Abstract
Background
Single nucleotide polymorphisms (SNPs) in lipoprotein lipase gene (LPL) have been shown to influence metabolism related to lipid phenotypes. Dietary factors have been shown to modify the association between LPL SNPs and lipids; however, to date, there are no studies in South Asians. Hence, we tested for the association of four common LPL SNPs with plasma lipids and examined the interactions between the SNPs and dietary factors on lipids in 1,845 Asian Indians.
Methods
The analysis was performed in 788 Type 2 diabetes cases and 1,057 controls randomly chosen from the cross-sectional Chennai Urban Rural Epidemiological Study. Serum triacylglycerol (TAG), serum total cholesterol, and high-density lipoprotein cholesterol (HDL-C) were measured using a Hitachi-912 autoanalyzer (Roche Diagnostics GmbH, Mannheim, Germany). Dietary intake was assessed using a semi-quantitative food frequency questionnaire. The SNPs (rs1121923, rs328, rs4922115 and rs285) were genotyped by polymerase chain reaction followed by restriction enzyme digestion and 20% of samples were sequenced to validate the genotypes obtained. Statistical Package for Social Sciences for Windows version 22.0 (SPSS, Chicago, IL) was used for statistical analysis.
Results
After correction for multiple testing and adjusting for potential confounders, SNPs rs328 and rs285 showed association with HDL-C (P = 0.0004) and serum TAG (P = 1×10−5), respectively. The interaction between SNP rs1121923 and fat intake (energy %) on HDL-C (P = 0.003) was also significant, where, among those who consumed a high fat diet (28.4 ± 2.5%), the T allele carriers (TT + XT) had significantly higher HDL-C concentrations (P = 0.0002) and 30% reduced risk of low HDL-C levels compared to the CC homozygotes. None of the interactions on other lipid traits were statistically significant.
Conclusion
Our findings suggest that individuals carrying T allele of the SNP rs1121923 have increased HDL-C levels when consuming a high fat diet compared to CC homozygotes. Our finding warrants confirmation in prospective studies and randomized controlled trials.
Similar content being viewed by others
Background
The Asian Indian population has a greater predisposition to non-communicable diseases such as type 2 diabetes (T2D) [1, 2] and cardiovascular disease (CVD) [3] compared to Europeans. Despite low body mass index (BMI), Indians are characterized by a higher frequency of hyperinsulinemia [4], insulin resistance [5], dyslipidemia with hypertriacylglycerolemia and low high-density lipoprotein cholesterol (HDL-C) levels [6] and increased visceral fat, which are referred to as ‘Asian Indian Phenotype’ or ‘Atherogenic Lipoprotein Phenotype’ [7, 8]. Blood lipid levels are heritable phenotypes and findings from previous studies show that the blood concentrations of HDL-C, low density lipoprotein cholesterol (LDL-C) and triacylglycerol (TAG) have a strong inheritance [9].
Genetic studies have implicated several gene loci in the predisposition to dyslipidemia in Asian Indians [10–13], one of which is lipoprotein lipase (LPL) [14–16]. LPL plays an important role in the metabolism of HDL-C, where it has been shown to hydrolyze TAG in TAG-rich lipoproteins such as chylomicrons and very low density lipoproteins [17]. It has been postulated that increased activity of LPL enzyme enhances the release of components of TAG-rich lipoproteins which are then transferred to HDL to raise HDL levels; conversely, lack of LPL can retard the transfer of these components to HDL [18]. Several candidate gene studies have shown an association between single nucleotide polymorphisms (SNPs) in LPL and lipid traits in various populations including Asian Indians [11, 12, 19–22]. Genome wide association studies have also demonstrated strong evidence for the association of LPL polymorphisms with HDL-C concentrations [23–25]. A few studies have examined the LPL gene–diet interactions in association with HDL-C [26–30]; however, the findings have been quite inconsistent due to variations in sample size, dietary factors and the selection of LPL polymorphisms.
Given that there are no gene-diet interaction studies, to date, in Asian Indian populations, we examined the association of four common LPL SNPs [Val135Val C/T (rs1121923), Ser447Ter C/G (rs328), G/A (rs4922115) and Pvu II C/T (rs285)] with HDL-C and investigated the interactions of these four polymorphisms with dietary carbohydrate, fat and protein percentage on HDL-C in up to 1,845 participants (788 T2D cases and 1,057 controls) from the cross-sectional Chennai Urban Rural Epidemiological Study (CURES). In addition, we examined the genetic associations and interactions for other lipid traits such as TAG, LDL-C and total cholesterol in these participants.
Methods
Study population
One thousand eight hundred and forty five participants comprising 788 cases with T2D and 1,057 controls with normal glucose tolerance (NGT) were randomly chosen from the urban component of the Chennai Urban Rural Epidemiological Study (CURES), an epidemiological study conducted on a representative population (age >20 years) of Chennai (formerly Madras), the fourth largest city in India. The detailed methodology of the study participants is published elsewhere [31]. Briefly, in Phase 1 of CURES, 26,001 individuals were recruited based on a systematic random sampling technique. Participants with self-reported diabetes taking drug treatment for diabetes were classified as “known diabetes subjects.” All known diabetes participants (n = 1,529) were invited to visit the center for detailed studies. In addition, every 10th individual of the 26,001 individuals without known diabetes was invited to undergo oral glucose tolerance tests using a 75-g oral glucose load (dissolved in 250 ml of water) (Phase 3 of CURES). Those who were confirmed by oral glucose tolerance test to have 2-h plasma glucose value ≥11.1 mmol/l based on World Health Organization (WHO) consulting group criteria were labeled as “newly detected diabetes subjects” and those with 2-h plasma glucose value <7.8 mmol/l as being NGT [32]. CURES participants who were on lipid lowering drugs such as statins, fibrates and niacin were excluded from the study (n = 134). On the basis of the National Cholesterol Education Program-Adult Treatment Panel III (NCEP-ATP III) guidelines [33] the study population was divided into those with normal HDL-C (≥1.03 mmol/l for men; ≥1.3 mmol/l for women) and low HDL-C (<1.03 mmol/l for men; <1.3 mmol/l for women). Written informed consent was obtained from each study participant, and the study was approved by the Madras Diabetes Research Foundation Institutional Ethics Committee.
Phenotype measurements
Anthropometric measurements including weight, height, and waist were obtained using standardized techniques. The BMI was calculated as weight (in kg) divided by the square of height (in m). Biochemical analyses were performed on a Hitachi-912 Auto Analyzer (Hitachi, Mannheim, Germany) using kits supplied by Roche Diagnostics (Mannheim). Fasting plasma glucose (glucose oxidase–peroxidase method), serum total cholesterol (cholesterol oxidase-phenol-4-amino-antipyrene peroxidase method), serum TAG (glycerol phosphatase oxidase-phenol-4-amino-antipyrene peroxidase method), and HDL-C (direct method; polyethylene glycol-pretreated enzymes) were measured. Low-density lipoprotein cholesterol was calculated using the Friedewald formula [34]. Glycated haemoglobin (HbA1c) was estimated by high-performance liquid chromatography using a Variant™ machine (Bio-Rad, Hercules, CA, USA). Serum insulin concentration was estimated using an enzyme-linked immunosorbent assay (Dako, Glostrup, Denmark).
Dietary assessment
Dietary intakes were assessed using a previously validated and published [35] interviewer administered semi-quantitative food frequency questionnaire (FFQ) containing 222 food items to estimate food intake over the past year. Briefly, individuals were asked to estimate the usual frequency (number of times per day, week, month or year/never) and the usual serving size of the portion of the various food items in the FFQ. Common household measures such as household cups, bowls, ladles, spoons (for the cooked foods like vegetables), wedges, circles of different diameter and visual atlas of different sizes of fruits (small, medium, large) were shown to assist the individuals in estimating portions. A detailed description of the development of FFQ and the data on reproducibility and validity had been published [35]. EpiNu, an in-house database was used to assess the average daily food and nutrient intake.
SNP selection and genotyping
Four common SNPs in the LPL gene (rs285, rs328, rs4922115 and rs1121923) were chosen for the present study. The SNPs rs328 and rs285 were chosen based on their previous associations with lipid outcomes in several populations [11, 12, 22, 36]. The SNPs rs1121923 and rs4922115 were identified from the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) based on their location in the exon 3 and 3’UTR regions, respectively, assuming that variations in the coding and regulatory regions might confer a functional effect on the gene expression. The SNPs were genotyped by polymerase chain reaction on a GeneAmp® PCR system 9700 thermal cycler (Applied Biosystems, Foster City, CA) followed by restriction enzyme digestion (New England Biolabs, Inc., Beverly, MA). The program usually had the following steps: initial denaturation at 95 °C for 10 min, 30–35 cycles of denaturation at 95 °C for 45 s, primer-annealing at 58 °C for rs1121923 and rs285 SNPs and 60 °C for rs328 and rs4922115 SNPs for 45 s, and primer extension at 72 °C for 45 s, followed by a final extension at 72 °C for 5 min. The restrictions enzymes used for genotyping the SNPs were Sau96I for rs1121923, Mnl I enzyme for rs328, the Eco RV enzyme for rs4922115 and Pvu II for rs285. Agarose gel electrophoresis was used to detect the amplification of PCR reaction and the restriction enzyme digested products. To ensure that the genotyping was of adequate quality, we performed random duplicates in 10% of the samples. The assays were performed by a technician who was masked to the phenotype, and there was 98% concordance in the genotyping. Variants were also confirmed by direct sequencing using an ABI 3500 genetic analyzer (Applied Biosystems, Foster City, CA). Population stratification was performed using a case–control approach at 6 unlinked marker loci believed to be unrelated to the disease under study, but known to have allelic diversity among different populations [37].
Statistical analysis
Statistical Package for Social Sciences for Windows version 22.0 (SPSS, Chicago, IL) was used for statistical analysis. The effects of the variants on quantitative and categorical variables were analyzed. Allele frequencies were estimated by gene counting. Agreement with Hardy–Weinberg equilibrium (HWE) expectations was tested using a χ2goodness-of-fit test. Comparison of the means between the two groups was analyzed by independent t-test. The χ2 test was used to compare the proportions of genotypes or alleles. Dominant model was used, given the low frequency of minor allele homozygotes. Linear regression was used to examine the association of the LPL SNPs with various lipid outcomes. The SNP-diet interactions on lipid traits were tested by including the interaction term in linear regression models. Models were adjusted for age, gender, BMI (as continuous), T2D status and total energy intake wherever appropriate. Multiple testing correction using Bonferroni method was applied separately for the testing of main and interaction effects (i.e., association of the four SNPs with HDL-C and interaction with dietary factors on HDL-C levels) [P ≤ 0.003 (=0.05/20) was considered statistically significant] and additional analyses (i.e., association of the four SNPs with other lipid traits and interaction with dietary factors on other lipid traits) [P ≤ 0.001 (=0.05/48) was considered statistically significant].
Results
Table 1 shows the anthropometric and biochemical characteristics of NGT and T2D participants. T2D cases had markedly increased levels of TAG, LDL-C and total cholesterol, while HDL-C was significantly lower in cases compared to controls (P < 0.003 for all comparisons). In our study population, the genotype distributions for the four LPL polymorphisms were GG: 68.5%, GA: 27.5% and AA: 4.0% (rs4922115); CC: 87.5%, CT: 12.0% and TT: 0.5% (rs1121923); CC: 72.8%, CG: 25.2% and GG: 2.00% (rs328); and CC: 40.9%, CT: 45.6% and TT: 13.5% (rs285). All the four SNPs were in HWE (P > 0.05).
The association between LPL SNPs and HDL-C levels is presented in Table 2. Of the four LPL variants, the SNP rs328 alone showed a significant and a consistent association with HDL-C concentrations [both as continuous and categorical variable (stratified based on NCEP ATP III guidelines)] under a dominant model after correction for multiple testing (P = 0.0004 for the continuous variable and P = 0.001 for the categorical variable). The minor allele (G) carriers of the SNP rs328 had 5% higher HDL-C compared to the homozygous carriers of the common ‘C’ allele.
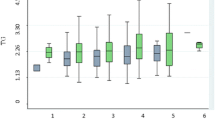
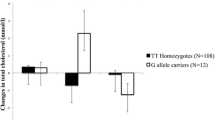
In the interaction analysis, after correction for multiple testing, none of the interactions were statistically significant except for the interaction between SNP rs1121923 and fat intake (energy %) on HDL-C (P = 0.003) (Table 3), where among those who consumed a high fat diet (3rd tertile: 28.4 ± 2.5%), the T allele carriers had significantly higher HDL-C concentrations compared to the CC homozygotes (P = 0.0002) (Fig. 1a). To test whether this interaction was significant on HDL-C as a categorical variable, we stratified the data based on normal and low HDL-C levels according to the NCEP ATP III guidelines for dyslipidemia [33] and found that among those who consumed a high fat diet, the individuals who carried the T allele had 30% reduced risk of low HDL-C levels compared to the CC homozygotes (P = 0.001) (Fig. 1b). We further investigated the interaction of the SNP with various fat subclass intakes on HDL-C but none of the interactions were statistically significant [monounsaturated (MUFA) (P = 0.36), polyunsaturated (PUFA) (P = 0.22) and saturated fatty acids (SFA) (P = 0.46)].
a Interaction between Lipoprotein lipase gene SNP rs1121923 and fat energy intake (%) on HDL-C concentrations (Pinteraction = 0.003). Among those who consumed a high fat diet, T allele carriers had significantly higher levels of HDL-C compared to the CC homozygotes (P = 0.0002). b Interaction between Lipoprotein lipase gene SNP rs1121923 and fat energy intake (%) on HDL-C as a categorical variable (Pinteraction = 0.01). Among those who consumed a high fat diet (28.4%), the individuals who carried the T allele have 30% reduced risk of low HDL-C levels compared to those who carry the CC genotype (P = 0.001). Data shown are represented as means ± SE. Pinteraction values adjusted for age, gender, body mass index, type 2 diabetes and total energy intake
The SNP rs285 alone showed a significant association with serum TAG (P = 1 × 10−5), where CC genotype carriers had higher TAG concentrations than T allele carriers (Table 4). In the interaction analysis, there was an interaction of the LPL SNP rs4922115 with fat intake (energy %) on TAG, where, among those who consumed a low (1st tertile: 18.1 ± 2.6%) or medium fat (2nd tertile: 23.4 ± 1.1%) diet, individuals carrying the GG genotype had significantly lower TAG concentrations compared to ‘A’ allele carriers (P = 0.01 for low fat intake; P = 0.02 for medium fat intake). However, after correction for multiple testing, this interaction was not statistically significant. None of the other interactions between the SNPs and dietary factors on total cholesterol, serum TAG and LDL-C were statistically significant (Additional file 1: Table S1).
Discussion
To our knowledge, this is the first genetic epidemiological study to investigate the interaction between SNPs at LPL gene and dietary factors on blood lipids in an Asian Indian population. Our study provides evidence for a novel interaction between SNP rs1121923 and fat intake (energy %) on HDL-C, where the T allele carriers had significantly higher levels of HDL-C compared to the CC homozygotes among those who consumed a high fat diet. Given that the total fat intake has increased in India in the last few decades and Asian Indians are characterized by altered lipid levels and at a higher risk of premature coronary artery disease (CAD) [38], our study findings have significant public health implications.
Several LPL polymorphisms have been extensively studied in association with various lipid traits [12, 22, 25, 26]. The most notable of these known functional common polymorphisms is rs328, also known as S447X (premature truncation at codon 447). SNP rs328 is a gain-of-function polymorphism that has been shown to be consistently associated with higher HDL-C [12]. Our study has also shown a significant association of the SNP rs328 with HDL-C concentrations, where the minor ‘G’ allele carriers had significantly higher HDL-C compared to those with common CC genotype. The rs285 (Pvu II) variant located in the intron 6 of the LPL gene has been shown to be associated with dyslipidemic phenotypes such as low HDL-C and high TAG among Caucasians [39], which is in accordance with our study findings in Asian Indians where the CC genotype carriers of the SNP rs285 had significantly higher TAG than T allele carriers. The ‘A’ allele of the SNP rs4922115 (located in the 3′ UTR region) and ‘C’ allele of the SNP rs1121923 (Val135Val) (located in the exon 3) were also associated with lower HDL-C levels in our study; however, after correction for multiple testing they were not significant. Our findings confirm the previously reported associations and reveal that LPL SNPs play an important role in lipid metabolism in this Asian Indian population.
Total fat intake has increased considerably in India in the last few decades [40]. The National Sample Survey Organization survey has reported that the fat intake of urban component of Indian populations has increased from 42.0 g/d/capita in 1993–1994 to 52.5 g/d/capita in 2011–2012 [41]. Interestingly our study in this South Indian population has identified an interaction between LPL SNP rs1121923 and fat intake (energy %) on HDL-C, where, among those who consumed a high fat diet (28.4%), individuals carrying the T allele had significantly higher HDL-C concentrations compared to the CC genotype carriers. Even though our study is the first to report this gene-diet interaction, previous studies in developing countries have shown that the quantity of dietary fats can affect the lipid profile and is directly related to the development of metabolic diseases such as obesity and diabetes [42, 43]. Quality of dietary fat has also shown to alter blood lipid levels [43] but the present study failed to identify an interaction of the LPL SNP with MUFA, PUFA and SFA, respectively, on lipid traits. Vegetable oils used in Indian cooking represent 80% of the visible fat consumed [44] and a study in an Indian population showed that the higher ratio of n6:n3 was attributed to the type and quantity of oil used [45]. Previous studies have shown that dietary fat increases HDL-C [46], which is partially explained by our study findings where the T allele carriers of the SNP rs1121923, who consumed a high fat diet, had higher HDL-C concentrations compared to CC genotype carriers. Even though the exact mechanism by which T allele contributes to the increase in HDL-C levels under conditions of high fat diet is unknown, the finding is suggestive of the complex inheritance pattern of the HDL-C levels [47], where several genes/polymorphisms are likely to contribute to the alteration of HDL-C levels through gene-gene and gene-diet interactions. Our findings are supported by animal studies [48] where mice that are challenged with a high fat diet showed a strong correlation between LPL activity and HDL cholesterol suggesting the link between LPL, fat intake and HDL levels.
Besides HDL-C, there were interactions of the LPL SNP rs4922115 with fat energy intake (%) on TAG, where, among those who consumed a low or medium fat diet, individuals carrying the GG genotype had significantly lower TAG concentrations compared to ‘A’ allele carriers. The SNP rs4922115 has not been studied previously in other populations except in Hispanics [49], where this SNP was identified by direct sequencing. However, the study did not explore the individual effect of the SNP on blood lipids and hence we are unable to compare our findings with the Hispanic study. Even though the interaction between SNP rs4922115 and fat energy intake (%) was not statistically significant after Bonferroni correction in our study, replication of these interactions in another large cohort is highly warranted.
One of the main limitations of the study is the small sample size. Given that there are no previously reported effect sizes for the LPL SNP-diet interaction on blood lipids, we are unable to calculate the statistical power for our study. However, we were still able to identify significant gene-diet interactions on HDL-C even after correction for multiple testing. The interaction was significant only with total fat intake (energy %) but not when split as PUFA, MUFA and SFA, which might be due to small sample size. Nevertheless, our study is well powered to identify the effect sizes for association between the SNPs and lipids. To increase the statistical power, individuals with and without T2D were included and hence it is possible that the T2D status could have introduced a bias in our study as the dietary pattern is likely to be changed among ‘known’ diabetic participants. However, T2D status was adjusted in all the analyses and the interaction findings were borderline significant even after excluding the ‘known’ diabetic participants from our analysis (data not shown), which could also be due to the small sample size after exclusion. Another limitation is that, our study was cross-sectional and therefore was unable to examine the casual relationship between fat intake and lowering of HDL-C levels; randomized controlled trials with prospective genotyping are required to explore the causality using genetic markers. The main strength of the present study is that a validated interviewer-administered FFQ was used to measure the usual long-term intake of the population. Furthermore, the sampling is representative of the overall population of Chennai. Indeed, the intake of major foods in our study was similar to the findings of the pooled urban data of the National Nutrition Monitoring Bureau for ten states in India [50] and hence, the results of the present study could be reasonably extrapolated to urban Indian population.
Conclusions
Our study confirms the association between LPL SNP rs328 and HDL-C concentrations and also provides an evidence for a novel interaction between SNP rs1121923 and fat intake (energy %) on HDL-C levels in this Asian Indian population. Given that Asian Indians have altered lipid profile and an increased predisposition to premature CAD [38, 40], our study suggesting that individuals carrying T allele of the SNP rs1121923 have increased HDL-C levels when consuming a high fat diet has significant public health implications. This finding warrants confirmation in prospective studies and randomized controlled trials.
Abbreviations
- BMI:
-
Body mass index
- CAD:
-
Coronary artery disease
- CURES:
-
Chennai urban rural epidemiological study
- CVD:
-
Cardiovascular disease
- FFQ:
-
Food frequency questionnaire
- HDL-C:
-
High-density lipoprotein cholesterol
- HWE:
-
Hardy–Weinberg equilibrium
- LDL-C:
-
Low density lipoprotein cholesterol
- LPL :
-
Lipoprotein lipase
- MUFA:
-
Monounsaturated
- NCEP-ATP III:
-
National Cholesterol Education Program-Adult Treatment Panel III, HbA1c, Glycated haemoglobin
- NGT:
-
Normal glucose tolerance
- PUFA:
-
Polyunsaturated
- SFA:
-
Saturated fatty acids
- SNPs:
-
Single nucleotide polymorphisms
- T2D:
-
Type 2 diabetes
- TAG:
-
Triacylglycerol
References
Ramachandran A, et al. Risk of noninsulin dependent diabetes mellitus conferred by obesity and central adiposity in different ethnic groups: a comparative analysis between Asian Indians, Mexican Americans and whites. Diabetes Res Clin Pract. 1997;36(2):121–5.
Pradeepa R, Mohan V. The changing scenario of the diabetes epidemic: implications for India. Indian J Med Res. 2002;116:121–32.
Garg R, et al. Association of atherosclerosis with dyslipidemia and co-morbid conditions: a descriptive study. J Nat Sci Biol Med. 2015;6(1):163–8.
McKeigue PM. Coronary heart disease in Indians, Pakistanis, and Bangladeshis: aetiology and possibilities for prevention. Br Heart J. 1992;67(5):341–2.
Mohan V, et al. Serum immunoreactive insulin responses to a glucose load in Asian Indian and European type 2 (non-insulin-dependent) diabetic patients and control subjects. Diabetologia. 1986;29(4):235–7.
Mohan V, et al. Association of low adiponectin levels with the metabolic syndrome--the Chennai urban rural epidemiology study (CURES-4). Metabolism. 2005;54(4):476–81.
Mohan V, et al. Epidemiology of type 2 diabetes: Indian scenario. Indian J Med Res. 2007;125(3):217–30.
Joshi SR. Metabolic syndrome--emerging clusters of the Indian phenotype. J Assoc Physicians India. 2003;51:445–6.
Namboodiri KK, et al. The collaborative lipid research clinics family study: biological and cultural determinants of familial resemblance for plasma lipids and lipoproteins. Genet Epidemiol. 1985;2(3):227–54.
Braun TR, et al. A replication study of GWAS-derived lipid genes in Asian Indians: the chromosomal region 11q23.3 harbors loci contributing to triglycerides. PLoS One. 2012;7(5):e37056.
Radha V, et al. Association of lipoprotein lipase gene polymorphisms with obesity and type 2 diabetes in an Asian Indian population. Int J Obes (Lond). 2007;31(6):913–8.
Radha V, et al. Association of lipoprotein lipase Hind III and Ser 447 Ter polymorphisms with dyslipidemia in Asian Indians. Am J Cardiol. 2006;97(9):1337–42.
Sanghera DK, et al. Genome-wide linkage scan to identify loci associated with type 2 diabetes and blood lipid phenotypes in the Sikh diabetes study. PLoS One. 2011;6(6):e21188.
Goldberg IJ. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J Lipid Res. 1996;37(4):693–707.
Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297(2):E271–88.
Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43(12):1997–2006.
Rader DJ. Molecular regulation of HDL metabolism and function: implications for novel therapies. J Clin Invest. 2006;116(12):3090–100.
de Vries R, Borggreve SE, Dullaart RP. Role of lipases, lecithin:cholesterol acyltransferase and cholesteryl ester transfer protein in abnormal high density lipoprotein metabolism in insulin resistance and type 2 diabetes mellitus. Clin Lab. 2003;49(11–12):601–13.
Marcais C, et al. Severe hypertriglyceridaemia in type II diabetes: involvement of apoC-III Sst-I polymorphism, LPL mutations and apo E3 deficiency. Diabetologia. 2000;43(11):1346–52.
Mattu RK, et al. Lipoprotein lipase gene variants relate to presence and degree of microalbuminuria in type II diabetes. Diabetologia. 2002;45(6):905–13.
Socquard E, et al. Association of HindIII and PvuII genetic polymorphisms of lipoprotein lipase with lipid metabolism and macrovascular events in type 2 diabetic patients. Diab Metab. 2006;32(3):262–9.
Shatwan IM, et al. Impact of lipoprotein lipase gene polymorphism, S447X, on postprandial triacylglycerol and glucose response to sequential meal ingestion. Int J Mol Sci. 2016;17(3):397.
Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41(1):47–55.
Heid IM, et al. Genome-wide association analysis of high-density lipoprotein cholesterol in the population-based KORA study sheds new light on intergenic regions. Circ Cardiovasc Genet. 2008;1(1):10–20.
Sagoo GS, et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am J Epidemiol. 2008;168(11):1233–46.
Lee J, et al. The lipoprotein lipase S447X polymorphism and plasma lipids: interactions with APOE polymorphisms, smoking, and alcohol consumption. J Lipid Res. 2004;45(6):1132–9.
Marques-Vidal P, et al. No interaction between alcohol consumption and HDL-related genes on HDL cholesterol levels. Atherosclerosis. 2010;211(2):551–7.
Nettleton JA, et al. Associations between HDL-cholesterol and polymorphisms in hepatic lipase and lipoprotein lipase genes are modified by dietary fat intake in African American and white adults. Atherosclerosis. 2007;194(2):e131–40.
Pyun JA, et al. Interaction effects of lipoprotein lipase polymorphisms with lifestyle on lipid levels in a Korean population: a cross-sectional study. Genomics Inform. 2012;10(2):88–98.
Garcia-Rios A, et al. Genetic variations at the lipoprotein lipase gene influence plasma lipid concentrations and interact with plasma n-6 polyunsaturated fatty acids to modulate lipid metabolism. Atherosclerosis. 2011;218(2):416–22.
Deepa M, et al. The Chennai urban rural epidemiology study (CURES)--study design and methodology (urban component) (CURES-I). J Assoc Physicians India. 2003;51:863–70.
Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–53.
Grundy SM, et al. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. J Am Coll Cardiol. 2004;44(3):720–32.
Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18(6):499–502.
Sudha V, et al. Reproducibility and validity of an interviewer-administered semi-quantitative food frequency questionnaire to assess dietary intake of urban adults in southern India. Int J Food Sci Nutr. 2006;57(7–8):481–93.
Cho YS, et al. Association of lipoprotein lipase (LPL) single nucleotide polymorphisms with type 2 diabetes mellitus. Exp Mol Med. 2008;40(5):523–32.
Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theor Popul Biol. 2001;60(3):155–66.
Pradeepa R, et al. Relationship of diabetic retinopathy with coronary artery disease in Asian Indians with type 2 diabetes: the Chennai urban rural epidemiology study (CURES) eye study--3. Diabetes Technol Ther. 2015;17(2):112–8.
Chamberlain JC, et al. DNA polymorphisms at the lipoprotein lipase gene: associations in normal and hypertriglyceridaemic subjects. Atherosclerosis. 1989;79(1):85–91.
Misra A, et al. Nutrition transition in India: secular trends in dietary intake and their relationship to diet-related non-communicable diseases. J Diabetes. 2011;3(4):278–92.
Narasimhan S, et al. Dietary fat intake and its association with risk of selected components of the metabolic syndrome among rural South Indians. Indian J Endocrinol Metab. 2016;20(1):47–54.
Freire RD, et al. Dietary fat is associated with metabolic syndrome in Japanese Brazilians. Diabetes Care. 2005;28(7):1779–85.
Misra A, Singhal N, Khurana L. Obesity, the metabolic syndrome, and type 2 diabetes in developing countries: role of dietary fats and oils. J Am Coll Nutr. 2010;29(3 Suppl):289S–301S.
Ghafoorunissa. Requirements of dietary fats to meet nutritional needs & prevent the risk of atherosclerosis--an Indian perspective. Indian J Med Res. 1998;108:191–202.
Vijayalaxmi MP, et al. Influence of fats and oils intake on the lipid profile of adults belonging to different income groups. Karnataka J Agric Sci. 2010;20:112–4.
Nettleton JA, et al. Associations between dietary macronutrient intake and plasma lipids demonstrate criterion performance of the multi-ethnic study of atherosclerosis (MESA) food-frequency questionnaire. Br J Nutr. 2009;102(8):1220–7.
Weissglas-Volkov D, Pajukanta P. Genetic causes of high and low serum HDL-cholesterol. J Lipid Res. 2010;51(8):2032–57.
Clee SM, et al. Relationship between lipoprotein lipase and high density lipoprotein cholesterol in mice: modulation by cholesteryl ester transfer protein and dietary status. J Lipid Res. 1997;38(10):2079–89.
Goodarzi MO, et al. Haplotypes in the lipoprotein lipase gene influence fasting insulin and discovery of a new risk haplotype. J Clin Endocrinol Metab. 2007;92(1):293–6.
National Institute of Nutrition, I.C.o.M.R. 25 Years of the National Nutrition Monitoring Bureau: 1972–1997. Hyderabad: NIN, ICMR; 2000.
Acknowledgements
Dr Karani S Vimaleswaran acknowledges support from the British Nutrition Foundation. The study was supported by Lady Tata Memorial Trust, Mumbai. The Chennai Wellingdon Corporate Foundation supported the CURES field studies (CURES-144).
Availability of data and materials
Not applicable.
Authors’ contributions
AKA performed the genotyping and statistical analysis; SI performed the statistical analysis and drafted the manuscript; BD, LRB and SV assisted with the statistical analysis; MV, ARM, SV and RV designed the CURES study; RV designed the genetic study; BD, ARM, SV, JAL, MV and RV critically reviewed the manuscript; VKS conceived the nutrigenetics study, performed the statistical analysis and wrote the manuscript. All authors contributed to and approved the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Written informed consent for publication was obtained from all the study participants.
Ethics approval and consent to participate
Written informed consent was obtained from each study participant, and the study was approved by the Madras Diabetes Research Foundation Institutional Ethics Committee.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Table S1.
Interaction between single nucleotide polymorphisms (SNPs) at lipoprotein lipase gene and dietary factors on lipids traits. (DOCX 17 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ayyappa, K.A., Shatwan, I., Bodhini, D. et al. High fat diet modifies the association of lipoprotein lipase gene polymorphism with high density lipoprotein cholesterol in an Asian Indian population. Nutr Metab (Lond) 14, 8 (2017). https://doi.org/10.1186/s12986-016-0155-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12986-016-0155-1