Abstract
The suppressor of cytokine signaling (SOCS) box consists of the BC box and the cullin 5 (Cul5) box, which interact with Elongin BC and Cul5, respectively. SOCS box-containing proteins have ubiquitin ligase activity mediated by the formation of a complex with the scaffold protein Cul5 and the RING domain protein Rbx2, and are thereby members of the cullin RING ligase superfamily. Cul5-type ubiquitin ligases have a variety of substrates that are targeted for polyubiquitination and proteasomal degradation. Here, we review the current knowledge on the identification of Cul5 and the regulation of its expression, as well as the signaling pathways regulated by Cul5 and how viruses highjack the Cul5 system to overcome antiviral responses.
Similar content being viewed by others
Identification and regulation of cullin 5
Cullin 5 (Cul5) was originally identified as a vasopressin-activated calcium-mobilizing (VACM-1) protein, an arginine vasopressin (AVP) receptor [1]. AVP is a nonapeptide that regulates body fluid and blood pressure homeostasis. VACM-1 is recognized as Cul5 because of its homology to the Caenorhabditis elegans gene Cul5 [2, 3]. Cul5 is expressed in many cells and organs, including endothelial cells, brain, kidney collecting tubule cells, and vascular endothelial cells [2, 4–6, 7]. Cul5 inhibits cyclic AMP production, and this effect is reversed by staurosporin, a protein kinase A (PKA) inhibitor, or by mutating S730A, the PKA-dependent phosphorylation site in the Cul5 sequence in COS-1 cells [8]. The inhibitory effect of Cul5 on AVP-stimulated cAMP production is enhanced by a protein kinase C inhibitor [8]. CUL-5 expression is downregulated in 82 % (41/50) of breast tumors compared with matched normal tissues [9]. Overexpression of Cul5 in T47D breast cancer cells decreases cell growth and mitogen activated protein kinase (MAPK) phosphorylation [10], and Cul5 overexpression downregulates early growth response 1 (EGR-1) protein expression and upregulates Fas-L mRNA expression [10]. The regulation of both MAPK and EGR-1 pathways by 17β-estradiol led to the examination of estrogen-dependent T47D cell growth, which showed that Cul5 inhibits basal and 17β-estradiol-dependent cell growth and MAPK phosphorylation [11].
Resveratrol (trans-3,5,4′-trihydroxystilbene), which inhibits tumor initiation and promotion, is a natural component of the human diet, and its wide range of biological activities has been demonstrated in vivo and in vitro [12–15]. The antiproliferative effect of resveratrol is significantly enhanced by Cul5 overexpression in T47D cells [16].
The expression of Cul5 is regulated by several stimuli and pathways (Fig. 1). Resveratrol upregulates Cul5 expression and decreases T47D cell growth, suggesting that the antiproliferative effect of resveratrol is mediated by Cul5 [16]. Cul5 is a flexible scaffold protein with a preferred distribution of conformational states [17], and NEDD8 modification (neddylation) alters the conformation of Cul5 and activates it [18]. Cul5(S730A) accelerates cellular proliferation and induces angiogenic growth in rat adrenal medullary endothelial cells (RAMECs) [19]. Cul5 neddylation is increased by the S730A mutation, and activation of PKA by forskolin suppresses the neddylation of Cul5 [20]. Furthermore, PKC-induced RAMEC proliferation is enhanced by Cul5(S730A) [20]. Cul5(S730A) expression in RAMECs increases the levels of phosphorylated MAPK and the translocation of the transcription factor EGR-1, a tumor suppressor, to the nucleus; it also causes morphological alterations mediated by actin rearrangement [19]. Furthermore, Cul5(S730A) downregulates maspin, a putative tumor suppressor [21] that is essential for early embryonic development [22], although these functions are controversial [23]. These reports suggest that Cul5 plays a role in endothelial cell growth and angiogenesis by regulating MAPK phosphorylation, the nuclear localization of EGR-1, maspin expression, and actin polymerization. Nevertheless, no mutation was found at the putative phosphorylation or neddylation site of Cul5 in T47D breast cancer cells, U138MG glioma cells, ACHN renal cancer cells, and OVCAR-3 ovarian cancer cells [24]. C. elegans oocyte septum formation and egg production were absent in Cul5- or ring box protein 2 (Rbx2)-depleted Cul2 homozygotes, whereas control Cul2 homozygotes laid approximately 50 eggs [25]. Additionally, Cul5-depleted Cul2 mutants and Cul2-depleted Cul5 mutants show decreased MPK-1 activity, suggesting that oocyte maturation from pachytene exit and MPK-1 activation are redundantly controlled by the Rbx2-Cul5- and Rbx1-Cul2-based complexes [25].
C-peptide [26, 27], the product of the cleavage of proinsulin, is a peptide hormone that acts through a G protein-coupled membrane receptor [28–30]. Given that C-peptide and vasopressin share similar intracellular effects, including the activation of calcium influx and endothelial nitric oxide (NO) synthase [31–36], the effect of C-peptide on Cul5 was examined [37]. Cul5 expression was increased by C-peptide, and the induction was prevented by pertussis toxin, a specific inhibitor of G proteins [37].
Rat Cul5 mRNA is expressed in the brain and its levels increase in the rat cerebral cortex, hypothalamus, and kidney in response to 48 h of water deprivation [38, 39]. Cul5 overexpression in COS-1 cells downregulated aquaporin-1 (AQP1), and Cul5 was upregulated in rat mesenteric arteries, skeletal muscle, and the heart ventricle in response to 24 h of water deprivation [40]. Cul5 neddylation was also increased by 24 h of water deprivation, and AQP1 levels were inversely correlated with the ratio of Cul5 to neddylated Cul5 [40]. Furthermore, overexpression of Cul5 downregulated AQP2, and Cul5 was decreased in renal collecting ducts in response to water deprivation [41]. Cul5 mRNA levels were increased in the brainstem and cerebellum, and decreased in the hypothalamus of rats by hemorrhagic shock [42].
Cul5 disappears during the cell cycle S phase; it localizes to the cytosol during cell division and to the cell membrane at the completion of cytokinesis, suggesting that it plays a role in cell division [43]. Cul5 mRNA and protein levels are decreased in the rat cerebral cortex and hippocampus in response to traumatic brain injury (TBI) [44]. Another report showed a 6.5-fold upregulation of Cul5 associated with granulocytic differentiation of HL-60 cells [45].
Hepatitis B virus infection downregulates microRNA-145 (miR-145), upregulates Cul5 expression, and enhances cell proliferation [46]. miR-7, which upregulates Cul5 expression, is downregulated in hepatocellular carcinoma (HCC) tissues compared with adjacent non-tumor tissue [47]. By contrast, overexpression of miR-7 prevents colony formation and induces G1/S phase arrest, suggesting that miR-7 is a tumor suppressor in HCC [47]. miR-19a and -19b (miR-19a/b), which negatively regulate Cul5 expression, are highly expressed in human cervical cancer cells [48]. Upregulation of miR-19a/b promotes cell growth and invasion, whereas overexpression of miR-19a/b-resistant Cul5 without its 3′-UTR abolishes the effect of miR-19a/b on cell proliferation and invasion [48].
Rbx2 is polyubiquitinated by NEDD4-1, a HECT domain-containing E3 ubiquitin ligase, and targeted for proteasome-mediated degradation, suggesting that NEDD4-1 suppresses Cul5 ubiquitin ligase activity [49]. Overexpression of NEDD4-1 increases etoposide-induced apoptosis, suggesting that Rbx2 has an anti-apoptotic role [49, 50].
Cul5-containing ubiquitin ligases
CIS/SOCS family
Suppressor of cytokine signaling (SOCS) proteins (SOCS1, SOCS2, SOCS3, SOCS4, SOCS5, SOCS6, and SOCS7) and cytokine-inducible Src homology 2 (SH2) domain-containing protein (CIS, also known as CISH) interact with Cul5 through its “Cul5 box” [51–53]. The amino acid sequence LPΦP (Φ represents a hydrophobic residue) in the Cul5 box is required for specific interaction with Cul5 [51, 53, 54]. Cul5 also interacts with Rbx2, enabling SOCS box-containing proteins to form a protein complex with Cul5 and Rbx2 (Fig. 2) [51, 53, 54] (Table 1).
All CIS/SOCS family proteins have a central SH2 domain and a C-terminally located SOCS box, which consists of an Elongin C-interacting BC box and a Cul5-interacting Cul5 box with an approximately 40-amino acid motif (Fig. 3) [51–58]. CIS/SOCS family proteins bind to janus kinases (JAKs), certain cytokine receptors, or signaling molecules to suppress downstream signaling events [52, 56, 59]. A small kinase inhibitory region (KIR) of SOCS1 and SOCS3 inhibits JAKs by acting as a pseudo-substrate, thereby suppressing further signal transduction [52, 56]. By contrast, CIS/SOCS family proteins inhibit signaling by competing with downstream proteins for binding to the activated receptors, suppressing signal transduction by inducing the polyubiquitination and proteasomal degradation of target substrates [52, 56]. For example, SOCS1 polyubiquitinates JAK2, Vav, IRS1 and IRS2, the GM-CSF receptor βc subunit, Cdh1, p65, Mal, and HPV E7 [60–67].
Domain organization of SOCS box proteins. a The SOCS box consists of a BC box and a Cul5 box in the order indicated. SH2 Src homology 2 phosphotyrosine-binding domain, WD40 WD40 repeats, SPRY sp1A/ryanodine receptor domain, Ank ankyrin repeats, LRR leucine-rich repeats, GTPase GTPase domain. b Alignment of amino acid sequences of Cul5 boxes present in selected SOCS box proteins. Consensus amino acids are highlighted by bold font. The GenBank™ accession numbers of each protein are indicated. Φ hydrophobic residue
SOCS1 contains an incompletely conserved Cul5 box, and no interaction between SOCS1 and Cul5 has been detected [51]. Given that SOCS1 polyubiquitinates several substrates as described above, it is possible that the interaction of SOCS1 with these substrates recruits other ubiquitin ligase(s) that actually mediate their polyubiquitination and degradation, or that the bond between SOCS1 and the Cul5/Rbx2 complex is unstable [51]. SOCS1 and SOCS3 bind relatively weakly to Cul5, with affinities 100-fold and 10-fold lower, respectively, than those to the rest of the family [68]. This might explain why only SOCS1 and SOCS3 suppress signal transduction through both SOCS box-dependent and -independent mechanisms [68].
Knockdown of Cul5 accelerates growth factor-independent cell growth, migration, membrane dynamics, and colony dysmorphogenesis, which are all dependent on the endogenous tyrosine kinase Src [69]. Mechanistically, Cul5 and Src stimulate the degradation of the Src substrate p130Cas (Crk-associated substrate) [69]. Tyrosine phosphorylation of Cas stimulates the interaction between SOCS6 and Cas and the proteasomal degradation of Cas [69]. Cas is necessary for the transformation of Cul5 knockdown cells, and Cul5 suppresses epithelial cell transformation by regulating several pathways, including inhibition of Src–Cas-induced ruffling through SOCS6 [69].
Src is a non receptor tyrosine kinase that mediates many signaling pathways involving various soluble and adhesive signaling molecules and regulates cell proliferation, survival, differentiation, and migration [70]. Cul5 downregulates active but not inactive Src, and knockdown of Cul5 increases protein tyrosine phosphorylation, induces morphological transformation, and deregulates cell growth [71].
The mammalian cortical plate assembles from the inside outwards [72, 73]. This organization requires a signaling pathway mediated by an extracellular protein, reelin (Reln), and an intracellular molecule, disabled-1 (Dab1) [74–77]. Reln stimulates the tyrosine phosphorylation of Dab1 by the Src family tyrosine kinases (SFKs) Fyn and Src [78–82]. Tyrosine-phosphorylated Dab1 is degraded in a Cul5 and SOCS protein-dependent manner [83–85]. Functionally, knockdown of Cul5 in migrating neurons shifts their location to a more superficial position, suggesting that Cul5 is crucial for the precise location of the termination of neuronal migration [83]. Furthermore, Rbx2 knockdown resulted in a shift in neuronal positioning to a more superficial location [86]. Rbx2 conditional knockout mice show neocortical and cerebellar ectopias dependent on Dab1 [86]. Finally, SOCS7 is a Dab1 recognition protein that promotes polyubiquitination and degradation [86].
Tuberous sclerosis complex (TSC) is associated with neurodevelopmental abnormalities resulting from mutations in one of two genes, TSC1 (encoding hamartin) or TSC2 (encoding tuberin) [87]. Cul5 is upregulated at the mRNA and protein levels by increased mammalian target of rapamycin (mTOR) signaling or in the absence of Tsc2, providing potential molecular mechanisms underlying the neuronal migration deficit induced by the degradation of Dab1 in TSC pathology [88].
SPRY domain-containing SOCS box protein (SPSB/SSB) complex
The SplA/ryanodine receptor (SPRY)/B30.2 domain has a role in protein–protein interactions, although its main functions remain poorly understood [89]. The SPRY/B30.2 domain is a sequence repeat in the dual specificity kinase SplA and ryanodine receptors [89].
The four members of the SPSB family (SPSB1–SPSB4) are characterized by a C-terminal SOCS box and a central SPRY/B30.2 domain [89–92]. SPSB1, 2, and 4 polyubiquitinate inducible nitric oxide synthase (iNOS/NOS2), targeting it for proteasomal degradation [93, 94]. The activity of iNOS is approximately tenfold greater than that of NOS1 and NOS3, suggesting that iNOS is a high-output NOS compared with NOS1 and NOS3 [95]. iNOS is not detectable under normal conditions, whereas it is induced in response to cytokines, microbes, or microbial products, resulting in the sustained production of NO [95]. As a result, reactive nitrogen intermediates (such as NO, nitrite, and nitrate) and the products of the interaction of NO with reactive oxygen species (such as peroxynitrite and peroxynitrous acid) accumulate and inhibit viruses or bacteria [95–97]. SPSB2-deficient macrophages show prolonged iNOS and NO production, resulting in the enhanced killing of L. major parasites [93]. By contrast, SPSB1 and SPSB4 are major ubiquitin ligases for iNOS that prevent the overproduction of NO, which could cause cytotoxicity [94, 98, 99].
The transforming growth factor-β (TGF-β) signaling pathway is a crucial signaling pathway that requires tight regulation, and dysregulation of this pathway strongly correlates with the progression of human cancers [100, 101]. SPSB1 negatively regulates the TGF-β signaling pathway by ubiquitinating and targeting TGF-β type II receptor (TβRII) for proteasomal degradation [102]. Knockdown of SPSB1 results in the accumulation of TβRII and enhanced TGF-β signaling, migration, and invasion of tumor cells [102].
Ankyrin repeat and SOCS box (ASB) family
The ASB family is composed of 18 members from ASB1 to ASB18. Several members interact with Cul5-Rbx2 and act as ubiquitin ligase complexes [103]. ASB-Cul5 complexes can oligomerize, and Cul5 can form heterodimeric complexes with the Cul4a-DDB1 complex [104].
Although ASB1 is expressed in multiple organs, including the hematopoietic compartment, ASB1-deficient mice develop normally and exhibit no phenotypes, with the exception of diminished spermatogenesis and incomplete filling of seminiferous tubules [105].
ASB2 is induced by retinoic acid (RA) in acute promyelocytic leukemia cells, and exogenous ASB-2 in myeloid leukemia cells results in growth inhibition and chromatin condensation, which recapitulate the early steps of induced differentiation of acute promyelocytic leukemia cells [106]. ASB2 targets the actin-binding proteins filamin A and B for proteasomal degradation [107–110]. Knockdown of ASB2 in leukemia cells delays RA-induced differentiation, which suggests that ASB2 regulates hematopoietic cell differentiation by targeting filamins for degradation, thereby modulating actin remodeling [107]. ASB2 enhances the adhesion of hematopoietic cells to fibronectin, the main ligand of β1 integrins, by promoting filamin A degradation [111]. ASB2 heterodimerizes with Skp2 and forms a noncanonical Cul1- and Cul5-containing dimeric ubiquitin ligase complex that promotes the polyubiquitination and degradation of Jak3 [112, 113]. A list of candidate substrates of ASB2 was reported in a recent study [114].
Tumor necrosis factor receptor type 2 (TNF-R2) is polyubiquitinated by ASB3 and targeted for proteasomal degradation [115]. Thereby, ASB3 negatively regulates TNF-R2-mediated cellular responses initiated by TNF-α [115].
Insulin receptor substrate 4 (IRS4) is expressed predominantly in the pituitary, thymus, and brain [116]. IRS4 is an adaptor molecule involved in signal transduction by both insulin and leptin, and is widely expressed throughout the hypothalamus [117]. ASB4 colocalizes and interacts with IRS4 in hypothalamic neurons and polyubiquitinates IRS4 for degradation to decrease insulin signaling [118]. Downregulation of ASB4 in HCC cells hinders cell migration and invasion, whereas overexpression of ASB4 increases the migration rate; ASB4 is downregulated by miR-200a [119]. ASB4, which is highly differentially expressed in the vascular lineage during development [120], is an oxygen-sensitive ubiquitin ligase that is abundantly expressed in the developing placenta and is upregulated during the differentiation of embryonic stem cells into endothelial cell lineages [121]. Inhibitor of DNA binding 2 (ID2) negatively regulates vascular differentiation during development [122, 123], and ASB4 promotes the ubiquitination and proteasomal degradation of ID2 [124]. ASB4-deficient mice phenocopy human pre-eclampsia, including hypertension and proteinuria in late-stage pregnant females, indicating that ASB4 mediates vascular differentiation in the placenta through the degradation of ID2 [124].
ASB6 is expressed in 3T3-L1 adipocytes but not in fibroblasts, and may regulate the insulin signaling pathway in adipocytes by promoting the degradation of adapter protein with a pleckstrin homology and SH2 domain (APS) [125].
The crystal structure of ASB9 with or without Elongin B and C has been determined [126–128]. ASB9 alone is unstable, whereas it forms a stable complex with Elongin B and C that also binds with high affinity to the Cul5N-terminal domain (Cul5NTD) but not to Cul2NTD [129]. ASB9 polyubiquitinates and decreases the levels of creatine kinase B (CKB) and ubiquitous mitochondrial creatine kinase (uMtCK) [130–132]. CK plays a major role in cellular energy metabolism in non-muscle cells [133]. CKB is overexpressed in a number of tumors, including neuroblastoma, small cell lung carcinoma, colon and rectal adenocarcinoma, and breast and prostate carcinoma [133, 134]. Furthermore, high ASB9 mRNA expression is correlated with good prognosis, and knockdown of ASB9 increases colorectal cancer (CRC) cell invasiveness [135]. ASB9 upregulation may result in a good prognosis for CRC by promoting the degradation of CKB and uMtCK.
The Notch signaling pathway is essential for the spatio-temporal regulation of cell fate [136–138]. The single-pass transmembrane protein delta acts as a ligand for the Notch receptor. Danio rerio Asb11 (d-Asb11) regulates compartment size in the endodermal and neuronal lineages by promoting the ubiquitination and degradation of deltaA but not deltaD, leading to the activation of the canonical Notch pathway [139, 140]. Knockdown of d-Asb11 downregulates specific delta-Notch elements and their transcriptional targets, whereas these are induced when d-Asb11 is misexpressed in zebrafish embryos [139]. These data indicate that d-Asb11 regulates delta- Notch signaling for the fine-tuning of lateral inhibition gradients between deltaA and Notch [139]. Mutant zebrafish lacking the Cul5 box, which results in the inability to degrade delta, are defective in Notch signaling, as indicated by the impaired expression of Notch target genes [141].
Forced expression of d-asb11 impairs terminal differentiation and increases proliferation in the myogenic progenitor compartment [142]. By contrast, mutation of d-asb11 causes premature differentiation of muscle progenitors and delays regenerative responses in adult injured muscle, suggesting that d-asb11 is a principal regulator of embryonic as well as adult regenerative myogenesis [142]. ASB11 is an endoplasmic reticulum (ER)-associated ubiquitin ligase that promotes the ubiquitination and degradation of Ribophorin 1, an integral protein of the oligosaccharyltransferase (OST) glycosylation complex, which N-glycosylates newly synthesized proteins in the rough ER [104, 143].
WD repeat and SOCS box-containing protein 1 (WSB1)
WSB1 polyubiquitinates homeodomain-interacting protein kinase 2 (HIPK2) [144]. HIPK2 interacts with a variety of transcription factors, the p300/CBP co-activator, and the Groucho/TLE co-repressor [145–152]. Functionally, HIPK2 prevents apoptosis mediated by p53, CtBP, Axin, Brn3, Sp100, TP53INP1, and PML [153–157]. The loss of HIPK2 reduces apoptosis and increases the numbers of trigeminal ganglia, whereas overexpression of HIPK2 in the developing sensory and sympathetic neurons promotes apoptosis [153, 158]. DNA damaging agents such as adriamycin or cisplatin prevent the WSB1-mediated degradation of HIPK2, which thereby remains active and stable for the induction of apoptosis [144].
WSB1 is induced by sonic hedgehog (Shh) in developing limb buds and other embryonic structures [159]. Thyroid hormone-activating enzyme type 2 iodothyronine deiodinase (D2) is polyubiquitinated by WSB1 [160]. Ubiquitination of Shh-induced D2 by WSB1 induces parathyroid hormone-related peptide (PTHrP), thereby regulating chondrocyte differentiation [160].
Although WSB1 binds to the interleukin-21 receptor (IL-21R), WSB1 inhibits the degradation of the mature form of IL-21R [161]. Mechanistically, WSB1 associates with the intracytoplasmic region of IL-21R and facilitates the maturation of IL-21R from an N-linked glycosylated form to a fully glycosylated mature form [161].
The von Hippel-Lindau tumor suppressor pVHL is a ubiquitin ligase that targets hypoxia-inducible factor-α (HIF-α) for proteasomal degradation in normoxia [162, 163]. Dysregulation and accumulation of HIF-α upregulates downstream target gene expression and contributes to tumor progression, promoting invasion, metastasis, and angiogenesis [162, 163]. WSB1 is induced under hypoxic conditions [164] and promotes pVHL ubiquitination and proteasomal degradation, thereby stabilizing HIF-α under both normoxic and hypoxic conditions [165]. WSB1 upregulates gene expression regulated by HIF-1α and promotes cancer invasion and metastasis [165]. In a recent study, quantitative proteomic screening and functional analyses revealed that WSB1 promotes the ubiquitination and proteasomal degradation of the Rho-binding protein RhoGDI2, thereby activating Rac1 to stimulate tumor cell motility and invasion in hypoxia-driven osteosarcoma [166].
Rab40 complex
Xenopus homolog of Rab40 (XRab40) is localized at the Golgi apparatus and interacts with Elongin B/C and Cul5 [167]. Although the XRab40 complex ubiquitinates the Rap2 GTPase, it may not destabilize Rap2 [167]. The XRab40 complex regulates the membrane localization of dishevelled (Dsh), a key signaling molecule in the Wnt pathway, through Rap2 and its effector misshapen/Nck-interacting kinase (XMINK) [167]. The XRab40 complex, Rap2, and XMINK are suggested to play a crucial role in the regulation of the noncanonical Wnt pathway.
MUF1 complex
MUF1 binds the Cul5/Elongin BC complex and has ubiquitin ligase activity; however, its substrate has not been identified to date [168]. MUF1 is a ubiquitously expressed nuclear protein that, upon coexpression with RhoBTB, a Cul3-type ubiquitin ligase, is partially retained in the cytoplasm, where both proteins colocalize [169].
Elongin ABC complex
The Elongin ABC complex interacts with Cul5 and Rbx2 and polyubiquitinates the large subunit of RNA polymerase II (Rpb1) in response to UV irradiation [170].
UV irradiation leads to the phosphorylation of Rpb1 at Ser5, which increases the interaction between Elongin A and Rpb1 [170]. UV irradiation-dependent ubiquitination and proteasomal degradation of Rpb1 are significantly suppressed in Elongin A-deficient cells [170].
Virus-related Cul5-containing ubiquitin ligases
Human immunodeficiency virus-1 (HIV-1)
Apolipoprotein B editing complex 3G (CEM15/APOBEC3G)(A3G), a human cytidine deaminase, is a broad antiviral factor against human HIV-1, simian immunodeficiency virus (SIV), mouse leukemia virus, and hepatitis B virus [171–179]. A3G induces C to U mutations in the viral minus DNA strand during reverse transcription, resulting in deleterious G to A mutations in the coding strand (Fig. 4) [171, 173–175, 179–181].
Degradation of APOBEC3 by the HIV Vif protein. APOBEC3 introduces nonsense and/or missense mutations in the HIV genome, thereby showing antivirus activity. The HIV-1 Vif protein forms a complex with Cul5, the Elongin B/C heterodimer, Rbx2, E2, ubiquitin (Ub), and CBF-β. The Vif complex targets APOBEC3 for polyubiquitination and proteasomal degradation
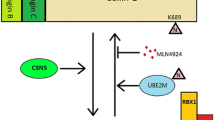
The HIV-1 virion infectivity factor (Vif) is essential for viral evasion of the host antiviral factor A3G [182, 183]. Vif interacts with Cul5, Elongins B and C, and Rbx1/Rbx2 [184–186]. This complex interacts with A3G and induces its ubiquitination and degradation (Fig. 4) [185–187]. HIV Vif can also bind APOBEC3F (A3F) and induce its polyubiquitination and degradation [188]. The SIV from rhesus macaques (SIVmac) Vif also forms a Cul5-containing ubiquitin ligase complex in human cells [186], and neddylation of Cul5 by the NEDD8-conjugating enzyme UBE2F is required for Vif-mediated degradation of A3G [189].
In the absence of the Vif protein, A3G is packaged into viral particles and functions by hypermutating viral DNA in the newly infected cell [171, 173–176, 179]. Lysine-free A3G (all lysine residues are mutated to arginine) is still degraded by the proteasome in a Vif-dependent manner [190], and polyubiquitination of Vif is critical for A3G proteasomal degradation [190].
Infection with HIV-1 causes cell cycle arrest or delay in the G2 phase, when the expression of the viral genome is optimal and long terminal repeat (LTR) is most active [191–193]. Several controversial reports suggest that viral protein R (Vpr) and/or Vif mediate cell cycle arrest. Vpr of HIV-1 alter the cell cycle by inhibiting the activation of Cdc2/Cdk1, a G2/M checkpoint regulating kinase, to prevent or delay entry into mitosis [194–196]. Vif and Vpr acting together, but not alone, cause G2 arrest [197]. However, Vif was reported to cause G2 arrest [198], and also to block Vpr-mediated G2 arrest [199]. Nevertheless, Vif-mediated G2 arrest is Cul5-dependent [200]. Vif also recruits the transcription cofactor CBF-β, which is required for Vif-mediated degradation of A3G but not A3A [201–203]. CBF-β is a subunit of a heterodimeric transcription factor without DNA-binding activity that regulates the folding and DNA-binding activity of partner RUNX family proteins, which is crucial for the development and differentiation of diverse cell types, including T lymphocytes [203–205].
Vif is phosphorylated on several serine and threonine residues, among which Ser144 plays a crucial role in regulating HIV-1 replication [206, 207]. Mutation of Ser144 to Ala suppresses Vif activity and causes >90 % inhibition of HIV-1 replication [206]. Mechanistically, phosphorylation at Ser144 negatively regulates the binding of the Vif BC box to Elongin C [208].
Vif contains a BC box and a SOCS box that are required for the interaction with ElonginB/C and Cul5, respectively [51, 209, 210]. Binding of Elongin B/C changes the conformation of Vif, facilitating its interaction with CBF-β and Cul5 [211]. Although both Rbx1 and Rbx2 can interact with Cul5, only the knockdown of Rbx2, but not that of Rbx1, impairs Vif-induced A3G degradation [212].
Susceptibility to HIV-1 and disease progression may be affected by variation in human genes [213, 214]. Cul5 is one of the genes in which signatures of selection have been reported [215]. Several single nucleotide polymorphisms (SNPs) in the CUL5 locus have been identified and shown to affect the rate of CD4+ T cell loss in patients infected with HIV-1 [216]. Cul5 haplotypes are grouped into two clusters with opposing effects, as cluster I delays and cluster II accelerates CD4+ T cell loss [216]. Reduced APOBEC3 activity is associated with the Cul5 SNP6 minor allele [217]; however, the Cul5 SNP6 has no effect on vertical transmission or progression to pediatric AIDS [218].
Epstein–Barr virus (EBV)
EBV, a human γ-herpesvirus, is associated with several B cell and epithelial cell malignancies, and there are two different infection states, latent and lytic [219]. BZLF1 (known as Zta, EB1, or ZEBRA) is a transcriptional transactivator that induces EBV early gene expression to promote an EBV lytic cycle cascade [220–223]. BZLF1 contains both a Cul2 box and a Cul5 box, thereby binding to both Cul2 and Cul5 [224]. BZLF1 polyubiquitinates and induces the degradation of p53, which inhibits apoptosis and is required for efficient viral propagation in the lytic replication stage [224, 225].
Human adenoviruses (Ad)
Human Ad are classified into six groups (A–F), and they comprise a large family of more than 50 different serotypes [226]. The human adenovirus type 5 (Ad5) early-region 4 34 kDa product from open reading frame 6 (E4orf6) contains three BC boxes [227–229]. Although Ad5 E4orf6 forms a complex containing Cul5, Elongin B, Elongin C, and Rbx1, a Cul5 box is not found in Ad5 E4orf6 (Fig. 5) [227, 229, 230]. Adenoviral early-region 1B 55 kDa protein (E1B55K) associates with E4orf6 and the complex targets substrates for proteasomal degradation [227, 228, 231]. Although efficient substrate degradation is dependent on the interaction with E1B55K in some cases, several substrates efficiently bind to E1B55K but are not degraded, whereas others are degraded without detectable interactions with E1B55K [232]. These results indicate that transient interactions with E1B55K may be sufficient for substrate degradation and that the orientation of the substrate in the ubiquitin ligase complex is probably crucial [232].
Degradation of substrate proteins by the adenoviral proteins E1B55K and E4orf6. The adenoviral protein E1B55K recognizes substrates to be polyubiquitinated, and also interacts with another adenoviral protein, E4orf6. E4orf6 further interacts with the Elongin B/C heterodimer, Cul5, and Rbx1, E2, and ubiquitin (Ub)
The E4orf6/E1B55K complex is essential for efficient viral replication, and some of its key substrates have been identified, such as p53 [233–239], meiotic recombination 11 (Mre11) [227, 240], DNA ligase IV [241, 242], integrin α3 [243], and the adeno-associated virus type 5 (AAV5) Rep52 and capsid proteins [244, 245].
The Mre11 complex, which consists of Mre11, RAD50, and Nijmegen breakage syndrome 1 (NBS1, also known as nibrin), detects DNA double strand breaks (DSBs) and induces p53-dependent apoptosis [246]. DNA ligase IV plays a pivotal role in repairing DSBs, and the mutation of this gene results in ligase IV (LIG4) syndrome, which is characterized by pronounced radiosensitivity, genome instability, malignancy, immunodeficiency, and bone marrow abnormalities [247]. The heterodimer of integrin α and β subunits functions as a transmembrane receptor that links external signals to intracellular signaling pathways. For example, integrin α3β1 binds a variety of extracellular matrix substrates, including fibronectin, collagen, vitronectin, and laminins [248]. Degradation of integrin α3 mediated by the E4orf6/E1B55K complex might be involved in cell detachment from the extracellular matrix, which may contribute to virus spread [243].
Although the human Ad5 E4orf6 complex binds Cul5, Cul2 is primarily present in the Ad12 and Ad40 E4orf6 complexes, as they contain a Cul2 box [229, 249]. The Ad16 E4orf6 complex binds Cul2 as well as Cul5 and is not able to degrade p53 and integrin α3 [229].
The anti-apoptotic protein Gam1 is an essential viral protein encoded by the avian adenovirus CELO (chicken embryo lethal orphan) [250, 251] that inhibits cellular sumoylation [252]. Gam1 contains a SOCS box-like domain and binds Cul2, Cul5, Elongin B/C, and Rbx1, targeting the SUMO E1 enzyme SAE1 for polyubiquitination and degradation [253].
LANA complex
Kaposi’s sarcoma-associated herpesvirus (KSHV)-encoded latency-associated nuclear antigen (LANA) contains a putative SOCS box and forms a complex with Elongin B/C and Cul5 [254]. This complex promotes the polyubiquitination and degradation of pVHL and p53 [254, 255]. Thus, LANA provides a favorable environment for the progression of KSHV-infected tumor cells by downregulating tumor suppressors.
Substrates of Cul5 (adaptor protein is unknown)
DEPTOR
DEPTOR binds mTOR and inhibits the mTOR complex 1 (mTORC1) and mTORC2 pathways [256]. DEPTOR accumulates upon nutrient deprivation and contributes to the induction of autophagy. In response to mitogens, DEPTOR is phosphorylated on three serine residues in a conserved degron and is recognized by F box protein βTrCP for polyubiquitination and consequent proteasomal degradation [257–259]. The Cul5/Elongin B complex also targets DEPTOR for ubiquitin-proteasomal degradation under nutrient-rich conditions, and knockdown of Cul5, but not of Cul2, results in autophagy induction [260]. Thus, Cul5 temporally controls the autophagy response.
Heat shock protein 90 (Hsp90) client proteins
Hsp90 is a molecular chaperone that facilitates the stabilization and activation of approximately 350 client proteins [261]. Pharmacologic inhibition of Hsp90 results in the Cul5 and Rbx2-dependent proteasomal degradation of client proteins including ErbB2, BRAF(V600E), AKT, CDK4, and HIF-1α, indicating the crucial role of Cul5 in the response to Hsp90 inactivation [262–266]. ErbB2 degradation mediated by Cul5 is independent of Elongin B/C function, as indicated by the fact that dominant negative Elongin C, which can bind Cul5 but not the SOCS box in the substrate receptor, has no effect on the degradation of ErbB2 [262].
TRIAD1
Two RING fingers and DRIL (double RING finger linked) 1 (TRIAD1) contains a RING-in-between-RING (RBR) domain and markedly inhibits myeloid colony formation [267]. TRIAD1-deficient mice die because of a severe multiorgan immune response [268]. Binding of neddylated Cul5 and Rbx2 to TRIAD1 enhances TRIAD1 ubiquitin ligase activity [269].
Conclusions
Cul5-containing ubiquitin ligases regulate a variety of signaling pathways by targeting particular substrates for proteasomal degradation or competing for protein–protein interactions. However, many Cul5-containing ubiquitin ligases remain to be studied, and a complete list of substrates or binding proteins of Cul5 is not available. Considering that some viruses hijack Cul5 to degrade antiviral proteins, it might be better to study the function of Cul5 during virus infection. Certain viruses target Elongin C-interacting Cul5 (and in some cases Cul2) for hijacking, although the cause remains undetermined. Studies focusing on Elongin C might shed light on the physiological functions of Cul5.
Abbreviations
- Ad:
-
adenoviruses
- APS:
-
adapter protein with a pleckstrin homology and SH2 domain
- AQP:
-
aquaporin
- ASB:
-
ankyrin repeat and SOCS box
- AVP:
-
arginine vasopressin
- A3F:
-
apolipoprotein B editing complex 3F
- A3G:
-
apolipoprotein B editing complex 3G
- Cas:
-
Crk-associated substrate
- CIS:
-
cytokine-inducible Src homology 2 (SH2) domain-containing protein
- CKB:
-
creatine kinase B
- Dab1:
-
disabled-1
- Dsh:
-
dishevelled
- D2:
-
thyroid hormone-activating enzyme type 2 iodothyronine deiodinase
- EBV:
-
Epstein–Barr virus
- EGR-1:
-
early growth response 1
- E1B55K:
-
early-region 1B 55 kDa protein
- E4orf6:
-
early-region 4 34 kDa product from open reading frame 6
- HIF:
-
hypoxia-inducible factor
- HIPK2:
-
homeodomain-interacting protein kinase 2
- HIV-1:
-
human immunodeficiency virus-1
- ID2:
-
inhibitor of DNA binding 2
- IL:
-
interleukin
- iNOS:
-
inducible nitric oxide synthase
- IRS:
-
insulin receptor substrate
- JAKs:
-
janus kinases
- KIR:
-
kinase inhibitory region
- LANA:
-
latency-associated nuclear antigen
- MAPK:
-
mitogen activated protein kinase
- miR:
-
microRNA
- NO:
-
nitric oxide
- PKA:
-
protein kinase A
- pVHL:
-
von Hippel-Lindau tumor suppressor
- RA:
-
retinoic acid
- RAMECs:
-
rat adrenal medullary endothelial cells
- Reln:
-
reelin
- SFKs:
-
Src family tyrosine kinases
- Shh:
-
sonic hedgehog
- SH2:
-
Src homology 2
- SIV:
-
simian immunodeficiency virus
- SIVmac:
-
SIV from rhesus macaques
- SOCS:
-
suppressor of cytokine signaling
- SPRY:
-
SplA/ryanodine receptor
- SPSB:
-
SPRY domain-containing SOCS box
- TBI:
-
traumatic brain injury
- TGF-β:
-
transforming growth factor-β
- TNF:
-
tumor necrosis factor
- TRIAD1:
-
two RING fingers and DRIL (double RING finger linked) 1
- TSC:
-
tuberous sclerosis complex
- VACM:
-
vasopressin-activated calcium-mobilizing
- Vif:
-
virion infectivity factor
- Vpr:
-
viral protein R
- WSB1:
-
WD repeat and SOCS box-containing protein 1
- XMINK:
-
Xenopus misshapen/Nck-interacting kinase
References
Burnatowskahledin MA, Spielman WS, Smith WL, Shi P, Meyer JM, Dewitt DL. Expression cloning of an Avp-activated, calcium-mobilizing receptor from rabbit kidney medulla. Am J Physiol Renal. 1995;268:F1198–210.
Byrd PJ, Stankovic T, McConville CM, Smith AD, Cooper PR, Taylor AMR. Identification and analysis of expression of human VACM-1, a cullin gene family member located on chromosome 11q22-23. Genome Res. 1997;7:71–5.
Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C-elegans and identifies a novel gene family. Cell. 1996;85:829–39.
Burnatowska-Hledin MA, Barney CC. New insights into the mechanism for VACM-1/cul5 expression in vascular tissue in vivo. Int Rev Cell Mol Biol. 2014;313:79–101.
Ceremuga TE, Yao XL, McCabe JT. Vasopressin-activated calcium, mobilizing (VACM-1) receptor mRNA is present in peripheral organs and the central nervous system of the laboratory rat. Endocr Res. 2001;27:433–45.
Hurbin A, Orcel H, Ferraz C, Moos FC, Rabie A. Expression of the genes encoding the vasopressin-activated calcium-mobilizing receptor and the dual angiotensin II/vasopressin receptor in the rat central nervous system. J Neuroendocrinol. 2000;12:677–84.
Burnatowska-Hledin M, Lazdins IB, Listenberger L, Zhao P, Sharangpani AVF, Card B. VACM-1 receptor is specifically expressed in rabbit vascular endothelium and renal collecting tubule. Am J Physiol Renal Physiol. 1999;276:F199–209.
Burnatowska-Hledin M, Zhao P, Capps B, Poel A, Parmelee K, Mungall C, Sharangpani A, Listenberger L. VACM-1, a cullin gene family member, regulates cellular signaling. Am J Physiol Cell Ph. 2000;279:C266–73.
Fay MJ, Longo KA, Karathanasis GA, Shope DM, Mandernach CJ, Leong JR, Hicks A, Pherson K, Husain A. Analysis of CUL-5 expression in breast epithelial cells, breast cancer cell lines, normal tissues and tumor tissues. Mol Cancer. 2003;2:40.
Burnatowska-Hledin MA, Kossoris JB, Van Dort CJ, Shearer RL, Zhao P, Murrey DA, Abbott JL, Kan CE, Barney CC. T47D breast cancer cell growth is inhibited by expression of VACM-1, a cul-5 gene. Biochem Bioph Res Co. 2004;319:817–25.
Johnson AE, Le IP, Buchwalter A, Burnatowska-Hiedin MA. Estrogen-dependent growth and estrogen receptor (ER)-alpha concentration in T47D breast cancer cells are inhibited by VACM-1, a cul 5 gene. Mol Cell Biochem. 2007;301:13–20.
Brownson DM, Azios NG, Fuqua BK, Dharmawardhane SF, Mabry TJ. Flavonoid effects relevant to cancer. J Nutr. 2002;132:3482s–9s.
Heiss EH, Schilder YDC, Dirsch VM. Chronic treatment with resveratrol induces redox stress- and ataxia telangiectasia-mutated (ATM)-dependent senescence in p53-positive cancer cells. J Biol Chem. 2007;282:26759–66.
Kotha A, Sekharam M, Cilenti L, Siddiquee K, Khaled A, Zervos AS, Carter B, Turkson J, Jove R. Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein. Mol Cancer Ther. 2006;5:621–9.
Pervaiz S. Resveratrol: from grapevines to mammalian biology. Faseb J. 2003;17:1975–85.
Lubbers J, Lewis S, Harper E, Hledin MP, Marquez GA, Johnson AE, Graves DR, Burnatowska-Hledin MA. Resveratrol enhances anti-proliferative effect of VACM-1/cul5 in T47D cancer cells. Cell Biol Toxicol. 2011;27:95–105.
Liu J, Nussinov R. Flexible cullins in cullin-RING E3 ligases allosterically regulate ubiquitination. J Biol Chem. 2011;286:40934–42.
Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006.
Buchwalter A, Van Dort C, Schultz S, Smith R, Le IP, Abbott JL, Oosterhouse E, Johnson AE, Hansen-Smith F, Burnatowska-Hledin M. Expression of VACM-1/cul5 mutant in endothelial cells induces MAPK phosphorylation and maspin degradation and converts cells to the angiogenic phenotype. Microvasc Res. 2008;75:155–68.
Bradley SE, Johnson AE, Le IP, Oosterhouse E, Hledin MP, Marquez GA, Burnatowska-Hledin M. Phosphorylation of VACM-1/Cul5 by protein kinase A regulates its neddylation and antiproliferative effect. J Biol Chem. 2010;285:4883–95.
Zou ZQ, Anisowicz A, Hendrix MJC, Thor A, Neveu M, Sheng SJ, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial-cells. Science. 1994;263:526–9.
Gao F, Shi HY, Daughty C, Cella N, Zhang M. Maspin plays an essential role in early embryonic development. Development. 2004;131:1479–89.
Teoh SS, Vieusseux J, Prakash M, Berkowicz S, Luu J, Bird CH, Law RH, Rosado C, Price JT, Whisstock JC, et al. Maspin is not required for embryonic development or tumour suppression. Nat Commun. 2014;5:3164.
Lewis SP, Willis AN, Johnson AE, Resau J, Burnatowska-Hledin MA. Mutational analysis of VACM-1/cul5 exons in cancer cell lines. Apmis. 2011;119:421–30.
Sasagawa Y, Sato S, Ogura T, Higashitani A. C-elegans RBX-2-CUL-5- and RBX-1-CUL-2-based complexes are redundant for oogenesis and activation of the MAP kinase MPK-1. FEBS Lett. 2007;581:145–50.
Rubenste Ah, Clark JL, Malani F, Steiner DF. Secretion of proinsulin C-peptide by pancreatic beta cells and its circulation in blood. Nature. 1969;224:697.
Steiner DF, Cunningh D, Spigelma L, Aten B. Insulin biosynthesis—evidence for a precursor. Science. 1967;157:697.
Johansson J, Ekberg K, Shafqat J, Henriksson M, Chibalin A, Wahren J, Jornvall H. Molecular effects of proinsulin C-peptide. Biochem Bioph Res Co. 2002;295:1035–40.
Rigler R, Pramanik A, Jonasson P, Kratz G, Jansson OT, Nygren PA, Stahl S, Ekberg K, Johansson BL, Uhlen S, et al. Specific binding of proinsulin C-peptide to human cell membranes. P Natl Acad Sci USA. 1999;96:13318–23.
Wahren J, Ekberg K, Johansson J, Henriksson M, Pramanik A, Johansson BL, Rigler R, Jornvall H. Role of C-peptide in human physiology. Am J Physiol Endoc M. 2000;278:E759–68.
BlotChabaud M, Coutry N, Laplace M, Bonvalet JP, Farman N. Role of protein phosphatase in the regulation of Na + –K + –ATPase by vasopressin in the cortical collecting duct. J Membrane Biol. 1996;153:233–9.
Feraille E, Mordasini D, Gonin S, Deschenes G, Vinciguerra M, Doucet A, Vandewalle A, Summa V, Verrey F, Martin PY. Mechanism of control of Na, K-ATPase in principal cells of the mammalian collecting duct. Ann Ny Acad Sci. 2003;986:570–8.
Kitamura T, Kimura K, Makondo K, Furuya DT, Suzuki M, Yoshida T, Saito M. Proinsulin C-peptide increases nitric oxide production by enhancing mitogen-activated protein-kinase-dependent transcription of endothelial nitric oxide synthase in aortic endothelial cells of Wistar rats. Diabetologia. 2003;46:1698–705.
Martin PY, Bianchi M, Roger F, Niksic L, Feraille E. Arginine vasopressin modulates expression of neuronal NOS in rat renal medulla. Am J Physiol Renal Physiol. 2002;283:F559–68.
Shafqat J, Juntti-Berggren L, Zhong Z, Ekberg K, Kohler M, Berggren PO, Johansson J, Wahren J, Jornvall H. Proinsulin C-peptide and its analogues induce intracellular Ca2+ increases in human renal tubular cells. Cell Mol Life Sci. 2002;59:1185–9.
Wallerath T, Kunt T, Forst T, Closs EI, Lehmann R, Flohr T, Gabriel M, Schafer D, Gopfert A, Pfutzner A, et al. Stimulation of endothelial nitric oxide synthase by proinsulin C-peptide. Nitric Oxide Biol Ch. 2003;9:95–102.
Maestroni A, Ruggieri D, Dell’Antonio G, Luzi L, Zerbini G. C-peptide increases the expression of vasopressin-activated calcium-mobilizing receptor gene through a G protein-dependent pathway. Eur J Endocrinol. 2005;152:135–41.
Ceremuga TE, Yao XL, McCabe JT. Cullin-5 is ubiquitous in the rat brain. Neurosci Lett. 2003;345:121–5.
Ceremuga TE, Yao XL, Xia Y, Mukherjee D, McCabe JT. Osmotic stress increases cullin-5 (cul-5) mRNA in the rat cerebral cortex, hypothalamus and kidney. Neurosci Res. 2003;45:305–11.
Johnson AE, Le IP, Andresen BT, Stodola J, Dewey GL, Dean SB, Resau J, Haak P, Ruch T, Sartor A, et al. VACM-1/cul5 expression in vascular tissue in vivo is induced by water deprivation and its expression in vitro regulates aquaporin-1 concentrations. Cell Tissue Res. 2012;349:527–39.
Le IP, Schultz S, Andresen BT, Dewey GL, Zhao P, Listenberger L, Deen PM, Buchwalter A, Barney CC, Burnatowska-Hledin MA. Aquaporin-2 Levels in vitro and in vivo are Regulated by VACM-1, a Cul 5 Gene. Cell Physiol Biochem. 2012;30:1148–58.
Ceremuga TE, Yao XL, Alam HB, McCabe JT. Alterations of cullin-5 mRNA levels in the rat central nervous system following hemorrhagic shock. Neurol Res. 2003;25:211–6.
Burnatowska-Hledin M, Zeneberg A, Roulo A, Grobe J, Zhao P, Lelkes PI, Clare P, Barney C. Expression of VACM-1 protein in cultured rat adrenal endothelial cells is linked to the cell cycle. Endothelium. 2001;8:49.
Yao XL, Liu J, Lee E, Ling GSF, McCabe JT. Cullin 5 gene expression in the rat cerebral cortex and hippocampus following traumatic brain injury (TBI). Neurosci Lett. 2006;409:65–9.
Baxter SS, Carlson LA, Mayer AMS, Hall ML, Fay MJ. Granulocytic differentiation of HL-60 promyelocytic leukemia cells is associated with increased expression of Cul5. In Vitro Cell Dev Anim. 2009;45:264–74.
Gao F, Sun XY, Wang LK, Tang SX, Yan CQ. Downregulation of MicroRNA-145 caused by Hepatitis B virus X protein promotes expression of CUL5 and contributes to pathogenesis of Hepatitis B virus-associated hepatocellular carcinoma. Cell Physiol Biochem. 2015;37:1547–59.
Ma CQ, Qi Y, Shao LP, Liu M, Li X, Tang H. Downregulation of miR-7 upregulates cullin 5 (CUL5) to facilitate G1/S transition in human hepatocellular carcinoma cells. IUBMB Life. 2013;65:1026–34.
Xu XM, Wang XB, Chen MM, Liu T, Li YX, Jia WH, Liu M, Li X, Tang H. MicroRNA-19a and -19b regulate cervical carcinoma cell proliferation and invasion by targeting CUL5. Cancer Lett. 2012;322:148–58.
Zhou WH, Xu J, Zhao YC, Sun Y. SAG/RBX2 is a novel substrate of NEDD4-1 E3 ubiquitin ligase and mediates NEDD4-1 induced chemosensitization. Oncotarget. 2014;5:6746–55.
Duan HJ, Wang YL, Aviram M, Swaroop M, Loo JA, Bian JH, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–55.
Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–65.
Kazi JU, Kabir NN, Flores-Morales A, Ronnstrand L. SOCS proteins in regulation of receptor tyrosine kinase signaling. Cell Mol Life Sci. 2014;71:3297–310.
Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–13.
Okumura F, Matsuzaki M, Nakatsukasa K, Kamura T. The role of elongin BC-containing ubiquitin ligases. Front Oncol. 2012;2:10.
Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, et al. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–4.
Linossi EM, Nicholson SE. Kinase inhibition, competitive binding and proteasomal degradation: resolving the molecular function of the suppressor of cytokine signaling (SOCS) proteins. Immunol Rev. 2015;266:123–33.
Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, et al. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–9.
Starr R, Willson TA, Viney EM, Murray LJL, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–21.
Cooper JA, Kaneko T, Li SSC. Cell regulation by phosphotyrosine-targeted ubiquitin ligases. Mol Cell Biol. 2015;35:1886–97.
Bunda S, Kommaraju K, Heir P, Ohh M. SOCS-1 mediates ubiquitylation and degradation of GM-CSF receptor. PLoS One. 2013;8:e76370.
De Sepulveda P, Ilangumaran S, Rottapel R. Suppressor of cytokine signaling-1 inhibits VAV function through protein degradation. J Biol Chem. 2000;275:14005–8.
Kamio M, Yoshida T, Ogata H, Douchi T, Nagata Y, Inoue M, Hasegawa M, Yonemitsu Y, Yoshimura A. SOC1 inhibits HPV-E7-mediated transformation by inducing degradation of E7 protein. Oncogene. 2004;23:3107–15.
Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, et al. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem. 2001;276:12530–8.
Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O’Neill LAJ, Hertzog PJ. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. 2006;7:148–55.
Parrillas V, Martinez-Munoz L, Holgado BL, Kumar A, Cascio G, Lucas P, Rodriguez-Frade JM, Malumbres M, Carrera AC, van Wely KHM, et al. Suppressor of cytokine signaling 1 blocks mitosis in human melanoma cells. Cell Mol Life Sci. 2013;70:545–58.
Rui LY, Yuan MS, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem. 2002;277:42394–8.
Strebovsky J, Walker P, Lang R, Dalpke AH. Suppressor of cytokine signaling 1 (SOCS1) limits NF kappa B signaling by decreasing p65 stability within the cell nucleus. Faseb J. 2011;25:863–74.
Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS box encodes a hierarchy of affinities for cullin 5: implications for ubiquitin ligase formation and cytokine signalling suppression. J Mol Biol. 2009;387:162–74.
Teckchandani A, Laszlo GS, Simo S, Shah K, Pilling C, Strait AA, Cooper JA. Cullin 5 destabilizes Cas to inhibit Src-dependent cell transformation. J Cell Sci. 2014;127:509.
Liu WS, Kovacevic Z, Peng ZH, Jin RS, Wang PXZ, Yue F, Zheng MH, Huang MLH, Jansson PJ, Richardson V, et al. The molecular effect of metastasis suppressors on Src signaling and tumorigenesis: new therapeutic targets. Oncotarget. 2015;6:35522–41.
Laszlo GS, Cooper JA. Restriction of Src activity by cullin-5. Curr Biol. 2009;19:157–62.
Kawauchi T. Cellullar insights into cerebral cortical development: focusing on the locomotion mode of neuronal migration. Front Cell Neurosci. 2015;9.
Kriegstein AR, Noctor SC. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 2004;27:392–9.
Bielas S, Higginbotham H, Koizumi H, Tanaka T, Gleeson JG. Cortical neuronal migration mutants suggest separate but intersecting pathways. Annu Rev Cell Dev Bi. 2004;20:593–618.
Frotscher M. Role for reelin in stabilizing cortical architecture. Trends Neurosci. 2010;33:407–14.
Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci. 2001;24:1005–39.
Tissir F, Goffinet AM. Reelin and brain development. Nat Rev Neurosci. 2003;4:496–505.
Howell BW, Hawkes R, Soriano P, Cooper JA. Neuronal position in the developing brain is regulated by mouse disabled-1. Nature. 1997;389:733–7.
Kuo G, Arnaud L, Kronstad-O’Brien P, Cooper JA. Absence of Fyn and Src causes a reeler-like phenotype. J Neurosci. 2005;25:8578–86.
Sekine K, Kubo K, Nakajima K. How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex? Neurosci Res. 2014;86:50–8.
Sheldon M, Rice DS, Darcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–3.
Ware ML, Fox JW, Gonzalez JL, Davis NM, deRouvroit CL, Russo CJ, Chua SC, Goffinet AM, Walsh CA. Aberrant splicing of a mouse disabled homolog, mdab1, in the scrambler mouse. Neuron. 1997;19:239–49.
Feng L, Allen NS, Simo S, Cooper JA. Cullin 5 regulates Dab1 protein levels and neuron positioning during cortical development. Genes Dev. 2007;21:2717–30.
Kerjan G, Gleeson JG. A missed exit: reelin sets in motion Dab1 polyubiquitination to put the break on neuronal migration. Genes Dev. 2007;21:2850–4.
Simo S, Jossin Y, Cooper JA. Cullin 5 regulates cortical layering by modulating the speed and duration of Dab1-dependent neuronal migration. J Neurosci. 2010;30:5668–76.
Simo S, Cooper JA. Rbx2 regulates neuronal migration through different cullin 5-RING ligase adaptors. Dev Cell. 2013;27:399–411.
Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. New Engl J Med. 2006;355:1345–56.
Moon UY, Park JY, Park R, Cho JY, Hughes LJ, McKenna J, Goetzl L, Cho SH, Crino PB, Gambello MJ, et al. Impaired reelin-Dab1 signaling contributes to neuronal migration deficits of tuberous sclerosis complex. Cell Rep. 2015;12:965–78.
Perfetto L, Gherardini PF, Davey NE, Diella F, Helmer-Citterich M, Cesareni G. Exploring the diversity of SPRY/B30.2-mediated interactions. Trends Biochem Sci. 2013;38:38–46.
Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. P Natl Acad Sci USA. 1998;95:114–9.
Masters SL, Yao SG, Willson TA, Zhang JG, Palmer KR, Smith BJ, Babon JJ, Nicola NA, Norton RS, Nicholson SE. The SPRY domain of SSB-2 adopts a novel fold that presents conserved Par-4-binding residues. Nat Struct Mol Biol. 2006;13:77–84.
Wang DK, Li ZB, Messing EM, Wu G. The SPRY domain-containing SOCS box protein 1 (SSB-1) interacts with MET and enhances the hepatocyte growth factor-induced Erk-Elk-1-serum response element pathway. J Biol Chem. 2005;280:16393–401.
Kuang ZH, Lewis RS, Curtis JM, Zhan YF, Saunders BM, Babon JJ, Kolesnik TB, Low A, Masters SL, Willson TA, et al. The SPRY domain-containing SOCS box protein SPSB2 targets iNOS for proteasomal degradation. J Cell Biol. 2010;190:129–41.
Nishiya T, Matsumoto K, Maekawa S, Kajita E, Horinouchi T, Fujimuro M, Ogasawara K, Uehara T, Miwa S. Regulation of inducible nitric-oxide synthase by the SPRY domain- and SOCS box-containing proteins. J Biol Chem. 2011;286:9009–19.
Lowenstein CJ, Padalko E. Inos (Nos2) at a Glance. J Cell Sci. 2004;117:2865–7.
Fang FC. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Invest. 1997;99:2818–25.
Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. P Natl Acad Sci USA. 2000;97:8841–8.
Lewis RS, Kolesnik TB, Kuang ZH, D’Cruz AA, Blewitt ME, Masters SL, Low A, Willson T, Norton RS, Nicholson SE. TLR regulation of SPSB1 controls inducible nitric oxide synthase induction. J Immunol. 2011;187:3798–805.
Matsumoto K, Nishiya T, Maekawa S, Horinouchi T, Ogasawara K, Uehara T, Miwa S. The ECS(SPSB) E3 ubiquitin ligase is the master regulator of the lifetime of inducible nitric-oxide synthase. Biochem Bioph Res Co. 2011;409:46–51.
Nalluri SM, O’Connor JW, Gomez EW. Cytoskeletal signaling in TGFbeta-induced epithelial-mesenchymal transition. Cytoskeleton. 2015;72:557–69.
Young JC, Wakitani S, Loveland KL. TGF-beta superfamily signaling in testis formation and early male germline development. Semin Cell Dev Biol. 2015;45:94–103.
Liu S, Nheu T, Luwor R, Nicholson SE, Zhu HJ. SPSB1, a novel negative regulator of the transforming growth Factor-beta signaling pathway targeting the type II receptor. J Biol Chem. 2015;290:17894–908.
Kohroki J, Nishiyama T, Nakamura T, Masuho Y. ASB proteins interact with cullin5 and Rbx2 to form E3 ubiquitin ligase complexes. FEBS Lett. 2005;579:6796–802.
Andresen CA, Smedegaard S, Sylvestersen KB, Svensson C, Iglesias-Gato D, Cazzamali G, Nielsen TK, Nielsen ML, Flores-Morales A. Protein interaction screening for the ankyrin repeats and suppressor of cytokine signaling (SOCS) Box (ASB) family identify Asb11 as a novel endoplasmic reticulum resident ubiquitin ligase. J Biol Chem. 2014;289:2043–54.
Kile BT, Metcalf D, Mifsud S, DiRago L, Nicola NA, Hilton DJ, Alexander WS. Functional analysis of Asb-1 using genetic modification in mice. Mol Cell Biol. 2001;21:6189–97.
Guibal FC, Moog-Lutz C, Smolewski P, Di Gioia Y, Darzynkiewicz Z, Lutz PG, Cayre YE. ASB-2 inhibits growth and promotes commitment in myeloid leukemia cells. J Biol Chem. 2002;277:218–24.
Heuze ML, Lamsoul I, Baldassarre M, Lad Y, Leveque S, Razinia Z, Moog-Lutz C, Calderwood DA, Lutz PG. ASB2 targets filamins A and B to proteasomal degradation. Blood. 2008;112:5130–40.
Lamsoul I, Erard M, van der Ven PFM, Lutz PG. Filamins but not janus kinases are substrates of the ASB2 alpha cullin-ring E3 ubiquitin ligase in hematopoietic cells. PLoS One. 2012;7:e43798.
Razinia Z, Baldassarre M, Cantelli G, Calderwood DA. ASB2alpha, an E3 ubiquitin ligase specificity subunit, regulates cell spreading and triggers proteasomal degradation of filamins by targeting the filamin calponin homology 1 domain. J Biol Chem. 2013;288:32093–105.
Zakaria R, Lamsoul I, Uttenweiler-Joseph S, Erard M, Monsarrat B, Burlet-Schiltz O, Moog-Lutz C, Lutz PG. Phosphorylation of serine 323 of ASB2 alpha is pivotal for the targeting of filamin A to degradation. Cell Signal. 2013;25:2823–30.
Lamsoul I, Burande CF, Razinia Z, Houles TC, Menoret D, Baldassarre M, Erard M, Moog-Lutz C, Calderwood DA, Lutz PG. Functional and structural insights into ASB2 alpha, a novel regulator of integrin-dependent adhesion of hematopoietic cells. J Biol Chem. 2011;286:30571–81.
Nie L, Zhao Y, Wu W, Yang YZ, Wang HC, Sun XH. Notch-induced Asb2 expression promotes protein ubiquitination by forming non-canonical E3 ligase complexes. Cell Res. 2011;21:754–69.
Wu W, Sun XH. A mechanism underlying NOTCH-induced and ubiquitin-mediated JAK3 degradation. J Biol Chem. 2011;286:41153–62.
Spinner CA, Uttenweiler-Joseph S, Metais A, Stella A, Burlet-Schiltz O, Moog-Lutz C, Lamsoul I, Lutz PG. Substrates of the ASB2 alpha E3 ubiquitin ligase in dendritic cells. Sci Rep Uk. 2015;5.
Chung AS, Guan YJ, Yuan ZL, Albina JE, Chin YE. Ankyrin repeat and SOCS box 3 (ASB3) mediates ubiquitination and degradation of tumor necrosis factor receptor II. Mol Cell Biol. 2005;25:4716–26.
Fantin VR, Lavan BE, Wang Q, Jenkins NA, Gilbert DJ, Copeland NG, Keller SR, Lienhard GE. Cloning, tissue expression, and chromosomal location of the mouse insulin receptor substrate 4 gene. Endocrinology. 1999;140:1329–37.
Numan S, Russell DS. Discrete expression of insulin receptor substrate-4 mRNA in adult rat brain. Mol Brain Res. 1999;72:97–102.
Li JY, Chai B, Zhang W, Wu X, Zhang C, Fritze D, Xia Z, Patterson C, Mulholland MW. Ankyrin repeat and SOCS box containing protein 4 (Asb-4) colocalizes with insulin receptor substrate 4 (IRS4) in the hypothalamic neurons and mediates IRS4 degradation. BMC Neurosci. 2011;12:95.
Au V, Tsang FH, Man K, Fan ST, Poon RTP, Lee NP. Expression of ankyrin repeat and SOCS box containing 4 (ASB4) confers migration and invasion properties of hepatocellular carcinoma cells. Biosci Trends. 2014;8:101–10.
Wang H, Charles PC, Wu YX, Ren RQ, Pi XC, Moser M, Barshishat-Kupper M, Rubin JS, Perou C, Bautch V, et al. Gene expression profile signatures indicate a role for Wnt signaling in endothelial commitment from embryonic stem cells. Circ Res. 2006;98:1331–9.
Ferguson JE, Wu Y, Smith K, Charles P, Powers K, Wang H, Patterson C. ASB4 is a hydroxylation substrate of FIH and promotes vascular differentiation via an oxygen-dependent mechanism. Mol Cell Biol. 2007;27:6407–19.
Janatpour MJ, McMaster MT, Genbacev O, Zhou Y, Dong JY, Cross JC, Israel MA, Fisher SJ. Id-2 regulates critical aspects of human cytotrophoblast differentiation, invasion and migration. Development. 2000;127:549–58.
Lasorella A, Rothschild G, Yokota Y, Russell RG, Iavarone A. Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol Cell Biol. 2005;25:3563–74.
Townley-Tilson WHD, Wu YX, Ferguson JE, Patterson C. The ubiquitin ligase ASB4 promotes trophoblast differentiation through the degradation of ID2. PLoS One. 2014;9:e89451.
Wilcox A, Katsanakis KD, Bheda F, Pillay TS. Asb6, an adipocyte-specific ankyrin and SOCS box protein, interacts with APS to enable recruitment of elongins B and C to the insulin receptor signaling complex. J Biol Chem. 2004;279:38881–8.
Fei XW, Gu X, Fan SL, Yang ZX, Li F, Zhang C, Gong WM, Mao YM, Ji CN. Crystal structure of human ASB9-2 and substrate-recognition of CKB. Protein J. 2012;31:275–84.
Fei XW, Zhang Y, Gu X, Qiu R, Mao YM, Ji CN. Crystallization and preliminary X-Ray analysis of the splice variant of human ankyrin repeat and suppressor of cytokine signaling box protein 9 (hASB9-2). Protein Peptide Lett. 2009;16:333–5.
Muniz JR, Guo K, Kershaw NJ, Ayinampudi V, von Delft F, Babon JJ, Bullock AN. Molecular architecture of the ankyrin SOCS box family of Cul5-dependent E3 ubiquitin ligases. J Mol Biol. 2013;425:3166–77.
Thomas JC, Matak-Vinkovic D, Van Molle I, Ciulli A. Multimeric complexes among ankyrin-repeat and SOCS-box protein 9 (ASB9), ElonginBC, and cullin 5: insights into the structure and assembly of ECS-type cullin-RING E3 ubiquitin ligases. Biochem Us. 2013;52:5236–46.
Balasubramaniam D, Schiffer J, Parnell J, Mir SP, Amaro RE, Komives EA. How the ankyrin and SOCS box protein, ASB9, binds to creatine kinase. Biochem Us. 2015;54:1673–80.
Debrincat MA, Zhang JG, Willson TA, Silke J, Connolly LM, Simpson RJ, Alexander WS, Nicola NA, Kile BT, Hilton DJ. Ankyrin repeat and suppressors of cytokine signaling box protein Asb-9 targets creatine kinase B for degradation. J Biol Chem. 2007;282:4728–37.
Kwon S, Kim D, Rhee JW, Park JA, Kim DW, Kim DS, Lee Y, Kwon HJ. ASB9 interacts with ubiquitous mitochondrial creatine kinase and inhibits mitochondrial function. Bmc Biol. 2010;8:1.
Wyss M, Kaddurah-Daouk R. Creatine and creatinine metabolism. Physiol Rev. 2000;80:1107–213.
Wallimann T, Hemmer W. Creatine kinase in non-muscle tissues and cells. Mol Cell Biochem. 1994;133–134:193–220.
Tokuoka M, Miyoshi N, Hitora T, Mimori K, Tanaka F, Shibata K, Ishii H, Sekimoto M, Doki Y, Mori M. Clinical significance of ASB9 in human colorectal cancer. Int J Oncol. 2010;37:1105–11.
Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–73.
Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102.
Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–65.
Diks SH, Bink RJ, van de Water S, Joore J, van Rooijen C, Verbeek FJ, den Hertog J, Peppelenbosch MP, Zivkovic D. The novel gene asb11: a regulator of the size of the neural progenitor compartment. J Cell Biol. 2006;174:581–92.
Diks SH, da Silva MAS, Hillebrands JL, Bink RJ, Versteeg HH, van Rooijen C, Brouwers A, Chitnis AB, Peppelenbosch MP, Zivkovic D. d-Asb11 is an essential mediator of canonical delta-notch signalling. Nat Cell Biol. 2008;10:1190–8.
da Silva MAS, Tee JM, Paridaen J, Brouwers A, Runtuwene V, Zivkovic D, Diks SH, Guardavaccaro D, Peppelenbosch MP. Essential role for the d-Asb11 cul5 box domain for proper notch signaling and neural cell fate decisions in vivo. PLoS One 2010;5.
Tee JM, da Silva MAS, Rygiel AM, Muncan V, Bink R, van den Brink GR, van Tijn P, Zivkovic D, Kodach LL, Guardavaccaro D, et al. asb11 is a regulator of embryonic and adult regenerative myogenesis. Stem Cells Dev. 2012;21:3091–103.
Kelleher DJ, Kreibich G, Gilmore R. Oligosaccharyltransferase activity is associated with a protein complex composed of ribophorin-I and ribophorin-Ii and a 48kd protein. Cell. 1992;69:55–65.
Choi DW, Seo YM, Kim EA, Sung KS, Ahn JW, Park SJ, Lee SR, Choi CY. Ubiquitination and degradation of homeodomain-interacting protein kinase 2 by WD40 repeat/SOCS box protein WSB-1. J Biol Chem. 2008;283:4682–9.
Aikawa Y, Nguyen LA, Isono K, Takakura N, Tagata Y, Schmitz ML, Koseki H, Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1-and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–65.
Choi CY, Kim YH, Kim YO, Park SJ, Kim EA, Riemenschneider W, Gajewski K, Schulz RA, Kim Y. Phosphorylation by the DHIPK2 protein kinase modulates the corepressor activity of groucho. J Biol Chem. 2005;280:21427–36.
D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–9.
Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1–10.
Kim EA, Noh YT, Ryu MJ, Kim HT, Lee SE, Kim CH, Lee C, Kim YH, Choi CY. Phosphorylation and transactivation of Pax6 by homeodomain-interacting protein kinase 2. J Biol Chem. 2006;281:7489–97.
Kim EJ, Park JS, Um SJ. Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo. J Biol Chem. 2002;277:32020–8.
Zhang OH, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–86.
Zhang QH, Nottke A, Goodman RH. Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. P Natl Acad Sci USA. 2005;102:2802–7.
Doxakis E, Huang EJ, Davies AM. Homeodomain-interacting protein kinase-2 regulates apoptosis in developing sensory and sympathetic neurons. Curr Biol. 2004;14:1761–5.
Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–29.
Moller A, Sirma H, Hofmann TG, Rueffer S, Klimczak E, Droge W, Will H, Schmitz ML. PML is required for homeodomain-interacting protein kinase 2 (HIPK2)-mediated p53 phosphorylation and cell cycle arrest but is dispensable for the formation of HIPK domains. Cancer Res. 2003;63:4310–4.
Moller A, Sirma H, Hofmann TG, Staege H, Gresko E, Ludi KS, Klimczak E, Droge W, Will H, Schmitz ML. Sp100 is important for the stimulatory effect of homeodomain-interacting protein kinase-2 on p53-dependent gene expression. Oncogene. 2003;22:8731–7.
Tomasini R, Samir AA, Carrier A, Isnardon D, Cecchinelli B, Soddu S, Malissen B, Dagorn JC, Iovanna JL, Dusetti NJ. TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity. J Biol Chem. 2003;278:37722–9.
Wiggins AK, Wei GW, Doxakis E, Wong C, Tang AA, Zang K, Luo EJ, Neve RL, Reichardt LF, Huang EJ. Interaction of Brn3a and HIPK2 mediates transcriptional repression of sensory neuron survival. J Cell Biol. 2004;167:257–67.
Vasiliauskas D, Hancock S, Stern CD. SWiP-1: novel SOCS box containing WD-protein regulated by signalling centres and by Shh during development. Mech Develop. 1999;82:79–94.
Dentice M, Bandyopadhyay A, Gereben B, Callebaut I, Christoffolete MA, Kim BW, Nissim S, Mornon JP, Zavacki AM, Zeold A, et al. The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate. Nat Cell Biol. 2005;7:698.
Nara H, Onoda T, Rahman M, Araki A, Juliana FM, Tanaka N, Asao H. WSB-1, a novel IL-21 receptor binding molecule, enhances the maturation of IL-21 receptor. Cell Immunol. 2011;269:54–9.
Ivan M, Kaelin WG. The von Hippel-Lindau tumor suppressor protein. Curr Opin Genet Dev. 2001;11:27–34.
Kaelin WG. von Hippel-Lindau disease. Annu Rev Pathol Mech. 2007;2:145–73.
Tong Y, Li QG, Xing TY, Zhang M, Zhang JJ, Xia Q. HIF1 regulates WSB-1 expression to promote hypoxia-induced chemoresistance in hepatocellular carcinoma cells. FEBS Lett. 2013;587:2530–5.
Kim JJ, Lee SB, Jang JS, Yi SY, Kim SH, Han SA, Lee JM, Tong SY, Vincelette ND, Gao BW, et al. WSB1 promotes tumor metastasis by inducing pVHL degradation. Genes Dev. 2015;29:2244–57.
Cao J, Wang YJ, Dong R, Lin GY, Zhang N, Wang J, Lin NM, Gu YC, Ding L, Ying MD, et al. Hypoxia-induced WSB1 promotes the metastatic potential of osteosarcoma cells. Cancer Res. 2015;75:4839–51.
Lee RHK, Iioka H, Ohashi M, Iemura S, Natsume T, Kinoshita N. XRab40 and XCullin5 form a ubiquitin ligase complex essential for the noncanonical Wnt pathway. EMBO J. 2007;26:3592–606.
Kamura T, Burian D, Yan Q, Schmidt SL, Lane WS, Querido E, Branton PE, Shilatifard A, Conaway RC, Conaway JW. MUF1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J Biol Chem. 2001;276:29748–53.
Schenkova K, Lutz J, Kopp M, Ramos S, Rivero F. MUF1/leucine-rich repeat containing 41 (LRRC41), a substrate of RhoBTB-dependent cullin 3 ubiquitin ligase complexes, is a predominantly nuclear dimeric protein. J Mol Biol. 2012;422:659–73.
Yasukawa T, Kamura T, Kitajima S, Conaway RC, Conaway JW, Aso T. Mammalian Elongin A complex mediates DNA-damage-induced ubiquitylation and degradation of Rpb1. EMBO J. 2008;27:3256–66.
Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:809. (Erratum in Cell 2004;116).
Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–77.
Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112.
Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103.
Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31.
Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–50.
Takeuchi H, Kao S, Miyagi E, Khan MA, Buckler-White A, Plishka R, Strebel K. Production of infectious SIVagm from human cells requires functional inactivation but not viral exclusion of human APOBEC3G. J Biol Chem. 2005;280:375–82.
Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829.
Zhang H, Yang B, Pomerantz RJ, Zhang CM, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–8.
Suspene R, Sommer P, Henry M, Ferris S, Guetard D, Pochet S, Chester A, Navaratnam N, Wain-Hobson S, Vartanian JP. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–9.
Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–42.
Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–7.
Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601.
Kim DY. The assembly of Vif ubiquitin E3 ligase for APOBEC3 degradation. Arch Pharm Res. 2015;38:435–45.
Malim MH. HIV Ringside views. Nature. 2014;505:167–8.
Yu XH, Yu YK, Liu BD, Luo K, Kong W, Mao PY, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–60.
Shirakawa K, Takaori-Kondo A, Kobayashi M, Tomonaga M, Izumi T, Fukunaga K, Sasada A, Abudu A, Miyauchi Y, Akari H, et al. Ubiquitination of APOBEC3 proteins by the Vif-Cullin5-ElonginB-ElonginC complex. Virology. 2006;344:263–6.
Liu BD, Sarkis PTN, Luo K, Yu YK, Yu XF. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J Virol. 2005;79:9579–87.
Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, Kwon E, Yen L, Cartozo NC, Li M, Jager S, Mason-Herr J, et al. Inhibition of a NEDD8 cascade restores restriction of HIV by APOBEC3G. PLoS Pathog. 2012;8:e1003085.
Dang Y, Siew LM, Zheng YH. APOBEC3G is degraded by the proteasomal pathway in a vif-dependent manner without being polyubiquitylated. J Biol Chem. 2008;283:13124–31.
Bartz SR, Rogel ME, Emerman M. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G(2) accumulation by a mechanism which differs from DNA damage checkpoint control. J Virol. 1996;70:2324–31.
Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat Med. 1998;4:65–71.
Rogel ME, Wu LI, Emerman M. The human-immunodeficiency-virus type-1 Vpr gene prevents cell-proliferation during chronic infection. J Virol. 1995;69:882–8.
He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–11.
Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–13.
Re F, Braaten D, Franke EK, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–64.
Sakai K, Dimas J, Lenardo MJ. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc Natl Acad Sci USA. 2006;103:3369–74.
Wang J, Shackelford JM, Casella CR, Shivers DK, Rapaport EL, Liu B, Yu XF, Finkel TH. The Vif accessory protein alters the cell cycle of human immunodeficiency virus type 1 infected cells. Virology. 2007;359:243–52.
Wang J, Shackelford JM, Selliah N, Shivers DK, O’Neill E, Garcia JV, Muthumani K, Weiner D, Yu XF, Gabuzda D, et al. The HIV-1 Vif protein mediates degradation of Vpr and reduces Vpr-induced cell cycle arrest. DNA Cell Biol. 2008;27:267–77.
Wang J, Reuschel EL, Shackelford JM, Jeang L, Shivers DK, Diehl JA, Yu XF, Finkel TH. HIV-1 Vif promotes the G(1)- to S-phase cell-cycle transition. Blood. 2011;117:1260–9.
Guo YY, Dong LY, Qiu XL, Wang YS, Zhang BL, Liu HN, Yu Y, Zang Y, Yang MJ, Huang ZW. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229.
Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, et al. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–5.
Zhang WY, Du J, Evans SL, Yu YK, Yu XF. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376.
de Bruijn MFTR, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–48.
Ito Y. RUNX genes in development and cancer: Regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33.
Yang X, Goncalves J, Gabuzda D. Phosphorylation of Vif and its role in HIV-1 replication. J Biol Chem. 1996;271:10121–9.
Yang XY, Gabuzda D. Mitogen-activated protein kinase phosphorylates and regulates the HIV-1 Vif protein. J Biol Chem. 1998;273:29879–87.
Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004;18:2861–6.
Kitamura S, Ode H, Iwatani Y. Structural features of antiviral APOBEC3 proteins are linked to their functional activities. Front Microbiol. 2011;2:3389.
Yu YK, Xiao ZX, Ehrlich ES, Yu XG, Yu XF. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–72.
Wang H, Lv G, Zhou X, Li Z, Liu X, Yu XF, Zhang W. Requirement of HIV-1 Vif C-terminus for Vif-CBF-beta interaction and assembly of CUL5-containing E3 ligase. BMC Microbiol. 2014;14:290.
Wang XD, Wang XY, Wang WR, Zhang JY, Wang JW, Wang C, Lv MY, Zuo T, Liu DL, Zhang HH, et al. Both Rbx1 and Rbx2 exhibit a functional role in the HIV-1 Vif-Cullin5 E3 ligase complex in vitro. Biochem Bioph Res Co. 2015;461:624–9.
An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet. 2010;26:119–31.
O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–74.
Zhao K, Ishida Y, Oleksyk TK, Winkler CA, Roca AL. Evidence for selection at HIV host susceptibility genes in a West Central African human population. BMC Evol Biol. 2012;12:237.
An P, Duggal P, Wang LH, O’Brien SJ, Donfield S, Goedert JJ, Phair J, Buchbinder S, Kirk GD, Winkler CA. Polymorphisms of CUL5 are associated with CD4(+) T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19.
De Maio FA, Rocco CA, Aulicino PC, Bologna R, Mangano A, Sen L. APOBEC3-mediated editing in HIV type 1 from pediatric patients and its association with APOBEC3G/CUL5 polymorphisms and Vif variability. Aids Res Hum Retrov. 2012;28:619–27.
De Maio FA, Rocco CA, Aulicino PC, Bologna R, Mangano A, Sen L. Effect of HIV-1 Vif variability on progression to pediatric AIDS and its association with APOBEC3G and CUL5 polymorphisms. Infect Genet Evol. 2011;11:1256–62.
Tsurumi T. EBV replication enzymes. Curr Top Microbiol. 2001;258:65–87.
Chevalliergreco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein–Barr-virus (Ebv)-encoded trans-acting factors, Eb1 and Eb2, are required to activate transcription from an Ebv early promoter. EMBO J. 1986;5:3243–9.
Countryman J, Jenson H, Seibl R, Wolf H, Miller G. Polymorphic proteins encoded within Bzlf1 of defective and standard Epstein–Barr viruses disrupt latency. J Virol. 1987;61:3672–9.
Hammerschmidt W, Sugden B. Identification and characterization of orilyt, a lytic Origin of DNA-replication of Epstein–Barr virus. Cell. 1988;55:427–33.
Sinclair AJ, Brimmell M, Shanahan F, Farrell PJ. Pathways of activation of the Epstein–Barr-virus productive cycle. J Virol. 1991;65:2237–44.
Sato Y, Kamura T, Shirata N, Murata T, Kudoh A, Iwahori S, Nakayama S, Isomura H, Nishiyama Y, Tsurumi T. Degradation of Phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Pathog. 2009;5:e1000530.
Sato Y, Shirata N, Kudoh A, Iwahori S, Nakayama S, Murata T, Isomura H, Nishiyama Y, Tsurumi T. Expression of Epstein–Barr virus BZLF1 immediate-early protein induces p53 degradation independent of MDM2, leading to repression of p53-mediated transcription. Virology. 2009;388:204–11.
Forrester NA, Sedgwick GG, Thomas A, Blackford AN, Speiseder T, Dobner T, Byrd PJ, Stewart GS, Turnell AS, Grand RJA. Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection. J Virol. 2011;85:2201–11.
Blanchette P, Cheng CY, Yan Q, Ketner G, Ornelles DA, Dobner T, Conaway RC, Conaway JW, Branton PE. Both BC-Box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol Cell Biol. 2004;24:9619–29.
Cheng CY, Blanchette P, Branton PE. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology. 2007;364:36–44.
Cheng CY, Gilson T, Dallaire F, Ketner G, Branton PE, Blanchette P. The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity. J Virol. 2011;85:765–75.
Harada JN, Shevchenko A, Shevchenko A, Pallas DC, Berk AJ. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J Virol. 2002;76:9194–206.
Luo K, Ehrlich E, Xiao ZX, Zhang WY, Ketner G, Yu XF. Adenovirus E4orF6 assembles with cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. Faseb J. 2007;21:1742–50.
Cheng CY, Gilson T, Wimmer P, Schreiner S, Ketner G, Dobner T, Branton PE, Blanchette P. Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes. J Virol. 2013;87:6232–45.
Cathomen T, Weitzman MD. A functional complex of adenovirus proteins E1B-55 kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J Virol. 2000;74:11407–12.
Moore M, Horikoshi N, Shenk T. Oncogenic potential of the adenovirus E4orf6 protein. P Natl Acad Sci USA. 1996;93:11295–301.
Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. Two distinct activities contribute to the oncogenic potential of the adenovirus type 5 E4orf6 protein. J Virol. 2000;74:5168–81.
Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism, involving a Cullin-containing complex. Genes Dev. 2001;15:3104–17.
Querido E, Marcellus RC, Lai A, Charbonneau R, Teodoro JG, Ketner G, Branton PE. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–98.
Shen YQ, Kitzes G, Nye JA, Fattaey A, Hermiston T. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J Virol. 2001;75:4297–307.
Steegenga WT, Riteco N, Jochemsen AG, Fallaux FJ, Bos JL. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–57.
Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–52.
Baker A, Rohleder KJ, Hanakahi LA, Ketner G. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J Virol. 2007;81:7034–40.
Jayaram S, Gilson T, Ehrlich ES, Yu XF, Ketner G, Hanakahi L. E1B 55k-independent dissociation of the DNA ligase IV/XRCC4 complex by E4 34k during adenovirus infection. Virology. 2008;382:163–70.
Dallaire F, Blanchette P, Groitl P, Dobner T, Branton PE. Identification of Integrin alpha 3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J Virol. 2009;83:5329–38.
Farris KD, Fasina O, Sukhu L, Li L, Pintel DJ. Adeno-associated virus small rep proteins are modified with at least two types of polyubiquitination. J Virol. 2010;84:1206–11.
Nayak R, Farris KD, Pintel DJ. E4Orf6-E1B-55k-dependent degradation of de novo-generated adeno-associated virus type 5 Rep52 and capsid proteins employs a cullin 5-containing E3 ligase complex. J Virol. 2008;82:3803–8.
Stracker TH, Petrini JHJ. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Bio. 2011;12:90–103.
Chistiakov DA, Voronova NV, Chistiakov AP. Ligase IV syndrome. Eur J Med Genet. 2009;52:373–8.
Dipersio CM, Shah S, Hynes RO. Alpha-3a-Beta-1 Integrin localizes to focal contacts in response to diverse extracellular-matrix proteins. J Cell Sci. 1995;108:2321–36.
Gilson T, Cheng CY, Hur WS, Blanchette P, Branton PE. Analysis of the cullin binding sites of the E4orf6 proteins of human adenovirus E3 ubiquitin ligases. J Virol. 2014;88:3885–97.
Chiocca S, Baker A, Cotten M. Identification of a novel antiapoptotic protein, GAM-1, encoded by the CELO adenovirus. J Virol. 1997;71:3168–77.
Chiocca S, Kurzbauer R, Schaffner G, Baker A, Mautner V, Cotten M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J Virol. 1996;70:2939–49.
Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S. A mechanism for inhibiting the SUMO pathway. Mol Cell. 2004;16:549–61.
Boggio R, Passafaro A, Chiocca S. Targeting SUMO E1 to ubiquitin ligases—a viral strategy to counteract sumoylation. J Biol Chem. 2007;282:15376–82.
Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2006;2:1002–12.
Suzuki T, Isobe T, Kitagawa M, Ueda K. Kaposi’s sarcoma-associated herpesvirus-encoded LANA positively affects on ubiquitylation of p53. Biochem Bioph Res Co. 2010;403:194–7.
Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–86.
Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol Cell. 2011;44:317–24.
Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, et al. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell. 2011;44:290–303.
Zhao YC, Xiong XF, Sun Y. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCF beta TrCP E3 ubiquitin ligase and regulates survival and autophagy. Mol Cell. 2011;44:304–16.
Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, Marsella C, Piselli P, Gretzmeier C, Dengjel J, et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell. 2014;31:734–46.
Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001.
Ehrlich ES, Wang T, Luo K, Xiao ZX, Niewiadomska AM, Martinez T, Xu WP, Neckers L, Yu XF. Regulation of Hsp90 client proteins by a cullin5-RING E3 ubiquitin ligase. P Natl Acad Sci USA. 2009;106:20330–5.
Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76.
Samant RS, Clarke PA, Workman P. E3 ubiquitin ligase Cullin-5 modulates multiple molecular and cellular responses to heat shock protein 90 inhibition in human cancer cells. Proc Natl Acad Sci USA. 2014;111:6834–9.
Schulte TW, An WG, Neckers LM. Geldanamycin-induced destabilization of Raf-1 involves the proteasome. Biochem Biophys Res Commun. 1997;239:655–9.
Whitesell L, Cook P. Stable and specific binding of heat shock protein 90 by geldanamycin disrupts glucocorticoid receptor function in intact cells. Mol Endocrinol. 1996;10:705–12.
Marteijn JAF, van Ernst L, Erpelinck-Verschueren CAJ, Nikoloski G, Menke A, de Witte T, Lowenberg B, Jansen JH, van der Reijden BA. The E3 ubiquitin-protein ligase Triad1 inhibits clonogenic growth of primary myeloid progenitor cells. Blood. 2005;106:4114–23.
Lin AE, Ebert G, Ow Y, Preston SP, Toe JG, Cooney JP, Scott HW, Sasaki M, Saibil SD, Dissanayake D, et al. ARIH2 is essential for embryogenesis, and its hematopoietic deficiency causes lethal activation of the immune system. Nat Immunol. 2013;14:27–33.
Kelsall IR, Duda DM, Olszewski JL, Hofmann K, Knebel A, Langevin F, Wood N, Wightman M, Schulman BA, Alpi AF. TRIAD1 and HHARI bind to and are activated by distinct neddylated cullin-RING ligase complexes. EMBO J. 2013;32:2848–60.
Authors’ contributions
FO undertook the background literature study and wrote most part of this review article. AJO and KN helped in extending the initial manuscript. TK supervised the entire project. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by JSPS KAKENHI Grant Numbers 25291023 (to F.O. and T.K.), 25860043 (to F.O.), and 24112006 (to T.K.), a Grant-in-Aid from Scientific Research on Innovative Areas, and Grants from the Ministry of Education, Science, Sports, and Culture of Japan.
Competing interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Okumura, F., Joo-Okumura, A., Nakatsukasa, K. et al. The role of cullin 5-containing ubiquitin ligases. Cell Div 11, 1 (2016). https://doi.org/10.1186/s13008-016-0016-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13008-016-0016-3