the 3D-MRI sizing. Of the 13,800 possibilities, only the best fitting allografts by this sizing method were selected. In a third step, validation was performed using the closest difference in measured width and length of the contralateral side in MRI (2D-MRI sizing) and 3D measured width and length. Thereby, width and length were equally weighted and selected by the lowest error sum of squares: (widthsized−widthallograft)2 + (lengthsized−lengthallograft)2. In a fourth step, the same procedure was repeated based on the closest difference in derived width and length of the ipsilateral side (2D-RX sizing).
Finally, the original meniscus was compared with the selected menisci resulting from the different sizing methods using the surface distance as well as the meniscal width, length, and height as similarity measurements.
Meniscus outliers were defined as those with a difference between the original and selected meniscus over 5 mm in width, over 5 mm in length, or over 4 mm in height.
Inter-rater/intra-rater reliability
The first 30 knee joints—in alphabetic order based on patient names—were used to calculate interclass correlation. 3D surface models (3D-MRI sizing), as well as width/length in MRI (2D-MRI sizing) and X-ray (2D-RX sizing), were calculated by two trained orthopedic surgeons, as described above. Intra-rater reliability was calculated by the repetition of these measurements by one of the surgeons.
Statistics
The statistical analysis was performed with SPSS (IBM Corp. IBM SPSS Statistics for Windows, version 24.0. Armonk, NY: IBM Corp.) Descriptive analyses and independent-sample t tests were performed to investigate the meniscal diversity. P values below 0.05 were considered statistically significant. The correlations between width/length and MeSD/MaSD were analyzed with a linear regression analysis (Pearson correlation coefficient). Inter-rater and intra-rater reliability was calculated using interclass correlation coefficients (ICCs) (two-way mixed, absolute agreement). We used scatterplots and boxplots for visual presentation.
Results
Meniscus diversity (of the allograft tissue bank)
Overall, the medial meniscus (n = 140) was on average 31.6-mm (standard deviation (Std) ± 3.3 mm) wide, 46.8-mm (Std ± 3.7 mm) long, and 9.3-mm (Std ± 1.4 mm) high. The correlation between meniscal width and length was poor (R2 = 0.310).
The MeSD for all 140 medial surface models (n = 140 × 139 = 19,460) was on average 1.3 mm (Std ± 0.41 mm; min–max 0.57–3.70 mm), and the MaSD was on average 7.6 mm (Std ± 2.72 ; min–max 2.3–19.9 mm). The correlation between MeSD and MaSD values was R2 = 0.430 (Fig. 5).
Correlations between the MeSD and MaSD values. Scatterplot of all 38,920 possibilities of medial and lateral meniscus combinations (38,920 = 140 × 139 for medial and 140 × 139 for lateral meniscus). MaSD = maximum surface distance; MeSD = mean surface distance. Six meniscus examples with increasing mismatch by increasing MaSD and MeSD values
The lateral meniscus (n = 140) was on average 31.7-mm (Std ± 3.7 mm) wide, 35.3-mm (Std ± 2.8 mm) long, and 9.9-mm (Std ± 1.4 mm) high. The correlation between meniscal width and length was poor (R2 = 0.304).
The MeSD for all lateral surface models (n = 140 × 139 = 19,460) was on average 1.4 mm (Std ± 0.45 mm; min–max 0.449–3.89 mm), and the MaSD was on average 7.1 mm (Std ± 2.55; min–max 2.00–20.2 mm). The correlation between MeSD and MaSD values was R2 = 0.495 (Fig. 5).
Sizing validation
All results are summarized in Tables 1 and 2 and Figs. 6 and 7.
Results for the medial meniscus. Boxplots for the medial menisci. The median (middle quartile) marks the midpoint of the data and is shown by the line that divides the box into two parts. The box represents the middle 50% of values, and the upper and lower whiskers represent the range. The outliers are marked as separate points and defined as values more than 1.5 times the median. MeSD = mean surface distance; MaSD = maximum surface distance; Diff width/length/height= difference between the selected allograft dimensions and the original meniscus dimensions
Results for the lateral meniscus. Boxplots for the lateral menisci. The median (middle quartile) marks the midpoint of the data and is shown by the line that divides the box into two parts. The box represents the middle 50% of values, and the upper and lower whiskers represent the range. The outliers are marked as separate points and defined as values more than 1.5 times the median. MeSD = mean surface distance; MaSD = maximum surface distance; Diff width/length/height= difference between the selected allograft dimensions and the original meniscus dimensions
None of the presented sizing methods always selected the best available fitting allograft (which corresponded to the original meniscus) out of the imaginary tissue bank. In particular, the 3D shape (MeSD and MaSD) in these cases was different between the selected and the original meniscus, while the width, length, and height were not distinct.
3D-MRI sizing, which takes the 3D shape into account, significantly improved the accuracy of allograft selection compared with 2D-MRI (p < 0.001) and 2D-RX (p < 0.001) sizing.
Furthermore, 3D-MRI sizing reduced the number of outliers. For the medial meniscus, there were 12% fewer outliers selected compared with 2D-MRI sizing and 48% fewer outliers selected compared with 2D-RX sizing. For the lateral meniscus, there were 7% fewer outliers selected compared with 2D-MRI sizing and 14% fewer outliers selected compared with 2D-RX sizing.
Inter-rater/intra-rater reliability
The interclass correlations for the medial and lateral menisci were good to excellent; see Table 3 for details (< 0.5 = poor, 0.5–0.75 = moderate, 0.75–0.9 = good, > 0.9 = excellent).
Discussion
The most appropriate methods for meniscus sizing are still debated. Even under laboratory conditions, the most familiar radiographic method of Pollard had a standard error of prediction of ± 2.9 mm for meniscal width and ± 3.8 mm for meniscal length [31], which corresponds to a standard error of approximately 8%. Considering that the proposed 10% graft mismatch could cause relevant articular problems (i.e., 3- or 4-mm size difference based on meniscal sizes reported in the literature), the Pollard method seems to not be completely safe [17, 37]. Although MRI sizing seems to be superior to the radiological methods, 17% of the measured menisci in a cadaver study by Shaffer had differences of over 5 mm compared with the anatomical measurements [36]. Other authors have shown better results with an average error rate of 4.0–5.3% [35]. Because sizing is performed based on two variables (i.e., width and length), inaccuracy is probably magnified by the combination of both.
In a previous study, a new 3D meniscus sizing method was proposed, which was based on a healthy contralateral meniscus template to reduce inaccuracy [40]. For this sizing method, there are some basic prerequisites, which must be fulfilled and will be discussed in the following section. First, an easily determined, pose-invariant measurement parameter is required to accurately compare freely moveable 3D bodies. Second, the segmented 3D meniscus has to correspond with the actual 3D anatomical shape. Third, the contralateral meniscus has to be similar. Fourth, the benefits of a 3D sizing method must be compared with those of the currently used sizing methods. Fifth, the additional benefit has to be validated with improved clinical results.
Basic prerequisites for 3D sizing
-
1)
The similarity of 3D models can be evaluated by the surface distance, as already described in a previous study [40]. However, it is not yet known whether 3D sizing should be performed by the closest MeSD or by the closest MaSD, or a combination of these measures. Depending on the chosen criterion, the best allograft would be the selected according to the MeSD or to MaSD. Because there were poor correlations between these two values (R2 = 0.430–0.495), similar to the poor correlations between meniscal width and length (R2 = 304–310), these values could provide different information. The MeSD was generally less variable as the MaSD could be affected by one single surface point with a wide distance between the two models—for example, due to a partly segmented transverse or meniscotibial ligament. Therefore, we assessed sizing primarily based on the MeSD.
The MaSD value was on average 5.3 times the MeSD value in our collection of 38,920 meniscus matched pairs. The best relationship was 2.6 times the value and reached up to 11.7 times the value. In conclusion, a very good fitting allograft could be expected by low (< 1.0 mm) MeSD combined with an MaSD value of less than 3 times the MeSD value. For examples, see Fig. 5.
-
2)
A direct comparison of the anatomical and segmented shapes remains unavailable. However, to date, MRI measurements are often used as the gold standard and seem very good at identifying the meniscus tissue [32, 33, 35, 37, 39].
-
3)
In a previous study, we showed that the contralateral side can be used as a very precise and reliable meniscus template for 3D sizing.
-
4)
Our results are not directly comparable to previous studies. 3D sizing is based on MeSD and not on meniscal width and length. Therefore, a comparison to previous studies was not possible. We solved this problem by a simulated allograft selection based on the three sizing methods from an imaginary tissue bank of 138 different menisci. The selected allografts could be easily compared. An important advantage of this evaluation was the fact that the multiplicative inaccuracy due to a combination of two imprecise parameters (i.e., width and length) was taken into account. Herein, we showed a significant improvement of the 3D shape of the selected meniscus allograft with sizing by the MeSD compared with sizing by the combined meniscal width and length. There were also improvements based on meniscal width, length, and height measures, but these improvements were not statistically significant for all values. Similar to many other studies, the mean width and length values of the selected menisci were good and mostly acceptable across the three sizing methods. However, the main problem of meniscus allograft sizing is related to the number of selected outliers. In our opinion, this is exactly the strength of the 3D sizing method. The number of outliers selected could be reduced by 12–29% compared with 2D-MRI sizing and by 41–53% compared with 2D-RX sizing. We defined outliers as a size difference between the original and selected meniscus of > 5 mm in width or length and > 4 mm in height. A cutoff value of 5 mm for width and length has previously been used by other authors [25, 36]. A cutoff value based on meniscal height had not been previously described. In our collection, the meniscal height had a mean value of 9.3–9.9 mm with a standard deviation of only ± 1.4 mm. Therefore, a cutoff value for meniscal height of > 5 mm would probably never become applicable. Because the clinical evidence regarding meniscal height is still unclear, we did not choose our cutoff value based on the 10% meniscal mismatch rule [17], but on > 4 mm, which was slightly higher than the slide thickness and detected only 5 mismatched menisci overall.
There are few limitations of this study. As already mentioned under the second point above, a direct comparison of the anatomical and segmented shapes is missing. However, the meniscus can be accurately identified in MRI, and MRI is therefore often used as the gold standard [32, 33, 35, 37, 39]. Because a mismatch of 3–4 mm (10%) appears to be clinically relevant, a slide thickness of up to 3 mm can significantly influence the results. Therefore, a biplanar segmentation of the meniscus with excellent inter-rater and intrarater reliability for width and length (ICCinter 0.913–0.973; ICCintra 0.955–0.987) was used [40]. Furthermore, a larger tissue bank could probably have improved our results. However, the number of allografts used is probably sufficient, as there are many tissue banks with (much) less than 138 different available allografts (on request).
3D meniscus sizing has some great advantages, as already mentioned in a previous study [40]. In the present study, we demonstrated—even with a limited number of 138 allografts—that 3D sizing can significantly improve the accuracy and substantially reduce the number of relevant outliers. In conclusion, the limited number of meniscus allografts due to increased demand in the world could be much better matched to the patients. Moreover, this method can be used to do more than find the best fitting 3D allograft. If necessary, a slightly smaller size due to presumed extrusion or slightly greater size due to presumed shrinkage can be easily found by down- or upscaling the 3D template [14]. Furthermore, the feasibility of “bone plugs” or “bone bridge” fixation methods, which are supposed to be superior to “soft-tissue” and “suture bone tunnel” fixation techniques, could also be improved due to the 3D method identifying the shape of the meniscus [14, 42]. This 3D template could also be used in the future for biomimetic 3D-printed scaffolds [43, 44].
However, 3D sizing involves additional costs. An MRI scan of the contralateral side is needed, and the tissue banks must be willing to offer this option. 3D surface models must be generated for all allografts. In the future, meniscus segmentation could probably be performed in a semiautomated or fully automated manner [45, 46], which could reduce additional costs. Automatized comparison of the meniscus template of the healthy contralateral meniscus and all available allografts is already possible.
Conclusion
3D-MRI sizing by MeSD using the contralateral meniscus as a reconstruction template can significantly improve the accuracy of meniscus allograft selection and reduce outliers compared with sizing that relies only on width and length in radiography and MRI. Using these methods, the limited number of available allografts could probably be more effectively distributed based on the rising demand in the world.
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Abbreviations
- 2D/3D:
-
Two-/three-dimensional
- AP:
-
Anteroposterior
- IPC:
-
Iterative closest point
- LAT:
-
Lateral
- MaSD:
-
Maximum surface distance
- MeSD:
-
Mean surface distance
- MIMICS:
-
Materialise Interactive Medical Control System
- MRI:
-
Magnetic resonance imaging
- RX:
-
Radiograph
- Std:
-
Standard deviation
References
Messner K, Gao J. The menisci of the knee joint. Anatomical and functional characteristics, and a rationale for clinical treatment. J Anatomy. 1998;193(Pt 2):161–78.
Shoemaker SC, Markolf KL. The role of the meniscus in the anterior-posterior stability of the loaded anterior cruciate-deficient knee. Effects of partial versus total excision. J Bone Joint Surg Am. 1986;68:71–9.
Cox JS, Nye CE, Schaefer WW, Woodstein IJ. The degenerative effects of partial and total resection of the medial meniscus in dogs’ knees. Clin Orthop Relat Res. 1975:178–83.
Papalia R, Del Buono A, Osti L, Denaro V, Maffulli N. Meniscectomy as a risk factor for knee osteoarthritis: a systematic review. Br Med Bull. 2011;99:89–106.
Ryu RK, Dunbar VW, Morse GG. Meniscal allograft replacement: a 1-year to 6-year experience. Arthroscopy. 2002;18:989–94.
Milachowski KA, Weismeier K, Wirth CJ. Homologous meniscus transplantation. Experimental and clinical results. Int Orthopaed. 1989;13:1–11.
Verdonk PC, Verstraete KL, Almqvist KF, et al. Meniscal allograft transplantation: long-term clinical results with radiological and magnetic resonance imaging correlations. Knee Surg Sports Traumatol Arthrosc. 2006;14:694–706.
McCormick F, Harris JD, Abrams GD, et al. Survival and reoperation rates after meniscal allograft transplantation: analysis of failures for 172 consecutive transplants at a minimum 2-year follow-up. Am J Sports Med. 2014;42:892–7.
Stone KR, Adelson WS, Pelsis JR, Walgenbach AW, Turek TJ. Long-term survival of concurrent meniscus allograft transplantation and repair of the articular cartilage: a prospective two- to 12-year follow-up report. J Bone Joint Surg Br. 2010;92:941–8.
Verdonk PC, Demurie A, Almqvist KF, Veys EM, Verbruggen G, Verdonk R. Transplantation of viable meniscal allograft. Survivorship analysis and clinical outcome of one hundred cases. J Bone Joint Surg Am. 2005;87:715–24.
Vundelinckx B, Bellemans J, Vanlauwe J. Arthroscopically assisted meniscal allograft transplantation in the knee: a medium-term subjective, clinical, and radiographical outcome evaluation. Am J Sports Med. 2010;38:2240–7.
Zaffagnini S, Grassi A, Marcheggiani Muccioli GM, et al. Survivorship and clinical outcomes of 147 consecutive isolated or combined arthroscopic bone plug free meniscal allograft transplantation. Knee Surg Sports Traumatol Arthrosc. 2016;24:1432–9.
Figueroa F, Figueroa D, Calvo R, Vaisman A, Espregueira-Mendes J. Meniscus allograft transplantation: indications, techniques and outcomes. EFORT Open Rev. 2019;4:115–20.
Rodeo SA. Meniscal allografts--where do we stand? Am J Sports Med. 2001;29:246–61.
Haut Donahue TL, Hull ML, Rashid MM, Jacobs CR. The sensitivity of tibiofemoral contact pressure to the size and shape of the lateral and medial menisci. J Orthop Res. 2004;22:807–14.
Huang A, Hull ML, Howell SM, Haut DT. Identification of cross-sectional parameters of lateral meniscal allografts that predict tibial contact pressure in human cadaveric knees. J Biomech Eng. 2002;124:481–9.
Dienst M, Greis PE, Ellis BJ, Bachus KN, Burks RT. Effect of lateral meniscal allograft sizing on contact mechanics of the lateral tibial plateau: an experimental study in human cadaveric knee joints. Am J Sports Med. 2007;35:34–42.
von Lewinski G, Kohn D, Wirth CJ, Lazovic D. The influence of nonanatomical insertion and incongruence of meniscal transplants on the articular cartilage in an ovine model. Am J Ssports Med. 2008;36:841–50.
Sekaran SV, Hull ML, Howell SM. Nonanatomic location of the posterior horn of a medial meniscal autograft implanted in a cadaveric knee adversely affects the pressure distribution on the tibial plateau. Am J Sports Med. 2002;30:74–82.
Alhalki MM, Hull ML, Howell SM. Contact mechanics of the medial tibial plateau after implantation of a medial meniscal allograft. A human cadaveric study. Am J Sports Med. 2000;28:370–6.
Maher SA, Wang H, Koff MF, Belkin N, Potter HG, Rodeo SA. Clinical platform for understanding the relationship between joint contact mechanics and articular cartilage changes after meniscal surgery. J Orthop Res. 2017;35:600–11.
Cameron JC, Saha S. Meniscal allograft transplantation for unicompartmental arthritis of the knee. Clin Orthop Relat Res. 1997:164–71.
Rath E, Richmond JC, Yassir W, Albright JD, Gundogan F. Meniscal allograft transplantation. Two- to eight-year results. Am J Sports Med. 2001;29:410–4.
van Arkel ER, de Boer HH. Survival analysis of human meniscal transplantations. J Bone Joint Surg Br. 2002;84:227–31.
Stevenson C, Mahmoud A, Tudor F, Myers P. Meniscal allograft transplantation: undersizing grafts can lead to increased rates of clinical and mechanical failure. Knee Surg Sports Traumatol Arthrosc. 2019.
Garrett JC, Steensen RN. Meniscal transplantation in the human knee: a preliminary report. Arthroscopy. 1991;7:57–62.
Garrett JC. Meniscal transplantation: a review of 43 cases with 2- to 7-year follow-up. 1993;1:164–7.
Johnson DL, Swenson TM, Livesay GA, Aizawa H, Fu FH, Harner CD. Insertion-site anatomy of the human menisci: gross, arthroscopic, and topographical anatomy as a basis for meniscal transplantation. Arthroscopy. 1995;11:386–94.
Johnson DL, Swenson TM, Harner CD. Meniscal reconstruction using allograft tissue: an arthroscopic technique. Oper Tech Sports Med. 1994;2:223–31.
Lee SM, Bin SI, Kim JM, et al. Long-term outcomes of meniscal allograft transplantation with and without extrusion: mean 12.3-year follow-up study. Am J Sports Med. 2019;47:815–21.
Pollard ME, Kang Q, Berg EE. Radiographic sizing for meniscal transplantation. Arthroscopy. 1995;11:684–7.
Donahue TL, Hull ML, Howell SM. New algorithm for selecting meniscal allografts that best match the size and shape of the damaged meniscus. J Orthop Res. 2006;24:1535–43.
Haut TL, Hull ML, Howell SM. Use of roentgenography and magnetic resonance imaging to predict meniscal geometry determined with a three-dimensional coordinate digitizing system. J Orthop Res. 2000;18:228–37.
Yoon JR, Kim TS, Lim HC, Lim HT, Yang JH. Is radiographic measurement of bony landmarks reliable for lateral meniscal sizing? Am J Sports Med. 2011;39:582–9.
Prodromos CC, Joyce BT, Keller BL, Murphy BJ, Shi K. Magnetic resonance imaging measurement of the contralateral normal meniscus is a more accurate method of determining meniscal allograft size than radiographic measurement of the recipient tibial plateau. Arthroscopy. 2007;23:1174–1179.e1171.
Shaffer B, Kennedy S, Klimkiewicz J, Yao L. Preoperative sizing of meniscal allografts in meniscus transplantation. Am J Sports Med. 2000;28:524–33.
Haen TX, Boisrenoult P, Steltzlen C, Pujol N. Meniscal sizing before allograft: comparison of three imaging techniques. Knee. 2018;25:841–8.
McConkey M, Lyon C, Bennett DL, et al. Radiographic sizing for meniscal transplantation using 3-D CT reconstruction. J Knee Surg. 2012;25:221–5.
Kaleka CC, Netto AS, Silva JC, et al. Which are the most reliable methods of predicting the meniscal size for transplantation? Am J Sports Med. 2016;44:2876–83.
Beeler S, Vlachopoulos L, Jud L, Sutter R, Furnstahl P, Fucentese SF. Contralateral MRI scan can be used reliably for three-dimensional meniscus sizing - retrospective analysis of 160 healthy menisci. Knee. 2019.
Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16:494–502.
Ambra LF, Mestriner AB, Ackermann J, Phan AT, Farr J, Gomoll AH. Bone-plug versus soft tissue fixation of medial meniscal allograft transplants: a biomechanical study. Am J Sports Med. 2019;363546519870179.
Szojka A, Lalh K, Andrews SHJ, Jomha NM, Osswald M, Adesida AB. Biomimetic 3D printed scaffolds for meniscus tissue engineering. Bioprinting. 2017;8:1–7.
Zhang ZZ, Wang SJ, Zhang JY, et al. 3D-printed poly(epsilon-caprolactone) scaffold augmented with mesenchymal stem cells for total meniscal substitution: a 12- and 24-week animal study in a rabbit model. Am J Sports Med. 2017;45:1497–511.
Paproki A, Engstrom C, Chandra SS, Neubert A, Fripp J, Crozier S. Automated segmentation and analysis of normal and osteoarthritic knee menisci from magnetic resonance images--data from the Osteoarthritis Initiative. Osteoarthritis Cartilage. 2014;22:1259–70.
Dam EB, Lillholm M, Marques J, Nielsen M. Automatic segmentation of high- and low-field knee MRIs using knee image quantification with data from the osteoarthritis initiative. J Med Imaging (Bellingham). 2015;2:024001.
Acknowledgements
None
Funding
None
Author information
Authors and Affiliations
Contributions
SB, SFF, and LV contributed the idea. SB and LJ collected and evaluated the data. MVA and PF worked the computer programming part. RS performed the radiological part. SB, LJ, MVA, RS, PF, SFF, and LV wrote the manuscript. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Kantonale Ethikkommission Zürich had given the approval for the study (BASEC-No. 2018-00856).
Consent for publication
None
Competing interests
The authors, their immediate families, and any research foundations with which they are affiliated have not received any financial payments or other benefits from any commercial entity related to the subject of this article. All authors have read and approved the final submitted manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Beeler, S., Jud, L., von Atzigen, M. et al. Three-dimensional meniscus allograft sizing—a study of 280 healthy menisci. J Orthop Surg Res 15, 74 (2020). https://doi.org/10.1186/s13018-020-01591-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-020-01591-z