Abstract
Background
New sequencing technologies have enabled the identification of mutations in Ten-eleven-translocation 2 (TET2), an enzyme that catalyzes the conversion of 5-methylcytosine into 5-hydroxymethylcytosine (5-hmC) in myeloid neoplasms. We have recently identified reduced TET2 mRNA expression in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML), which is associated with a poor overall survival in MDS. We herein aimed to investigate TET2 mutations and their impact on TET2 expression in a cohort of patients with myeloid neoplasms, including MDS and AML patients.
Findings
TET2 mutations were observed in 8 out of 19 patients (42 %) with myeloid neoplasms. The TET2 expression profile was similar between in wild type and in TET2 mutated patients.
Conclusion
Our results suggest that TET2 expression is reduced in MDS/AML patients, independently of mutational status.
Similar content being viewed by others
Findings
Introduction
New sequencing technologies have enabled the identification of several mutations in epigenetic regulator genes, such as Ten-eleven-translocation 2 (TET2), which encodes an enzyme that catalyzes the conversion of 5-methylcytosine into 5-hydroxymethylcytosine (5-hmC). Mutations in the coding regions of TET2 lead to loss of TET2 protein function, reduce 5-hmc levels and cause a significant shift towards monocyte/granulocyte differentiation in detriment of erythroid/lymphoid differentiation [1, 2]. TET2 mutations confer a worse prognosis in myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) [3, 4]. We have recently reported that TET2 mRNA expression is downregulated in MDS and AML, and predicts poor overall survival in MDS patients [5]. However, the correlation between the presence of TET2 mutations and TET2 transcript levels has rarely been addressed [4, 6]. We, herein, expanded our previous observations and aimed to investigate TET2 mutational status and the impact of this mutational status on TET2 expression in a cohort of MDS and AML patients. For this purpose, TET2 mutation analysis and TET2 gene expression were tested by Sanger sequencing and quantitative PCR, respectively, in bone marrow samples from healthy donors and myeloid neoplasm patients.
Materials and methods
Patients’ characteristics
Bone marrow samples were obtained from a total of 22 healthy donors, and 19 patients with myeloid neoplasms (MDS = 10 and AML = 9) followed at the outpatient clinics of the University of Campinas, who had also been included in a previous study [5]. The present study was approved by the Institutional and National Review Board in accordance with the Helsinki Declaration. Written informed consent was obtained from all patients who participated in this study. Patients were diagnosed according to the 2008 Word Health Organization criteria [7]. Patients’ characteristics are described in Table 1.
Polymerase chain reaction (PCR) and DNA sequencing
The screening of TET2 mutations was performed on coding exons 3, 4, 5, 6, 7, 8, 9, 10 and 11 (GenBank reference NM_0175628.4). Primer sequences and PCR conditions were previously described [8]. Amplicons were sequenced with an ABI 3500 Genetic Analyzer (Life Technologies, Carlsbad, CA, USA) using the Big Dye terminator V1.1 cycle sequencing kit and analyzed using CLC Main Workblench v.7.6.2 software (Qiagen, Aarhus, Denmark). All alterations found were searched in SNP (dbSNP; http://www.ncbi.nlm.nih.gov/projects/SNP), Ensembl Genome Browser databases (http://asia.ensembl.org/index.html), and in the Catalogue of Somatic Mutations in Cancer (COSMIC; http://cancer.sanger.ac.uk/cosmic).
Quantitative PCR (qPCR)
Total RNA was extracted from cells using the TRIzol reagent (Life Technologies, Carlsbad, CA, USA). The reverse transcription reaction was performed using RevertAid™ First Strand cDNA Synthesis Kit (MBI Fermentas, St. Leon-Rot, Germany). TET2 mRNA level was detected by Maxima Sybr green qPCR master mix (MBI Fermentas, St. Leon-Rot, Germany) in the ABI 7500 Sequence Detection System (Life Technologies) using specific primers: forward 5′- ACGCAAGCCAGGCTAAACA -3′, reverse 5′- GCTGGGACTGCTGCATGA -3′; HPRT1 (hypoxanthine phosphoribosyltransferase 1) was used as the endogenous control: forward 5′-GAACGTCTTGCTCGAGATGTGA-3′, reverse 5′-TCCAGCAGGTCAGCAAAGAAT-3′. The relative quantification value was calculated using the equation, 2-ΔΔCT [9]. A negative ‘No Template Control’ was included for each primer pair. The dissociation protocol was performed at the end of each run to check for non-specific amplifications. Three replicas were run on the same plate for each sample.
Statistical analyses
Statistical analyses were performed using GraphPad Instat 5 (GraphPad Software, Inc., San. Diego, CA, USA). For comparisons, Mann-Whitney test was used for measured factors with two levels. A p value <0.05 was considered as statistically significant.
Results and discussion
We analyzed the mutation status of TET2 exons and the impact of this status on TET2 expression in bone marrow cells from myeloid neoplasm patients. In total, 17 TET2 variants were detected. After excluding confirmed SNPs (P29R [rs12498609], V218M [rs6843141], P363L [rs17253672], G355D [rs61744960], H1778R [rs62621450], I1762V [rs2454206] and L1721W [rs34402524]), TET2 mutations were observed in 8/19 (42 %) patients with myeloid neoplasms [4/10 (40 %) MDS and 4/9 (44 %) AML], including six missense (E709K, Y867H, H924R, S1109P, P1723S and H1868L) and three stop codon (E1073X, S1516X and S1518X) mutations (Fig. 1). A total of nine TET2 mutations were found in exon 3 (n = 5) or exon 11 (n = 3) in eight patients; one patient had two mutations. Four TET2 mutations (E1073X, S1109P, S1518X and H1868L) found in our study had not been previously described in the COSMIC database. Among the six missense mutations, three mutations presented a high probably of damage to protein function, according to Polyphen2 analysis. The PolyPhen2 scores for predicted damage of TET2 Y867H, S1109P and H1868L mutations were 0.999, 0.995 and 1, respectively (http://genetics.bwh.harvard.edu/pph2/). TET2 mutations in exons have been implicated in protein loss-of-function and myeloid neoplasm mechanism of disease [1, 2], which supports our focus on sequencing TET2 coding regions only. The relevance of mutations in the TET2 promoter region has not been explored yet in myeloid neoplasms.
TET2 mutations identified in myelodysplastic syndromes and acute myeloid leukemia patients. In a cohort of 19 patients, nine TET2 mutations were identified in eight patients. Genomic sequencing of protein-coding regions revealed missense (black arrows), and stop codon (red arrows) mutations in TET2; Sanger sequencing analysis is illustrated in the figure. TET2 protein primary structure indicating the domains and specific known conserved motifs are shown: cysteine-rich region (C-rich), double strand beta helix (DSBH). The aminoacid position is indicated
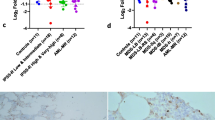
TET2 expression was reduced in MDS/AML patients (median 0.8 [minimum 0.01- maximum 6.41]), compared with healthy donors (2.72 [0.43–31.49]); p <0.005 (Fig. 2a). The standard deviation of TET2 expression for the healthy donors was high in our cohort of patients, which is in agreement with reports from other authors [4, 10]. TET2 expression was similar in TET2 wild type (0.8 [0.01–3.79]) and in mutated patients (1.82 [0.14–6.41]), for the entire cohort, and among patients with MDS and AML only, all p >0.05 (Fig. 2b–d). These findings are in agreement with Jankowska and colleagues [4] and Coutinho and colleagues [6], who reported TET2 downregulation in 16 patients with MDS/myeloproliferative neoplasms and in 12 patients with pediatric MDS, respectively, regardless of TET2 mutational status. Reduced TET2 levels in MDS patients have also been previously reported by our group [5], Li and collaborators [10], and Zhang and collaborators [11]. However, the TET2 mutation was not evaluated in these studies.
TET2 expression, according to TET2 mutational status, in myelodysplastic syndromes and acute myeloid leukemia patients. a qPCR analysis of Ten-eleven-translocation 2 (TET2) mRNA levels in total bone marrow cells from healthy donors and from patients with the diagnosis of myelodysplastic syndromes (MDS)/acute myeloid leukemia (AML). TET2 expression in patients stratified according to TET2 mutational status for the entire cohort of MDS/AML patients (b), for patients with MDS (c) or AML diagnosis (d). The “y” axis represents the relative TET2 mRNA expression. The numbers of subjects studied and the p values (Mann-Whitney) are indicated in the graph. Abbreviations: WT wild type, MUT mutated
Somatic mutations in TET2 have been described in healthy elderly individuals with clonal hematopoiesis and have been associated with alterations in DNA methylation [12]. TET2 mutations have been indicated as early events in clonal evolution and potently lead to the development of preleukemic hematopoietic stem cells [13, 14]. For this reason, growing attention has been given to the hypothesis that TET2 is a gate keeper in hematological malignancies. The occurrence of TET2 mutations may indirectly lead to the disruption of other important genes by inducing epigenetic changes, as recently described for JAK2 [15]. TET2 mutations alter the transcriptional consequences of JAK2 V617F in a cell-intrinsic manner and prevent JAK2 V617F from up-regulating a proliferative program [15]. In addition, Kameda et al. [16] and Rasmussen and collaborators [17] demonstrated, using mouse models, that the loss of Tet2 contributes towards the progression of myeloid neoplasms. Thus, reduced TET2 function, due to loss-of-function mutations, reduced mRNA expression due to epigenetic silencing or not yet elucidated mechanisms may have a great contribution to the malignant phenotype of hematological cancers.
Taken together, our findings suggest that decreased TET2 expression is observed in myeloid neoplasms and does not correlate with TET2 mutation status. Other molecular mechanisms, including mutations of additional genes and/or epigenetic regulation may affect TET2 expression in myeloid neoplasms [18]. TET2 mRNA expression levels, in addition to TET2 mutation status, may also play a role in myeloid neoplasm physiopathology. Future studies to further investigate the mechanisms that lead to reduced TET2 expression in hematological malignancies will be of importance.
Abbreviations
- 5-hmC:
-
5-hydroxymethylcytosine
- AML:
-
acute myeloid leukemia
- COSMIC:
-
catalogue of somatic mutations in cancer
- HPRT1:
-
hypoxanthine phosphoribosyltransferase 1
- JAK2:
-
Janus kinase 2
- MDS:
-
myelodysplastic syndromes
- PCR:
-
polymerase chain reaction
- qPCR:
-
quantitative PCR
- SNPs:
-
single nucleotide polymorphisms
- TET2:
-
ten-eleven-translocation 2
References
Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–43.
Pronier E, Almire C, Mokrani H, Vasanthakumar A, Simon A, da Costa Reis Monte Mor B, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulomonocytic differentiation of human hematopoietic progenitors. Blood. 2011;118:2551–5.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–301.
Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–10.
Scopim-Ribeiro R, Machado-Neto JA, Campos Pde M, Silva CA, Favaro P, Lorand-Metze I, et al. Ten-Eleven-Translocation 2 (TET2) is downregulated in myelodysplastic syndromes. Eur J Haematol. 2015;94:413–8.
Coutinho DF, Monte-Mor BC, Vianna DT, Rouxinol ST, Batalha AB, Bueno AP, et al. TET2 expression level and 5-hydroxymethylcytosine are decreased in refractory cytopenia of childhood. Leuk Res. 2015;39:1103–8.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. 4th ed. Lyon: IARC; 2008.
Traina F, Visconte V, Elson P, Tabarroki A, Jankowska AM, Hasrouni E, et al. Impact of molecular mutations on treatment response to DNMT inhibitors in myelodysplasia and related neoplasms. Leukemia. 2014;28:78–87.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Li S, Fan R, Zhao XL, Wang XQ. CXXC4 mRNA levels are associated with clinicopathological parameters and survival of myelodysplastic syndrome patients. Leuk Res. 2014;38:1072–8.
Zhang W, Shao ZH, Fu R, Wang HQ, Li LJ, Wang J, et al. TET2 expression in bone marrow mononuclear cells of patients with myelodysplastic syndromes and its clinical significances. Cancer Biol Med. 2012;9:34–7.
Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–81.
Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4:149ra118.
Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121:2186–98.
Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, et al. Effect of mutation order on myeloproliferative neoplasms. N Engl J Med. 2015;372:601–12.
Kameda T, Shide K, Yamaji T, Kamiunten A, Sekine M, Taniguchi Y, et al. Loss of TET2 has dual roles in murine myeloproliferative neoplasms: disease sustainer and disease accelerator. Blood. 2015;125:304–15.
Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–22.
Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, et al. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell. 2015;57:662–73.
Acknowledgments
The authors would like to thank Dr. Nicola Conran and Raquel S Foglio for English review, and Tereza Salles for her valuable technical assistance. This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). Grant numbers: 610021/2009-5; 2011/51959-0; 2011/15905-3; 2012/09982-8.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RSR designed the study and experiments, performed all the experiments, statistical analyses, patient database, manuscript preparation, completion and final approval. JAMN participated in the analysis of the data, quantitative PCR experiments, manuscript preparation and final approval. PMC participated in the interpretation of manuscript data, clinical data collection, manuscript editing, and final approval. FSN participated in sequencing experiments. ILM revised the diagnoses and together with FFC participated in the manuscript editing and final approval. STOS participated in patient follow up, manuscript editing and final approval. FT participated in the overall design of the study and experiments, statistical analyses, patient follow up, manuscript preparation, editing, completion and final approval. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Scopim-Ribeiro, R., Machado-Neto, J.A., de Melo Campos, P. et al. Low Ten-eleven-translocation 2 (TET2) transcript level is independent of TET2 mutation in patients with myeloid neoplasms. Diagn Pathol 11, 28 (2016). https://doi.org/10.1186/s13000-016-0476-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13000-016-0476-4