Abstract
Background
Xp11.2 or TFE3 translocation renal cell carcinomas (RCC) and alveolar soft part sarcoma (ASPS) are characterized by chromosome translocations involving the Xp11.2 breakpoint resulting in transcription factor TFE3 gene fusions. The most common translocations documented in TFE3 RCCs are t(X;1) (p11.2;q21) and t(X;17) (p11.2;q25) which leads to fusion of TFE3 gene on Xp11.2 with PRCC or ASPL respectively. TFE3 immunohistochemistry (IHC) has been inconsistent over time due to background staining problems in part related to fixation issues. Karyotyping to detect TFE3 gene rearrangement requires typically unavailable fresh tissue. Reverse transcriptase-polymerase chain reaction (RT-PCR) is generally very challenging due to degradation of RNA in archival material. The study objective was to develop and validate a TFE3 break-apart fluorescence in situ hybridization (FISH) assay to confirm Xp11 translocation RCCs and ASPS.
Methods
Representative sections of formalin-fixed paraffin-embedded tissue blocks were selected in 40 possible cases. Approximately 60 tumor cells were analyzed in the targeted region. The validation of TFE3 FISH was done with 11 negative and two positive cases. Cut off for a positive result was validated as >7.15 % positive nuclei with any pattern of break-apart signals. FISH evaluation was done blinded of the immunohistochemical or karyotype data.
Results
Three out of forty cases were positive for the TFE3 break-apart signals by FISH. The negative cases were reported as clear cell RCC with papillary features (10), clear cell RCC with sarcomatoid areas (2), Papillary RCC with clear cell areas (9), Chromophobe RCC (2), RCC, unclassified type (3) and renal medullary carcinoma (1). 3 of the negative cases were consultation cases for renal tumor with unknown histology. Seven negative cases were soft tissue tumor suspicious for ASPS.
Conclusion
Our study validates the utility of TFE3 break-apart FISH on formalin-fixed paraffin-embedded tissue sections for diagnosis and confirmation of Xp11.2 translocation RCCs and ASPS.
Similar content being viewed by others
Background
Xp11.2 or TFE3 translocation renal cell carcinoma (RCC) is one of the new entities added in the 2004 World Health Organization (WHO) classification of renal tumors [1]. They are characterized by translocations involving the TFE3 transcription factor located at Xp11.2 locus. The five known gene fusion partners of TFE3 are papillary 1 renal cell carcinoma (PRCC), alveolar soft part sarcoma locus (ASPL), polypyrimidine tract-binding protein-associated splicing factor (PSF), non-POU domain-containing octamer-binding (NonO, p54nrb), and clathrin heavy chain (CLTC) genes, situated on chromosome loci 1q21 [2–5], 17q25 [6–8], 1p34 [9], Xq12 [9], and 17q23 [10] respectively. The t(X; 17) (p11.2; q25) or TFE3-ASPL translocation in RCC and alveolar soft part sarcoma (ASPS) contain the identical TFE3-ASPL fusion transcript; however, the t(X; 17) translocation is consistently balanced (reciprocal) in the Xp11.2 translocation RCC and unbalanced in the ASPS [11]. In translocation RCC involving t(X; 10) (p11; q23) or t(X;3) (p11.2;q23), the participating gene which fuses to TFE3 remains unknown [12, 13].
Xp11.2 RCCs are classically recognized as pediatric RCC affecting children and young adults [14–20]. These tumors are considered aggressive with early age of onset and variable morphologic features including clear cell or eosinophilic morphology, and papillary or alveolar architecture [21]. This entity seems to be underdiagnosed and misclassified as clear cell or papillary RCC in adults, because of overlapping morphologic features. Definite diagnosis in suspicious cases requires confirmation of the presence of TFE3 protein by immunohistochemistry (IHC) or TFE3 gene rearrangement by karyotyping or reverse transcriptase-polymerase chain reaction (RT-PCR) to detect chimeric TFE3 mRNA fusion transcripts. TFE3 IHC, though less time consuming and relatively less expensive, has been inconsistent over time due to background staining problems [22]. Besides variable fixation time, especially common in consultation cases, gives variable results [23]. False positive may often be seen due to titration problem. Karyotyping requires fresh tissue which is generally not sent for cytogenetic analysis of adult renal masses in most institutes. RT-PCR on formalin-fixed, paraffin-embedded (FFPE) tissue is infrequently used as a diagnostic tool. It is also very challenging as fresh tissue is rarely available and there is degradation of RNA in the archival material. Moreover, it may necessitate multiple PCRs to cover all the known partners of TFE3.
ASPS is a rare soft tissue tumor which has ASPL-TFE3 gene fusion as a result of unbalanced translocation der (17) t(X;17) (p11;q25) or rarely a balanced translocation t(X;17) (p11;q25). The classical alveolar pattern surrounded by fibrous septa and large round to oval tumor cells is fairly non-specific requiring help from ancillary studies [11].
As the morphology of both Xp11.2 RCCs and ASPS are non-specific and there are a lot of technical difficulties with the available ancillary tools – IHC limited by equivocal results, karyotyping limited by availability of viable tumor cells and RT-PCR limited by RNA quality, we tried to validate and utilize TFE3 break-apart fluorescence in-situ hybridization (FISH) assay in FFPE tissue to confirm the diagnosis of an Xp11.2 RCC and ASPS. Eventually, we find that a break-apart FISH assay is an excellent diagnostic and confirmatory test in the evaluation of TFE3 gene rearrangement in primary as well as metastatic Xp11.2 RCCs and other TFE3 tumors.
Methods
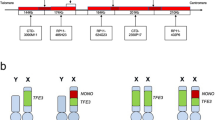
FFPE tissue blocks were serially sectioned at 4 μ intervals. Hematoxylin and eosin (H&E) sections were used to determine the area of the tissue to be targeted for analysis. FISH slides were deparaffinized in xylene twice for 10 min, dehydrated twice with 100 % ethanol and then pretreated using the Vysis Paraffin Pretreatment Kit (Abbott Molecular, Des Plaines, IL). Slides were digested for 36 min in protease solution (0.5 mg/ml) at 37 °C. TFE3 FISH was performed using a dual-color break apart probe labeled in Texas Red and FITC (Abnova Co., Taipei, Taiwan). The target slide was denatured in 70 % Formamide at 75 °C for 5 min and dehydrated in 70, 85, and 100 % ethanol. Slides were incubated with probe overnight at 42 °C in a humidified chamber. Post-hybridization washes were performed using 2 × SSC/0.3 % Igepal at 73 °C for 2 min (Sigma, St. Louis, MO). Slides were air-dried in the dark and counterstained with 4,6-diamidino-2-phenylindole (DAPI)/antifade (Abbott Molecular). All slides were kept at 4 °C in the dark after hybridization. Analysis was performed using a Leica DM5500 B fluorescence microscope (Leica Microsystems) and CytoVision Workstation (Applied Imaging, Santa Clara, CA) equipped with Chroma Technology 83,000 filter set with single and dual band excitors for Texas Red, Spectrum Green, and DAPI (uv 360 nm) (Abbott Molecular). Only individual and well delineated cells were scored. Overlapping cells were excluded from the analysis. Approximately 60 tumor cell nuclei were analyzed in the targeted region by each of the 2 experienced technicians. The expected normal nuclei had 2 fusion signals reflecting intact TFE3 alleles in a female individual and 1 fusion signal reflecting an intact TFE3 allele in a male individual. The signal pattern 1 red 1 green 1 fusion (yellow) was the most common positive pattern for a balanced TFE3 translocation in a female individual, whereas the signal pattern 1 red 1 green was the most typical positive pattern for a balanced TFE3 rearrangement in a male individual (Fig. 1). Unbalanced translocations in a female individual yielded a 1 red 2 fusion pattern. To be scored as a break apart and to avoid false positive, the signals had to be separated by >2 signal diameters. To avoid false negative in a 4 μ section where red or green signal may be out of the visible plane of section, a minimum of 60 nuclei were evaluated per case.
To determine a cutoff for a positive result, 11 negative and 2 positive cases were evaluated using statistical criteria. Cut off for a positive result was calculated as >7.15 % positive nuclei with any pattern of break-apart signals when 60 nuclei were scored thoroughly. FISH evaluation was done blinded of the IHC or karyotype data.
Results
A total of 40 cases were evaluated for a TFE3 gene rearrangement using a break-apart FISH which comprised 33 renal neoplasms and 7 soft tissue sarcomas suspicious for Xp11.2 RCC and ASPS respectively. The evaluation was done blinded of morphologic possibility, karyotype, and TFE3 IHC results. Of the 40 cases, 3 cases were positive for the TFE3 break-apart signals by FISH. On retrospective analysis of these 3 cases, one was a 26-year-old female with unilateral renal mass diagnosed morphologically as chromophobe RCC arising in a cyst, stage pT1b, positive for CD-10, E-cadherin, caveolin and parvalbumin; focally positive for AE1/AE3, CAM5.2, CK7, EMA, RCC antigen and colloidal iron; and negative for carbonic anhydrase IX and BerEP4. This case was later subjected to TFE3 IHC which was positive. The cytogenetic analysis of this case revealed a novel translocation involving t(X;19) (p11.2;q13.1). The other was a renal tumor consultation case in a 10-year-old male child. The third positive case was a 39-year-old male with 12.8 cm retroperitoneal mass diagnosed as pT2b ASPS in 2002. He had a left upper lobe lung metastasis in 2006 and a jejunal metastasis in 2014. This jejunal metastasis revealed the TFE3 gene rearrangement when subjected to break apart FISH. The negative cases were reported as clear cell RCC with papillary features (10), clear cell RCC with sarcomatoid areas (2), Papillary RCC with clear cell areas (9), Chromophobe RCC (2), RCC, unclassified type (3) and renal medullary carcinoma (1). Three of the negative cases were consultation cases for renal tumor with unknown histology. Seven negative cases were soft tissue tumor suspicious for ASPS.
FISH evaluation of tissue used for validation purpose showed break-apart signals in both positive cases, each of TFE3 RCC and ASPS, but in none of the non-Xp11.2 RCCs and normal tissue.
Discussion
Xp11.2 translocation carcinoma is a recently recognized entity in the 2004 WHO renal tumor classification [1]. It is an uncommon tumor generally arising in children and young adults but recently has been recognized in adults as old as 58 years [24–36]. To date, 5 distinct reciprocal gene fusion partners of the TFE3 gene located at Xp11.2 have been recognized in RCCs which include ASPL, PRCC, PSF, NonO, and CLTC situated on chromosome loci 17q25, 1q21, 1p34, Xq12, and 17q23 respectively [2–10]. Another rare group of renal carcinomas showing the translocation t(6; 11) (p21; q12) involving transcription factor EB (TFEB) has also been reported [37, 38]. TFE3 and TFEB belong to the microphthalmia transcription factor (MiTF) subfamily, which also includes MiTF and transcription factor EC. Argani and Ladanyi have proposed regrouping these neoplasms into the category of MiTF/TFE family translocation carcinomas [21].
The most characteristic morphologic pattern of the Xp11.2 translocation RCC is that of an epithelioid neoplasm with predominance of clear cells, papillary architecture and psammoma bodies. TFE3 translocation RCCs regularly express CD10 and the RCC marker, and most express the renal transcription factors PAX2 and PAX8. In contrast, they are less immunoreactive for epithelial markers such as cytokeratins and EMA [29]. Few of them are reactive for melanocytic markers such as Melan A and HMB45. Infrequently, they may also express papain-like cystein protease cathepsin K. These tumors show characteristically strong nuclear TFE3 immunostaining [39, 40].
ASPS is a rare soft tissue tumor that harbors the ASPL-TFE3 gene fusion. These tumors are most often seen in the deep soft tissues of the extremities. Classically, the tumors show the distinctive alveolar growth pattern and strong nuclear immunostaining for TFE3 [41].
The morphologic differential diagnosis of Xp11.2 translocation RCC is quite broad. The most common renal neoplasms that mimic TFE3 RCCs are those with clear cells and papillary architecture. These include clear cell RCC with focal papillary/pseudopapillary areas, papillary RCC with focal clear cell areas and clear cell papillary RCC. Diffuse carbonic Anhydrase-IX (CA-IX) immune-labeling of clear cell RCC and cytokeratin 7 labeling of papillary RCC may help differentiate the TFE3 RCCs, which will show strong nuclear immunoreactivity for TFE3 IHC. Clear cell papillary RCC typically shows a branching tubular architecture, has apically aligned nuclei with subnuclear clearing, and reveals a low nuclear grade. Other neoplasms that may be confused with Xp11.2 translocation RCC include chromophobe RCC. Diffuse CD117 labeling favors chromophobe RCC over Xp11 translocation RCC.
Besides Xp11 translocation RCC and ASPS, perivascular epithelioid cell tumors (PEComas) have also shown immunoreactivity for TFE3. Folpe et al. showed that five of 17 PEComas were TFE3 positive [42]. Recently, a case of PEComa with PSF-TFE3 gene fusion proven by FISH and RT-PCR has been reported [43]. Interestingly, Argani et al. reported a distinctive type of renal cancer with overlapping features of PEComa, Xp11 translocation carcinoma, and melanoma [44].
The diagnosis of Xp11 translocation carcinoma can be problematic in cases of unusual clinical or morphologic presentation or technical difficulty with TFE3 IHC staining. TFE3 IHC may be inconsistent due to many reasons like variation in staining in different lot because of polyclonal antibody, fixation dependence of the antibody and the subjectivity in interpretation of the TFE3 IHC. As such, cytogenetic examination of fresh tissue remains the gold standard. When fresh tissue is unavailable, FISH and/or RT-PCR can be performed on FFPE tissues. However, RT-PCR of archival tissue is difficult due to degradation of RNA, insufficient extraction efficiency and difficulty with the availability of adequate material in small biopsy samples. In addition, Xp11.2 translocation RCC has at least five known fusion partners with TFE3 making RT-PCR more time consuming and labor intensive. Potential unknown translocation(s) involving the TFE3 gene may yield false negative results. FISH on the other hand represents a cost and time-efficient method that uses FFPE tissue. Aulmann et al. has reported the feasibility of detecting the ASPS-TFE3 gene fusion in ASPS with both split and fusion probes [45]. Their paper is the first development of a FISH assay for the detection of TFE3 gene translocation in paraffin-embedded tissues. The advantage of the break apart probe type FISH assay is that though the fusion partner with TFE3 is not identified, but potentially all translocations involving TFE3 can be detected. Because all the probes target the X chromosome, the patient’s sex is important for FISH interpretation. The male patient should have one pair of signals (1 X chromosome) and the female patient should have two pairs of signals (2 X chromosomes).
Conclusion
In this study, we validated a TFE3 break apart FISH assay to aid as a relatively rapid test for detecting Xp11.2 translocation in cases of TFE3 RCC and ASPS. This FISH assay can be utilized as an adjunct to morphology and immunohistochemistry to diagnose TFE3-associated carcinomas and other neoplasms.
References
Eble JN, Sauter G, Epstein JI, Sesterhenn IA. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. Lyon: IARC Press; 2004.
de Jong B, Molenaar IM, Leeuw JA, Idenberg VJ, Oosterhuis JW. Cytogenetics of a renal adenocarcinoma in a 2-year-old child. Cancer Genet Cytogenet. 1986;21:165–9.
Meloni AM, Dobbs RM, Pontes JE, Sandberg AA. Translocation (X;1) in papillary renal cell carcinoma. A new cytogenetic subtype. Cancer Genet Cytogenet. 1993;65:1–6.
Sidhar SK, Clark J, Gill S, Hamoudi R, Crew AJ, Gwilliam R, et al. The t(X;1)(p11.2;q21.2) translocation in papillary renal cell carcinoma fuses a novel gene PRCC to the TFE3 transcription factor gene. Hum Mol Genet. 1996;5:1333–8.
Weterman MA, Wilbrink M, Geurts van Kessel A. Fusion of the transcription factor TFE3 gene to a novel gene, PRCC, in t(X;1)(p11;q21)-positive papillary renal cell carcinomas. Proc Natl Acad Sci U S A. 1996;93:15294–8.
Tomlinson GE, Nisen PD, Timmons CF, Schneider NR. Cytogenetics of a renal cell carcinoma in a 17-month-old child. Evidence for Xp11.2 as a recurring breakpoint. Cancer Genet Cytogenet. 1991;57:11–7.
Argani P, Antonescu CR, Illei PB, Lui MY, Timmons CF, Newbury R, et al. Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Pathol. 2001;159:179–92.
Heimann P, El Housni H, Ogur G, Weterman MA, Petty EM, Vassart G. Fusion of a novel gene, RCC17, to the TFE3 gene in t(X;17)(p11.2;q25.3)-bearing papillary renal cell carcinomas. Cancer Res. 2001;61:4130–5.
Clark J, Lu YJ, Sidhar SK, Parker C, Gill S, Smedley D, et al. Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene. 1997;15:2233–9.
Argani P, Ladanyi M. Distinctive neoplasms characterised by specific chromosomal translocations comprise a significant proportion of paediatric renal cell carcinomas. Pathology. 2003;35:492–8.
Ladanyi M, Lui MY, Antonescu CR, Krause-Boehm A, Meindl A, Argani P, et al. The der(17)t(X;17)(p11;q25) of human alveolar soft part sarcoma fuses the TFE3 transcription factor gene to ASPL, a novel gene at 17q25. Oncogene. 2001;20:48–57.
Argani P, Ladanyi M. The evolving story of renal translocation carcinomas. Am J Clin Pathol. 2006;126:332–4.
Dijkhuizen T, van den Berg E, Wilbrink M, Weterman M, Geurts van Kessel A, Störkel S, et al. Distinct Xp11.2 breakpoints in two renal cell carcinomas exhibiting X; autosome translocations. Genes Chromosomes Cancer. 1995;14:43–50.
Altinok G, Kattar MM, Mohamed A, Poulik J, Grignon D, Rabah R. Pediatric renal carcinoma associated with Xp11.2 translocations/TFE3 gene fusions and clinicopathologic associations. Pediatr Dev Pathol. 2005;8:168–80.
Leuschner I, Harms D, Schmidt D. Renal cell carcinoma in children: histology, immunohistochemistry, and follow-up of 10 cases. Med Pediatr Oncol. 1991;19:33–41.
Ramphal R, Pappo A, Zielenska M, Grant R, Ngan BY. Pediatric renal cell carcinoma: clinical, pathologic, and molecular abnormalities associated with the members of the mit transcription factor family. Am J Clin Pathol. 2006;126:349–64.
Rao Q, Chen JY, Wang JD, Ma HH, Zhou HB, Lu ZF, et al. Renal cell carcinoma in children and young adults: clinicopathological, immunohistochemical, and VHL gene analysis of 46 cases with follow-up. Int J Surg Pathol. 2011;19:170–9.
Renshaw AA, Granter SR, Fletcher JA, Kozakewich HP, Corless CL, Perez-Atayde AR. Renal cell carcinomas in children and young adults: increased incidence of papillary architecture and unique subtypes. Am J Surg Pathol. 1999;23:795–802.
Winarti NW, Argani P, De Marzo AM, Hicks J, Mulyadi K. Pediatric renal cell carcinoma associated with Xp11.2 translocation/TFE3 gene fusion. Int J Surg Pathol. 2008;16:66–72.
Wu A, Kunju LP, Cheng L, Shah RB. Renal cell carcinoma in children and young adults: analysis of clinicopathological, immunohistochemical and molecular characteristics with an emphasis on the spectrum of Xp11.2 translocation-associated and unusual clear cell subtypes. Histopathology. 2008;53:533–44.
Argani P, Ladanyi M. Translocation carcinomas of the kidney. Clin Lab Med. 2005;25:363–78.
Mosquera JM, Dal Cin P, Mertz KD, Perner S, Davis IJ, Fisher DE, et al. Validation of a TFE3 break-apart FISH assay for Xp11.2 translocation renal cell carcinomas. Diagn Mol Pathol. 2011;20:129–37.
Green WM, Yonescu R, Morsberger L, Morris K, Netto GJ, Epstein JI, et al. Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol. 2013;37:1150–63.
Mansouri D, Dimet S, Couanet D, Terrier-Lacombe MJ, Vasiliu V, Khalifa C, et al. Renal cell carcinoma with an Xp11.2 translocation in a 16-year-old girl: a case report with cytological features. Diagn Cytopathol. 2006;34:757–60.
Schinstine M, Filie AC, Torres-Cabala C, Abati A, Linehan WM, Merino M. Fine-needle aspiration of a Xp11.2 translocation/TFE3 fusion renal cell carcinoma metastatic to the lung: report of a case and review of the literature. Diagn Cytopathol. 2006;34:751–6.
Meyer PN, Clark JI, Flanigan RC, Picken MM. Xp11.2 translocation renal cell carcinoma with very aggressive course in five adults. Am J Clin Pathol. 2007;128:70–9.
Rais-Bahrami S, Drabick JJ, De Marzo AM, Hicks J, Ho C, Caroe AE, et al. Xp11 translocation renal cell carcinoma: delayed but massive and lethal metastases of a chemotherapy-associated secondary malignancy. Urology. 2007;70:178.
Suzigan S, Drut R, Faria P, Argani P, De Marzo AM, Barbosa RN, et al. Xp11 translocation carcinoma of the kidney presenting with multilocular cystic renal cell carcinoma-like features. Int J Surg Pathol. 2007;15:199–203.
Argani P, Olgac S, Tickoo SK, Goldfischer M, Moch H, Chan DY, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol. 2007;31:1149–60.
Armah HB, Parwani AV. Renal cell carcinoma in a 33-year-old male with an unusual morphology and an aggressive clinical course: possible Xp11.2 translocation. Pathology. 2008;40:306–8.
Camparo P, Vasiliu V, Molinie V, Couturier J, Dykema KJ, Petillo D, et al. Renal translocation carcinomas: clinicopathologic, immunohistochemical, and gene expression profiling analysis of 31 cases with a review of the literature. Am J Surg Pathol. 2008;32:656–70.
Geller JI, Argani P, Adeniran A, Hampton E, De Marzo A, Hicks J, et al. Translocation renal cell carcinoma: lack of negative impact due to lymph node spread. Cancer. 2008;112:1607–16.
Hintzy MC, Camparo P, Vasiliu V, Peyromaure M, Vieillefond A. Renal carcinoma associated with MiTF/TFE translocation: report of six cases in young adults. Prog Urol. 2008;18:275–80.
Komai Y, Fujiwara M, Fujii Y, Mukai H, Yonese J, Kawakami S, et al. Adult Xp11 translocation renal cell carcinoma diagnosed by cytogenetics and immunohistochemistry. Clin Cancer Res. 2009;15:1170–6.
Bovio IM, Allan RW, Oliai BR, Hampton T, Rush DS. Xp11.2 translocation renal carcinoma with placental metastasis: a case report. Int J Surg Pathol. 2011;19:80–3.
Koie T, Yoneyama T, Hashimoto Y, Kamimura N, Kusumi T, Kijima H, et al. An aggressive course of Xp11 translocation renal cell carcinoma in a 28-year-old man. Int J Urol. 2009;16:333–5.
Argani P, Hawkins A, Griffin CA, Goldstein JD, Haas M, Beckwith JB, et al. A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol. 2001;158:2089–96.
Argani P, Laé M, Hutchinson B, Reuter VE, Collins MH, Perentesis J, et al. Renal carcinomas with the t(6;11)(p21;q12): clinicopathologic features and demonstration of the specific alpha-TFEB gene fusion by immunohistochemistry, RTPCR, and DNA PCR. Am J Surg Pathol. 2005;29:230–40.
Martignoni G, Pea M, Gobbo S, Brunelli M, Bonetti F, Segala D, et al. Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol. 2009;22:1016–22.
Martignoni G, Gobbo S, Camparo P, Brunelli M, Munari E, Segala D, et al. Differential expression of cathepsin-K in neoplasms harbouring TFE3 gene fusions. Mod Pathol. 2011;24:1313–9.
Folpe AL, Deyrup AT. Alveolar soft-part sarcoma: a review and update. J Clin Pathol. 2006;59:1127–32.
Folpe AL, Mentzel T, Lehr HA, Fisher C, Balzer BL, Weiss SW. Perivascular epithelioid cell neoplasms of soft tissue and gynecologic origin: a clinicopathologic study of 26 cases and review of the literature. Am J Surg Pathol. 2005;29:1558–75.
Tanaka M, Kato K, Gomi K, Matsumoto M, Kudo H, Shinkai M, et al. Perivascular epithelioid cell tumor with SFPQ/PSF-TFE3 gene fusion in a patient with advanced neuroblastoma. Am J Surg Pathol. 2009;33:1416–20.
Argani P, Aulmann S, Karanjawala Z, Fraser RB, Ladanyi M, Rodriguez MM. Melanotic Xp11 translocation renal cancers: a distinctive neoplasm with overlapping features of PEComa, carcinoma, and melanoma. Am J Surg Pathol. 2009;33:609–19.
Aulmann S, Longerich T, Schirmacher P, Mechtersheimer G, Penzel R. Detection of the ASPSCR1-TFE3 gene fusion in paraffin-embedded alveolar soft part sarcomas. Histopathology. 2007;50:881–6.
Funding
Authors do not have any financial associations or any source of funding to disclose.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DP drafted the manuscript, acquired the data, analyzed and interpreted the results and data and also participated in its design and coordination. SR, KC, ALM carried out the FISH studies and did the FISH evaluation. GQG, SB, RD participated in the design of the study and reviewed the manuscript and results and revised it critically for important intellectual content. AVP conceived of the study and helped to draft the manuscript and also revised it critically for important intellectual content. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Pradhan, D., Roy, S., Quiroga-Garza, G. et al. Validation and utilization of a TFE3 break-apart FISH assay for Xp11.2 translocation renal cell carcinoma and alveolar soft part sarcoma. Diagn Pathol 10, 179 (2015). https://doi.org/10.1186/s13000-015-0412-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13000-015-0412-z