Abstract
Background
Loquat (Eriobotrya japonica) is a subtropical tree bearing fruit that ripens during late spring and early summer, which is the off-season for fruit production. The specific flowering habit of loquat, which starts in fall and ends in winter, has attracted an increasing number of researchers who believe that it may represent an ideal model for studying flowering shift adaptations to climate change in Rosaceae. These studies require an understanding of gene expression patterns within the fruit and other tissues of this plant. Although ACTINs (ACTs) have previously been used as reference genes (RGs) for gene expression studies in loquats, a comprehensive analysis of whether these RGs are optimal for normalizing RT-qPCR data has not been performed.
Results
In this study, 11 candidate RGs (RIBOSOMAL-LIKE PROTEIN4 (RPL4), RIBOSOMAL-LIKE PROTEIN18 (RPL18), Histone H3.3 (HIS3), Alpha-tubulin-3 (TUA3), S-Adenosyl Methionine Decarboxylase (SAMDC), TIP41-like Family Protein (TIP41), (UDP)-glucose Pyrophosphorylase (UGPase), 18S ribosomal RNA (18S), Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH), Plasma Intrinsic Protein 2 (PIP2) and ACTIN(ACT)) were assessed to determine their expression stability in 23 samples from different tissues or organs of loquat. Integrated expression stability evaluations using five computational statistical methods (GeNorm, NormFinder, ΔCt, BestKeeper, and RefFinder) suggested that a RG set, including RPL4, RPL18, HIS3 and TUA3, was the most stable one across all of the tested loquat samples. The expression pattern of EjCDKB1;2 in the tested loquat tissues normalized to the selected RG set demonstrated its reliability.
Conclusions
This study reveals the reliable RGs for accurate normalization of gene expression in loquat. In addition, our findings demonstrate an efficient system for identifying the most effective RGs for different organs, which may be applied to related rosaceous crops.
Similar content being viewed by others
Introduction
Loquat (Eriobotrya japonica Lindl.) is a subtropical evergreen fruit tree belonging to the apple subfamily Maloideae of Rosaceae. Loquat is primarily cultivated in Southeast Asia and Mediterranean regions [1] and differs from temperate-zone relatives, such as apple, pear and peach, which normally flower in the spring, by flowering in fall or early winter [2]. Loquat fruits mature during the slack season of the fresh fruit market in late spring or early summer, and the nutritious and succulent fruits are attracting an increasing number of consumers worldwide [3]. In addition, loquat is also used in traditional Chinese medicine due to an abundance of therapeutic compounds and secondary metabolites in its leaves and other tissues, as well as the presence of anti-cancer compounds [4, 5]. Both its delicious fruit and ability to adapt to various subtropical climates have convinced breeders and researchers that loquat cultivation should be expanded [6]. However, certain disadvantages, such as the large seeds and tender flesh, may hinder loquat production due to a reduced edibility or shorter shelf life [7].
Many classic breeding approaches to improve this crop are underway [8, 9]. Moreover, considerable research has focused on the genetics and molecular mechanisms underlying specific biological traits/phenomena in loquat for exploiting molecular markers for assisted selection [10, 11] or identifying important genes that may be directly or indirectly used in future breeding endeavors [2, 12,13,14,15]. Overall, gene expression analysis is one of the most important approaches for identifying candidate genes related to specific biological characteristics. Multiple molecular techniques, including northern blotting, semiquantitative reverse transcription PCR, in situ hybridization and quantitative reverse transcription-PCR, are effective methods of investigating the expression characteristics of specific genes. Among these, RT-qPCR (Quantitative Real-Time Polymerase Chain Reaction) is the most widely used technique due to its advantages of precision, high sensitivity, flexibility and scalability [16, 17]. However, confounding results are usually obtained when a reference gene (RG) without stable expression is used in gene expression assays [18]; therefore, the stability of a RG is critical for generating reliable and accurate qPCR results [19, 20].
Although an optimal RG was once considered to be a gene stably expressed in various tissues under different experimental conditions [19], many researchers have suggested that the transcript levels of commonly used housekeeping genes can vary considerably under different conditions [21, 22]. Thus, an optimal RG might not always be suitable for all materials under different experimental conditions [21]. Accordingly, an increasing number of studies have been conducted to identify reliable RGs for various plant materials or different developmental stages [22], such as vascular development [23], somatic embryogenic culture [24], root development [25] and flower development [26]. In addition, multiple attempts have been made to select suitable housekeeping genes for materials under biotic and abiotic stresses in different plant species, such as potato [27], Arabidopsis [28], rice [29] and Pyropia yezoensis [30]. Considering that fruit is one of the most important commercial products of most crop plants, numerous stable RGs have been identified for quantitative expression analyses of fruit development in various crops, such as grape [31], banana [32], papaya [33], Chinese wolfberry [34], olive [35], orange [36], strawberry [37], plum [38], watermelon [39], peach [40] and tomato [41].
Considerable efforts have been made to clarify the molecular regulatory mechanisms of specific biological traits/phenomena related to loquat fruit, such as sorbitol metabolism [42], ethylene synthesis and signal transduction [43], flesh coloration [12], postharvest fruit development [15], flowering time regulation [2], volatile component formation in fruit [13] and fruit size formation [14]. Conversely, few RGs have been identified and applied to loquat. Of all known loquat housekeeping genes, ACTINs (ACTs, including JN004223, JX089589, AB710173.1 and FJ481118) have been widely used as RGs in research on fruit pulp coloration [12], floral initiation [2] and fruit cell division [14]. Nonetheless, previous gene expression analyses of fruit development lasting approximately four months have occasionally revealed the fluctuating ACT expression levels. Such results suggest that ACTs are not always suitable for detecting expression related to long-duration fruit development or even other experimental conditions. However, reports focused on RG selection in loquat have not been reported. Thus, there is a need to identify the appropriate RGs for gene expression studies based on RT-qPCR assays of the development of fruit or other tissues/organs in loquat.
In this study, partial 3′-terminal sequences of 7 new candidate RGs were cloned and identified. RT-qPCR assays of 23 samples belonging to five sample sets (fruit, floral tissues, seed development, vegetative development and all samples) and specific primer pairs for 11 candidate RGs were performed, and expression stability was evaluated with five methods: geNorm, Normfinder, BestKeeper, ΔCt and RefFinder. Comprehensive reports of the five sample sets based on these five methods were generated. The expression levels of EjCDKB1;2 (Cyclin-dependent kinase B1;2) in fruit during the cell division phase were normalized to the most stable RG set (RPL4, RPL18, HIS3 and TUA3) and the least stable RG set (SAMDC, PIP2 and 18S), with results demonstrating the reliability of the newly selected optimal RGs. RGs identified in this study provide a useful and reliable resource for the accurate quantification of gene expression in loquat and furthermore illustrate an effective system for identifying the best RGs for different tissues/organs of this crop.
Materials and methods
Plant materials
Eriobotrya japonica cv. ‘Zaozhong-6' cultivated in the Eriobotrya germplasm resource preservation garden at the South China Agricultural University under normal management was used as the plant material. A total of 23 samples were collected for RG selection, including fruits belonging to five developmental stages, 7 floral tissues, ovules belonging to three developmental stages, seeds belonging to three developmental stages and 5 vegetative tissues (shown in Additional file 1: Table S1). Receptacles or fruits were collected every 7 days from 4 DBA (days before anthesis) to 77 DPA (days past anthesis) for cell division gene expression assays.
Total RNA extraction and cDNA synthesis
The total RNA of the samples was extracted separately using an EASYspin Plus Plant RNA Extraction Kit (Aidlab, Beijing, China) according to the manufacturer’s instructions. cDNA synthesis was performed using a PrimeScript™ RT Reagent Kit (TaKaRa Bio, Shiga, Japan) according to the manufacturer’s instructions.
Identification of candidate reference genes
Eleven RGs were selected to identify the most stably expressed housekeeping gene(s). Sequence information of four reported candidate RGs (18S Ribosomal RNA (18S), Glyceraldehyde-3-phosphate (GAPDH), Plasma Intrinsic Protein 2 (PIP2) and ACT, under accession numbers AB636342.1, JQ731608.1, JX041626.1 and AB710173.1) were obtained from the National Center for Biotechnology Information (NCBI, Bethesda, MD, USA) database. Another seven candidate RGs, including Ribosomal Protein L4 (RPL4), Ribosomal Protein L18 (RPL18), Histone H3.3 (HIS3), Alpha-tubulin-3 (TUA3), S-Adenosyl Methionine Decarboxylase (SAMDC), TIP41-like Family Protein (TIP41) and (UDP)-glucose Pyrophosphorylase (UGPase), were selected based on previous reports in other plant species [24, 32, 34, 44,45,46,47]. Preliminary sequence information of the seven newly selected RGs was queried using our unpublished loquat genomic data. To verify and obtain detailed cDNA sequence information, specific primers were designed to amplify the sequence of the 3′ end. All primer pairs were supplied commercially (Sangon, Guangzhou, China). The target sequence fragments were isolated using a cDNA mixture of diverse ‘Zaozhong-6’ tissues as previously performed [14] and the primers were listed in Additional file 1: Table S2. The sequence information of these seven newly selected genes was submitted to GenBank under accession numbers MH196506-MH196512.
RT-qPCR primer design and validation
Quantitative primers for the 11 genes were designed using the BatchPrimer3 program [48], and primer specificities were confirmed as previously described [18]. A standard curve using a series of gradient-diluted cDNAs was generated to calculate the gene-specific PCR amplification efficiency (E) and correlation coefficients (R2) for each gene, and four replications were performed. The amplification efficiencies of the candidate RGs were calculated using the following formula: E (%) = (10−1/slope − 1) × 100. Detailed sequence information for all primer pairs is listed in Table 1. To verify the stabilities of the screened RGs, the expression pattern of EjCDKB1;2 (Forward: 5′-CTCGGTTCGGCTCACTACTC-3′, Reverse: 5′-GCCAATCTCGCAAAGAAGAA-3′) in loquat fruit was detected using the most and least stable RGs as internal genes.
Quantitative real-time polymerase chain reaction analysis
All RT-qPCRs were performed as described previously [14] using a LightCycler480 system with LC480 software (Roche Diagnostics, Penzberg, Germany). Each reaction contained 1 μL of diluted cDNA, 5 μL of iTaq™ universal SYBR Green Supermix (Bio-Rad, Foster City, CA, USA), 0.5 μL of each primer and 3 μL of ultrapure water to a final volume of 10 μL. The reactions were performed in 384-well reaction plates using the following conditions: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. The melting curves were analyzed at 60–95 °C after amplification. Four independent replicates were performed when assessing the expression stability of the RGs, whereas three biological replicates were performed when examining the expression patterns of fruit quality-related genes.
Data analysis
The Ct values of each RG were used to evaluate their expression levels. Expression stability was analyzed using the geNorm [49], NormFinder [50], BestKeeper [51] and ΔCt methods [52]. A comprehensive ranking report of the reliability of the genes was ultimately obtained using the RefFinder tool (http://150.216.56.64/ referencegene.php). The comprehensive stability value according to RefFinder was based on the NormFinder, BestKeeper, GeNorm, and comparative ΔCt results. The overall final ranking of each gene was computed as the geometric mean, and a lower geometric mean of the ranking value indicated a higher stability. SigmaPlot 12.5 software was used to prepare figures indicating the gene expression patterns.
Results
Isolation of loquat candidate reference genes
A total of 11 genes were selected for suitable RG identification for use in the expression analyses of different loquat materials. Four RGs, 18S, GAPDH, PIP2 and ACTIN, which have been reported in previous studies, were included. Considering the deficiency of appropriate RG information for loquat, RPL4, RPL18, HIS3, TUA3, SAMDC, TIP41 and UGPase, which are often used as RGs for other plants, were also chosen for assessment as candidate loquat RGs in this study. Partial mRNA sequences (279–381 bp) from the 3′ end of these newly selected candidate genes were cloned and sequenced using primer pairs listed in Additional file 1: Table S2. Detailed sequence information can be found in GenBank under accession numbers MH196506 to MH196512.
Verification of amplification efficiency
To ensure the specificity of our RT-qPCR analysis, primers for quantitative PCR of the 11 candidate RGs were designed to amplify the 3′ end of each gene, and the products were examined via agarose gel electrophoresis. The results showed that only the expected product was amplified for each of the tested RGs. The product sizes ranged from 80 bp (18S) to 215 bp (RPL4) (Fig. 1, Table 1). In addition, a single peak in the melting curve further supported specific amplification by each primer pair (Additional file 1: Figure S1). The amplification efficiencies of the genes ranged from 93.33% (UGPase) to 126.43% (TIP41). The primer sequences and amplification characteristics of all genes tested are summarized in Table 1.
Expression stability of candidate RGs based on expression profiles
A total of 23 samples, including fruits, floral tissues, ovules, seeds and vegetative tissues at diverse developmental stages, were used for quantitative detection (Fig. 2, Additional file 1: Table S1). Our former expression assays of genes related to flowering time and fruit growth in loquat suggested that the use of one RG (ACT2 or ACT4) for gene expression detection in multiple materials may not be suitable, none of ACTs exhibited stable expression in different tissues or the same tissues under various experimental conditions [2, 14]. Thus, to evaluate whether the candidate RGs are suitable for expression analysis in various developmental processes of loquat, the samples were divided into four subsets: fruit growth and quality development (Fruits), floral tissue development (Floral tissues), seed development (Ovules and seeds) and vegetative growth (Vegetative tissues).
Loquat samples used in this study. Rt, root; Yst, young stem; Mst, mature stem; Ml, mature leave; If, inflorescence; Pe, petal; An, anther; St, stigma; Fi, filament; Ca, callus; Ov, ovule; Ys, young seed; Ms, mature seed; Fl, flower; Fi, filament; Fr1, fruit1, receptacle 21 days before anthesis; Fr2, fruit2, receptacle at anthesis; Fr3, fruit3, 56 days past anthesis; Fr4, fruit4, 102 days past anthesis; and Fr5, fruit5, mature fruit 126 days past anthesis
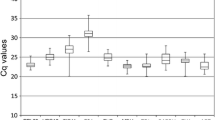
The expression levels of the 11 candidate RGs were first evaluated according to Ct values across all samples in an integrated set. As shown in the boxplot (Fig. 3a), the Ct values of these genes varied from 16.89 (18S) to 25.34 (TUA3). TUA3 displayed the lowest expression, whereas PIP2 showed the highest. ACT, the RG commonly used in former studies [2, 12, 14], showed a moderate expression level.
With respect to loquat fruit development, the Ct values of the different RGs ranged from 17.03 (18S) to 26.57 (TIP41). SAMDC and GAPDH showed the highest and lowest expression, respectively (Fig. 3b). According to the ΔCt values, RPL18 had a minimum average STDEV, followed by GAPDH and RPL4; in contrast, SAMDC had the highest STDEV at 1.48 (Table 2). The NormFinder analysis found that RPL18 was the most stable RG and SAMDC was the least stable (Table 2). Conversely, TIP41/ACT and GAPDH were the optimal RGs according to both the geNorm and BestKeeper analyses (Table 2). Finally, a comprehensive analysis considering all the methods using RefFinder suggested that RPL18, GAPDH and TIP41 were the most stable RGs in loquat fruits (Table 2).
Of the 7 floral tissues examined, TIP41 had the greatest Ct value at 25.55, whereas GAPDH had the lowest Ct value at 20.16 (Fig. 3c). Compared to other material sets, all candidate genes exhibited relatively higher Ct values in the floral tissues. The results of all evaluation methods indicated that SAMDC was the least stably expressed RG. UGPase was the most stable in the ΔCt and NormFinder assays, whereas HIS3 or HIS3/RPL18 was the most reliable housekeeping gene in the BestKeeper and geNorm evaluations. A comprehensive analysis of all evaluation methods using RefFinder revealed that RPL18, HIS3 and UGPase were the top three most reliable RGs for expression studies of floral tissues (Table 2).
Regarding seed development, TIP41 was found to be the most stable RG, and it had M values of 0.74, 0.12 and 0.38 in the ΔCt, NormFinder and geNorm assessments, respectively, whereas SAMDC was the best candidate according to BestKeeper (Tables 1, 2). In this sample set, TUA3 showed the highest Ct value and 18S showed the lowest value. The largest and smallest Ct value ranges were observed for PIP2 and SAMDC, respectively (Fig. 3d). Overall, the comprehensive ranking order indicated that TIP41, SAMDC and ACT were the best RGs for studies on seed development.
When assessing materials related to vegetative growth, RPL18 was the most stable RG, and it presented M values of 0.78, 0.51 and 0.20 according to the ΔCt, BestKeeper and NormFinder analysis, respectively, whereas TIP41/ACT was the most reliable RG according to the geNorm analysis. In most cases, RPL4 was the second most reliable RG (Table 2), and SAMDC and 18S were the least stable RGs according to all of the evaluation systems. An integration of the results of all four software analyses in RefFinder indicated that RPL18, RPL4, TIP41 and TUA3 were the most stably expressed RGs for studies on vegetative development in loquat.
Across all 23 samples, RPL4 and RPL18 were the most stable RGs using ΔCt and geNorm, whereas HIS3 and TUA3 were suggested to be the most reliable RGs using BestKeeper and NormFinder (Table 2). The comprehensive ranking by RefFinder further showed that RPL4, RPL18, HIS3 and TUA3 were the top four most stably expressed RGs, while SAMDC, PIP2 and 18S were the least stable RGs across all tested loquat samples. Former researches suggested that the use of a RG set, instead of one single housekeeping gene, would guarantee more accurate gene expression analyses under multiple experimental conditions [22], a RG set including the above selected stable RGs was recommended for the gene normalization in different materials of loquat.
Validation of candidate reference genes
Although ACT has been commonly used as an RG in previous gene expression analyses in loquat [2, 12, 14], our results suggested that the RG set RPL4, RPL18, HIS3 and TUA3 is more suitable for gene expression analyses in loquat (Table 2). To confirm the reliability of this RG set, the expression pattern of EjCDKB1;2, which is described as a cell division marker gene [14], was assessed in diverse tissue samples.
When using the most stable RG set (RPL4, RPL18, HIS3 and TUA3) for gene expression normalization, a single expression peak of EjCDKB1;2 was observed during the fruit cell division phase. The transcription abundance of EjCDKB1;2 dramatically increased soon after flowering, peaked at 28 DPA and then declined sharply until 42 DPA, when cell division of the pericarp was almost complete [14].
In contrast, distorted EjCDKB1;2 expression results were obtained when using the least stable RGs as internal standards. When normalized to PIP2, 18S or SAMDC + PIP2 + 18S, an unanticipated decrease in EjCDKB1;2 expression occurred from 7 to 14 DPA, and when normalized to SAMDC, a peak of EjCDKB1;2 expression occurred at the end of the cell division phase (Fig. 4a).
Relative expression levels of EjCDKB1;2 normalized to the most and least stable RG sets. a Fruit development and b seed development. OV1 and OV2 were ovules in flowers of 4 and 1 day before anthesis. The most stable RG set included RPL4, RPL18, HIS3 and TUA3. The least stable RGs included SAMDC, PIP2 and 18S
Because of the limited number of reports focused on cell division in loquat tissue samples, only the seeds were used as another sample set for further verification. When the RPL4 + RPL18 + HIS3 + TUA3 combination was used as the RG, moderate EjCDKB1;2 expression was obtained with small error bars. In addition, based on this set of the most stable RGs, an increase of EjCDKB1;2 transcription abundance from OV1 to OV2 during prefertilization ovule development could be detected (Fig. 4b). Taken together, the results of EjCDKB1;2 expression during fruit and seed development confirmed that RPL4, RPL18, HIS3 and TUA3 constitute the most appropriate set of RGs for accurate RT-qPCR analyses in loquat.
Discussion
Loquat sets fruit in fall or early winter and as such provides appealing, nutritious and succulent pome fruits in the off-season during spring to early summer [1]; thus, it is a promising fruit tree for cultivation worldwide in the next decades [3]. In addition to classical breeding, molecular studies aim to develop or identify molecular markers/genes associated with specific traits that contributes to improving this fruit. ACTs have been commonly used as housekeeping genes [12,13,14,15]; however, stable RGs in loquat have not been comprehensively identified. In addition, an increasing amount of evidence has revealed that typical house-keeping genes exhibit variations among long-duration developmental samples of fruit and shoot apical meristem tissues [2, 14], which indicates that unclear results may be obtained when only one internal gene is applied under all experimental conditions. Thus, to avoid such an issue, selecting suitable RGs for these development phases and other tissues is critical.
In the present study, seven novel candidate house-keeping genes that have been commonly used in previous research on other plant species were selected as candidate RGs in loquat [24, 32, 34, 44,45,46,47]. Another four previously assessed housekeeping genes, 18S, GAPDH, PIP2 and ACTIN, were also evaluated. The expression stability of these genes in five sample subsets, which included 23 samples, was analyzed. Although NormFinder and geNorm are widely used to identify suitable RGs [32, 33, 53, 54], the computational method of RefFinder [55] was also employed in this study as formerly performed [26, 56, 57].
Reports have suggested that researchers should exercise caution when using ribosomal genes as RGs because these genes can display tissue specific expression (e.g. RPL39-like and RPL3-like [58]). However, many other ribosomal genes are also confirmed to be suitable housekeeping genes for their stable expression in all types of cells [59]. The stability of RPLs should be confirmed individually. The results in this study revealed that the loquat RPL4/18 genes were stably expressed in all tissues examined (Table 2), which is consistent with the results of previous studies; for example, the stable expression of RPL18 has been reported in grape and olive [60, 61]. In addition, many other plant ribosomal protein genes have been identified as suitable housekeeping genes for leaf and fruit samples, such as MdRPL2 in apple [62], CmRPL in melon [54] and MaRPS2 in banana [32]. Consistent with the stably expressed HISTONE in strawberry fruit [37] and Taihangia flower [26] and CitTUA3 in citrus somatic embryo [24], the HIS3 and TUA3 genes in loquat were the 3rd and 4th most stable RGs across all the tested loquat samples (Table 2). TIP41 encodes a tonoplast intrinsic protein, and its homologs in cucumber tissues [63] and melon fruit [54] display remarkably stable expression patterns, and the results of the present study indicated that it was the most suitable internal gene for seed development in loquat (Table 2).
To validate the accuracy of the stably expressed RGs identified in this study, the expression profile of EjCDKB1;2, which is regarded as a marker gene representing the cell division capacity of loquat [14], was detected during early fruit and seed development. The application of two or more stable reference genes might be more reliable than one single housekeeping gene for gene quantification [22]. In this study, the expression profile of EjCDKB1;2 in fruit samples at different development stages was more consistent with the fruit cell division dynamics [14] when normalized to our newly identified RG set than when normalized to the least stable RGs (SAMDC, PIP2 and 18S) (Fig. 4a). Moreover, the most suitable RG set was able to detect a slight increase in EjCDKB1;2 expression from OV1 to OV2 (Fig. 4b). Taken together, these results further confirmed that the most stable RGs, including RPL4, RPL18, HIS3 and TUA3, identified by the comprehensive analyses are appropriate reference genes for accurate expression normalization in loquat.
Conclusions
In this study, we identified a stably expressed RG set (including RPL4, RPL18, HIS3 and TUA3) from 11 candidate internal genes with 23 loquat (E. japonica) tissue samples. An expression analysis of EjCDKB1;2 in fruits and seeds further confirmed the suitability of the novel RG set. The findings provide a foundation for more accurate gene expression studies based on RT-qPCR in loquat. The findings can also be potentially transferred to closely related rosaceous crops and other agronomically important crops.
Abbreviations
- 18S:
-
18S Ribosomal RNA
- ACT:
-
Actin
- DBA:
-
Days before anthesis
- DPA:
-
Days past anthesis
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- HIS3:
-
Histone H3.3
- PIP2:
-
Plasma intrinsic protein 2
- RG:
-
Reference gene
- RPL4:
-
Ribosomal protein L4
- RPL18:
-
Ribosomal protein L18
- RT-qPCR:
-
Quantitative real-time polymerase chain reaction
- SAMDC:
-
S-Adenosyl methionine decarboxylase
- TIP41:
-
TIP41-like family protein
- TUA3:
-
Alpha-tubulin-3
- UGPase:
-
(UDP)-glucose pyrophosphorylase
References
Lin S, Sharpe RH, Janick J. Loquat: botany and horticulture. In: Janick J, editor. Horticlutral reviews, vol. 23. Hoboken: Wiley; 1999. p. 233–76.
Zhang L, Yu H, Lin S, Gao Y. Molecular characterization of FT and FD Homologs from Eriobotrya deflexa Nakai forma koshunensis. Front Plant Sci. 2016;7:8.
Badenes ML, Lin S, Yang X, Liu C, Huang X. Loquat (Eriobotrya Lindl.). In: Folta KM, Gardiner SE, editors. Genetics and genomics of rosaceae. Plant genetics and genomics: crops and models, vol. 6. New York: Springer; 2009. p. 525–38.
Yuan Y, Gao Y, Song G, Lin S. Ursolic acid and oleanolic acid from Eriobotrya fragrans inhibited the viability of A549 cells. Nat Prod Commun. 2015;10(2):239–42.
Hong Y, Qiao Y, Lin S, Jiang Y, Chen F. Characterization of antioxidant compounds in Eriobotrya fragrans Champ leaf. Sci Hortic Amst. 2008;118(4):288–92.
Lin S, Huang X, Cuevas J, Janick J. Loquat: an ancient fruit crop with a promising future. Chron Hortic. 2007;47(2):12–5.
Badenes ML, Janick J, Lin S, Zhang Z, Liang G, Wang W. Breeding loquat. In: Janick J, editor. Plant breeding reviews, vol. 37. Hoboken: Wiley; 2013. p. 259–96.
Tepe S, Turgutoğlu E, Arslan MA, Polat AA. Improvement of loquat by conventional breeding. Acta Hortic. 2011;887:89–94.
Zheng S. Achievement and prospect of loquat breeding in China. Acta Hortic. 2007;750:85–92.
Fukuda S, Ishimoto K, Terakami S, Yamamoto T, Hiehata N. Genetic mapping of the loquat canker resistance gene pse-c in loquat (Eriobotrya japonica). Sci Hortic Amst. 2016;200:19–24.
Carrera L, Sanzol J, Herrero M, Hormaza JI. Genomic characterization of self-incompatibility ribonucleases (S-RNases) in loquat (Eriobotrya japonica Lindl.) (Rosaceae, Pyrinae). Mol Breed. 2009;23(4):539–51.
Fu X, Kong W, Peng G, Zhou J, Azam M, Xu C, Grierson D, Chen K. Plastid structure and carotenogenic gene expression in red- and white-fleshed loquat (Eriobotrya japonica) fruits. J Exp Bot. 2012;63(1):341–54.
Koeduka T, Kajiyama M, Suzuki H, Furuta T, Tsuge T, Matsui K. Benzenoid biosynthesis in the flowers of Eriobotrya japonica: molecular cloning and functional characterization of p-methoxybenzoic acid carboxyl methyltransferase. Planta. 2016;244(3):725–36.
Su W, Zhu Y, Zhang L, Yang X, Gao Y, Lin S. The cellular physiology of loquat (Eriobotrya japonica Lindl.) fruit with a focus on how cell division and cell expansion processes contribute to pome morphogenesis. Sci Hortic Amst. 2017;224:142–9.
Zeng J, Li X, Xu Q, Chen J, Yin X, Ferguson IB, Chen K. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol J. 2015;13(9):1325–34.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22.
Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30(PII S0301-472X(02)00806-86):503–12.
Udvardi MK, Czechowski T, Scheible W. Eleven golden rules of quantitative RT-PCR. Plant Cell. 2008;20(7):1736–7.
Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6(4):279–84.
Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–82.
Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29(2):332–7.
Duc Quan N, Eamens AL, Grof CPL. Reference gene identification for reliable normalisation of quantitative RT-PCR data in Setaria viridis. Plant Methods. 2018;14:24.
Cassan-Wang H, Soler M, Yu H, Camargo ELO, Carocha V, Ladouce N, Savelli B, Paiva JAP, Leple J, Grima-Pettenati J. Reference genes for high-throughput quantitative reverse transcription-PCR analysis of gene expression in organs and tissues of Eucalyptus grown in various environmental conditions. Plant Cell Physiol. 2012;53(12):2101–16.
Liu Z, Ge X, Wu X, Kou S, Chai L, Guo W. Selection and validation of suitable reference genes for mRNA qRT-PCR analysis using somatic embryogenic cultures, floral and vegetative tissues in citrus. Plant Cell Tiss Org. 2013;113(3):469–81.
Zhou Z, Cong P, Tian Y, Zhu Y. Using RNA-seq data to select reference genes for normalizing gene expression in apple roots. PLoS ONE. 2017;12:e01852889.
Li W, Zhang L, Zhang Y, Wang G, Song D, Zhang Y. Selection and validation of appropriate reference genes for quantitative real-time PCR normalization in staminate and perfect flowers of andromonoecious Taihangia rupestris. Front Plant Sci. 2017;8:729.
Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005;56(421):2907–14.
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227(6):1343–9.
Auler PA, Benitez LC, Do Amaral MN, Vighi IL, Rodrigues GDS, Da Maia LC, Bolacel Braga EJ. Evaluation of stability and validation of reference genes for RT-qPCR expression studies in rice plants under water deficit. J Appl Genet. 2017;58(2):163–77.
Gao D, Kong F, Sun P, Bi G, Mao Y. Transcriptome-wide identification of optimal reference genes for expression analysis of Pyropia yezoensis responses to abiotic stress. BMC Genom. 2018;19:251.
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6:27.
Chen L, Zhong H, Kuang J, Li J, Lu W, Chen J. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta. 2011;234(2):377–90.
Zhu X, Li X, Chen W, Chen J, Lu W, Chen L, Fu D. Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE. 2012;7:e444058.
Wang L, Wang Y, Zhou P. Validation of reference genes for quantitative real-time PCR during Chinese wolfberry fruit development. Plant Physiol Biochem. 2013;70:304–10.
Resetic T, Stajner N, Bandelj D, Javornik B, Jakse J. Validation of candidate reference genes in RT-qPCR studies of developing olive fruit and expression analysis of four genes involved in fatty acids metabolism. Mol Breed. 2013;32(1):211–22.
Wu J, Su S, Fu L, Zhang Y, Chai L, Yi H. Selection of reliable reference genes for gene expression studies using quantitative real-time PCR in navel orange fruit development and pummelo floral organs. Sci Hortic Amst. 2014;176:180–8.
Galli V, Borowski JM, Perin EC, Messias RS, Labonde J, Pereira IS, Silva SD, Rombaldi CV. Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene. 2015;554(2):205–14.
Kim H, Saha P, Farcuh M, Li B, Sadka A, Blumwald E. RNA-seq analysis of spatiotemporal gene expression patterns during fruit development revealed reference genes for transcript normalization in plums. Plant Mol Biol Rep. 2015;33(6):1634–49.
Kong Q, Yuan J, Gao L, Zhao L, Cheng F, Huang Y, Bie Z. Evaluation of appropriate reference genes for gene expression normalization during watermelon fruit development. PLoS ONE. 2015;10:e01308656.
Kou X, Zhang L, Yang S, Li G, Ye J. Selection and validation of reference genes for quantitative RT-PCR analysis in peach fruit under different experimental conditions. Sci Hortic Amst. 2017;225:195–203.
Cheng Y, Bian W, Pang X, Yu J, Ahammed GJ, Zhou G, Wang R, Ruan M, Li Z, Ye Q, et al. Genome-wide identification and evaluation of reference genes for quantitative RT-PCR analysis during tomato fruit development. Front Plant Sci. 2017;8:1440.
Bantog NA, Yamada K, Niwa N, Shiratake K, Yamaki S. Gene expression of NAD +-dependent sorbitol dehydrogenase and NADP +-dependent sorbitol-6-phosphate dehydrogenase during development of loquat (Eriobotrya japonica Lindl.) fruit. J Jpn Soc Hortic Sci. 2000;69(3):231–6.
Wang P, Zhang B, Li X, Xu C, Yin X, Shan L, Ferguson I, Chen K. Ethylene signal transduction elements involved in chilling injury in non-climacteric loquat fruit. J Exp Bot. 2010;61(1):179–90.
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139(1):5–17.
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008;8:131.
Tong Z, Gao Z, Wang F, Zhou J, Zhang Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol. 2009;10:71.
Bu J, Zhao J, Liu M. Expression stabilities of candidate reference genes for RT-qPCR in Chinese Jujube (Ziziphus jujuba Mill.) under a variety of conditions. PLoS ONE. 2016;11:e01542124.
You FM, Huo N, Gu YQ, Luo M, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD. BatchPrimer3: A high throughput web application for PCR and sequencing primer design. BMC Bioinform. 2008;9(1):253.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):H34.
Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–50.
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–15.
Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33.
Cheng Y, Pang X, Wan H, Ahammed GJ, Yu J, Yao Z, Ruan M, Ye Q, Li Z, Wang R, et al. Identification of optimal reference genes for normalization of qPCR analysis during pepper fruit development. Front Plant Sci. 2017;8:1128.
Kong Q, Gao L, Cao L, Liu Y, Saba H, Huang Y, Bie Z. Assessment of suitable reference genes for quantitative gene expression studies in melon fruits. Front Plant Sci. 2016;7:1178.
Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012;80:75–84.
Zhou B, Chen P, Khan A, Zhao Y, Chen L, Liu D, Liao X, Kong X, Zhou R. Candidate reference genes selection and application for RT-qPCR analysis in kenaf with cytoplasmic male sterility background. Front Plant Sci. 2017;8:1520.
Duan M, Wang J, Zhang X, Yang H, Wang H, Qiu Y, Song J, Guo Y, Li X. Identification of optimal reference genes for expression analysis in radish (Raphanus sativus L) and its relatives based on expression stability. Front Plant Sci. 2017;8:1605.
Thorrez L, Van Deun K, Tranchevent L, Van Lommel L, Engelen K, Marchal K, Moreau Y, Van Mechelen I, Schuit F. Using ribosomal protein genes as reference: a tale of caution. PLoS ONE. 2008;3:e18543.
Hsiao LL, Dangond F, Yoshida T, Hong R, Jensen RV, Misra J, Dillon W, Lee KF, Clark KE, Haverty P, et al. A compendium of gene expression in normal human tissues. Physiol Genomics. 2001;7(2):97–104.
Gamm M, Heloi RM, Kelloniemi J, Poinssot B, Wendehenne D, Adrian M. Identification of reference genes suitable for qRT-PCR in grapevine and application for the study of the expression of genes involved in pterostilbene synthesis. Mol Genet Genomics. 2011;285(4):273–85.
Ray DL, Johnson JC. Validation of reference genes for gene expression analysis in olive (Olea europaea) mesocarp tissue by quantitative real-time RT-PCR. BMC Res Notes. 2014;7:304.
Kumar G, Singh AK. Reference gene validation for qRT-PCR based gene expression studies in different developmental stages and under biotic stress in apple. Sci Hortic Amst. 2015;197:597–606.
Migocka M, Papierniak A. Identification of suitable reference genes for studying gene expression in cucumber plants subjected to abiotic stress and growth regulators. Mol Breed. 2011;28(3):343–57.
Authors’ contributions
SL, YL and WS designed the research. SL and YL obtained the funding. WS, YY, YJ, XG, YB, JP and JW performed the experiments. WS, YL, LZ, and SL analyzed the data and prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study was supported by the State Key Laboratory for the Conservation and Utilization of Subtropical Agro-Bioresources (201504010028) and the Open Fund of the Key Laboratory of Loquat Germplasm Innovation and Utilization of Putian University (2017002).
Competing interests
The authors have no competing interests to disclose. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Table S1.
Samples used for reference gene selection. Table S2. Primer sequences for reference gene isolation. Figure S1. Melt curve analysis of the selected 11 candidate reference genes. 23 distinct tissues of loquat were tested to show single peak for each primer pair at a specific annealing temperature.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Su, W., Yuan, Y., Zhang, L. et al. Selection of the optimal reference genes for expression analyses in different materials of Eriobotrya japonica. Plant Methods 15, 7 (2019). https://doi.org/10.1186/s13007-019-0391-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13007-019-0391-2